Nickel Carcinogenesis Mechanism: DNA Damage
Abstract
1. Introduction
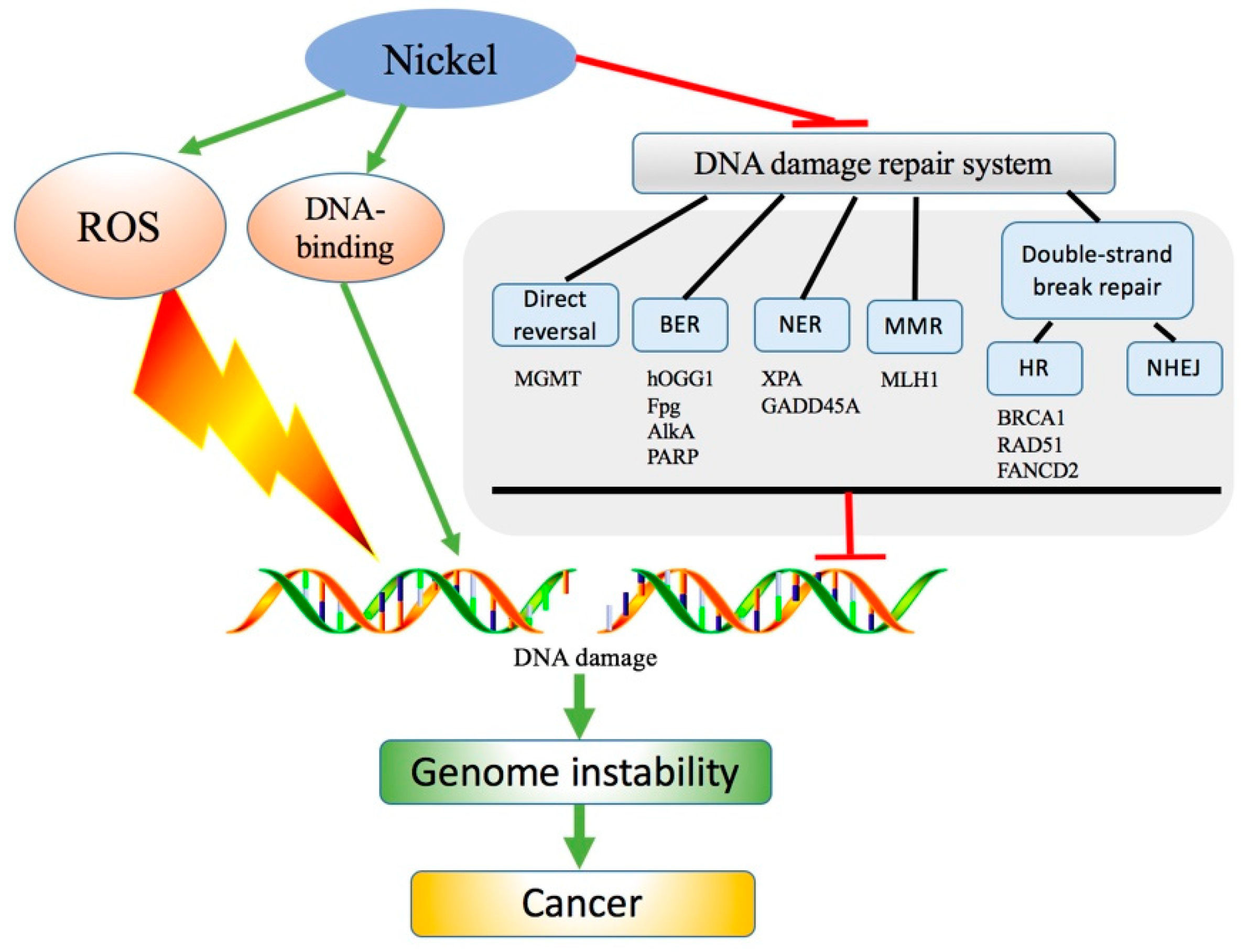
2. Ni-Induced DNA Damage
3. Binding of Ni to DNA and Nuclear Proteins in Ni-Induced DNA Damage
4. Reactive Oxygen Species (ROS) in Ni-Induced DNA Damage
4.1. Ni-Induced ROS Accumulation
4.2. ROS-Dependent Ni-Induced DNA Damage
4.3. ROS-Independent Ni-Induced DNA Damage
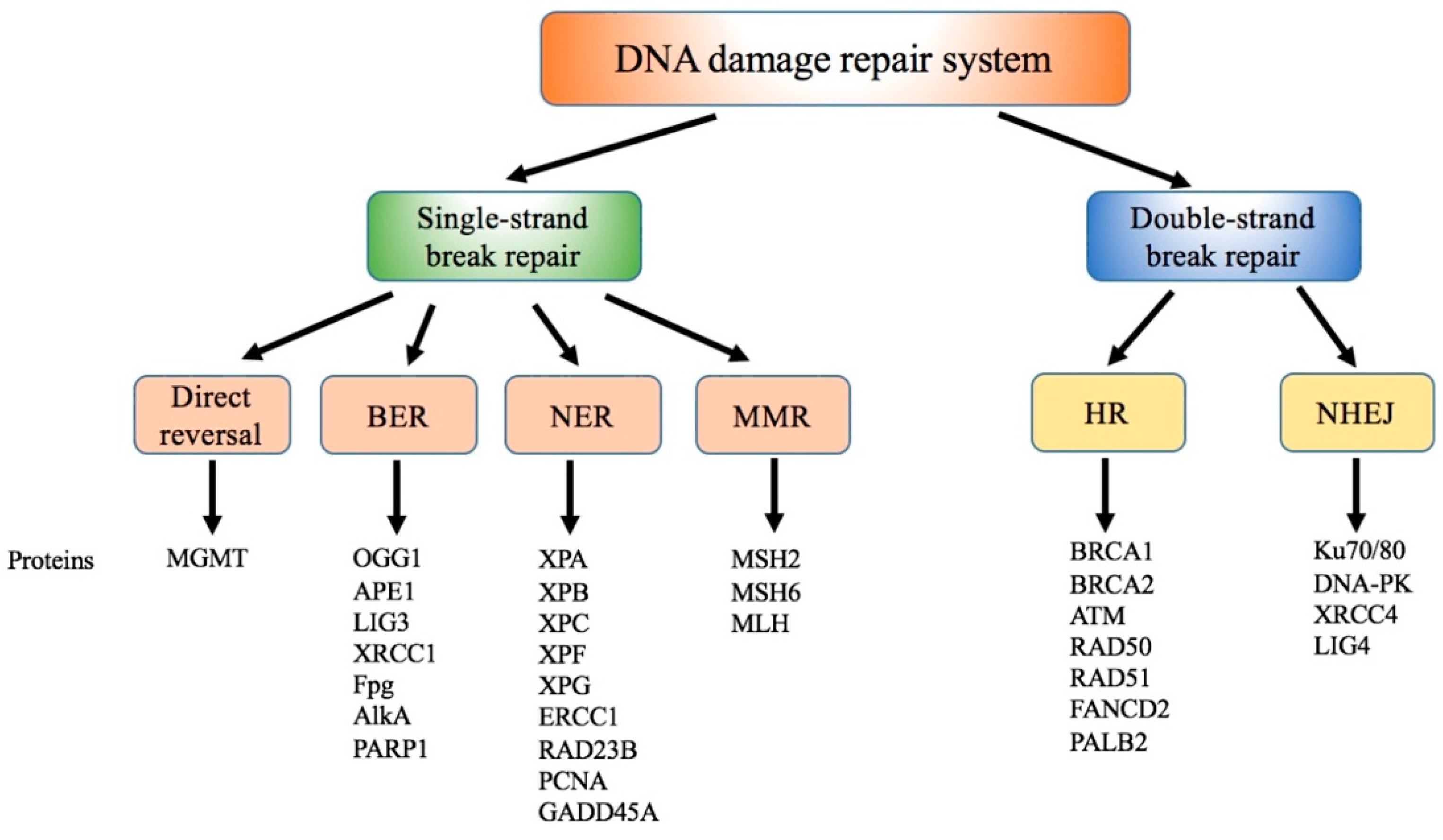
5. Interference of Ni with DNA Damage Repair Systems
5.1. Effect of Ni on Direct Reversal
5.2. Effect of Ni on BER
5.3. Effect of Ni on NER
5.4. Effect of Ni on MMR
5.5. Effect of Ni on Double-Strand Breaks Repair Pathways
5.6. Others
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ROS | reactive oxygen species |
| NER | nucleotide repair |
| BER | base excision repair |
| MMR | mismatch repair |
| HR | homologous recombination repair |
| NHEJ | non-homologous end joining |
| ER | endoplasmic reticulum |
| SOD | superoxide dismutase |
| CAT | catalase |
| GSH-Px | glutathione peroxidase |
| GSH | glutathione |
| 8-OHdG | 8-hydroxy-2′-deoxyguanosine |
| MNNG | N-methyl-N′-nitro-N-nitrosoguanidine |
| MNU | N-methyl-N-nitrosourea |
| MMS | methyl methanesulfonate |
| MGMT | O6-Methylguanine DNA methyltransferase |
| FeKGDs | ALKBH α-ketoglutarate Fe(II) dioxygenases |
| NiS | nickel sulfide |
| DNMT1 | DNA methyltransferase 1 |
| OGG1 | 8-oxoguanine DNA glycosylase |
| APE1 | AP endonuclease |
| LIG1 | DNA ligase 1 |
| XRCC1 | X-ray repair cross-complementing protein 1 |
| Fpg | formamidopyrimidine-DNA glycosylase |
| AlkA | 3-methyladenine-DNA glycosylase II |
| XP | xeroderma pigmentosum |
| ERCC1 | excision repair cross-complementation group 1 |
| PCNA | proliferating cell nuclear antigen |
| PARP | poly ADP-ribose polymerase |
| GADD45A | DNA-damage-inducible protein 45 alpha |
| MLH | mutL homolog |
| DSB | double-strand breaks |
| BRCA1 | breast cancer 1 |
| RAD51 | human homolog of S. cerevisiae RAD50 |
| f FANCD2 | anconi anemia group D2 protein |
| PALB2 | partner and localizer of BRCA2 |
| XRCC4 | X-ray repair cross complementing 4 |
| DNA-PK | DNA-dependent protein kinase |
| CHO | Chinese hamster ovary cells |
References
- Zambelli, B.; Uversky, V.N.; Ciurli, S. Nickel impact on human health: An intrinsic disorder perspective. Biochim. Biophys. Acta 2016, 1864, 1714–1731. [Google Scholar] [CrossRef] [PubMed]
- Zdrojewicz, Z.; Popowicz, E.; Winiarski, J. Nickel-role in human organism and toxic effects. Pol. Merkur. Lek. 2016, 41, 115–118. [Google Scholar]
- Zambelli, B.; Ciurli, S. Nickel and human health. Met. Ions Life Sci. 2013, 13, 321–357. [Google Scholar] [PubMed]
- Afridi, H.I.; Talpur, F.N.; Kazi, T.G.; Brabazon, D. Estimation of aluminum, arsenic, lead and nickel status in the samples of different cigarettes and their effect on human health of irish smoker hypertensive consumers. Clin. Lab. 2015, 61, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Boer, J.L.; Mulrooney, S.B.; Hausinger, R.P. Nickel-dependent metalloenzymes. Arch. Biochem. Biophys. 2014, 544, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Das, K.K.; Reddy, R.C.; Bagoji, I.B.; Das, S.; Bagali, S.; Mullur, L.; Khodnapur, J.P.; Biradar, M.S. Primary concept of nickel toxicity—An overview. J. Basic Clin. Physiol. Pharmacol. 2018, 30, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Arakaki, R.; Yamada, A.; Tsunematsu, T.; Kudo, Y.; Ishimaru, N. Molecular mechanisms of nickel allergy. Int. J. Mol. Sci. 2016, 17, 202. [Google Scholar] [CrossRef] [PubMed]
- Dukes, M.P.; Rowe, R.K.; Harvey, T.; Rangel, W.; Pedigo, S. Nickel reduces calcium dependent dimerization in neural cadherin Electronic supplementary information (ESI) available. Metallomics 2019, 11, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, B.; Tanveer, M.; Rehman, A.; Cheema, S.A.; Fahad, S.; Rehman, S.; Sharma, A. Nickel; whether toxic or essential for plants and environment—A review. Plant Physiol. Biochem. 2018, 132, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Doll, R. Nickel exposure: A human health hazard. IARC Sci. Publ. 1984, 53, 3–21. [Google Scholar]
- Zeinali, T.; Salmani, F.; Naseri, K. Dietary intake of cadmium, chromium, copper, nickel, and lead through the consumption of meat, liver, and kidney and assessment of human health risk in birjand, southeast of Iran. Biol. Trace Elem. Res. 2019, 191, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Das, K.K.; Das, S.N.; Dhundasi, S.A. Nickel, its adverse health effects & oxidative stress. Indian J. Med. Res. 2008, 128, 412–425. [Google Scholar] [PubMed]
- Shi, Z. Nickel carbonyl: Toxicity and human health. Sci. Total Environ. 1994, 148, 293–298. [Google Scholar] [CrossRef]
- Haber, L.T.; Erdreicht, L.; Diamond, G.L.; Maier, A.M.; Ratney, R.; Zhao, Q.; Dourson, M.L. Hazard identification and dose response of inhaled nickel-soluble salts. Regul. Toxicol. Pharmacol. 2000, 31, 210–230. [Google Scholar] [CrossRef] [PubMed]
- Bolek, E.C.; Erden, A.; Kulekci, C.; Kalyoncu, U.; Karadag, O. Rare occupational cause of nasal septum perforation: Nickel exposure. Int. J. Occup. Med. Environ. Health 2017, 30, 963–967. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salnikow, K.; Zhitkovich, A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: Nickel, arsenic, and chromium. Chem. Res. Toxicol. 2008, 21, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.G.; Ren, T.; Xiao, C.Y.; Li, H.Y.; Wu, T.C. Nickel promotes the invasive potential of human lung cancer cells via TLR4/MyD88 signaling. Toxicology 2011, 285, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Beyersmann, D.; Hartwig, A. Carcinogenic metal compounds: Recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008, 82, 493–512. [Google Scholar] [CrossRef]
- Denkhaus, E.; Salnikow, K. Nickel essentiality, toxicity, and carcinogenicity. Crit. Rev. Oncol. Hematol. 2002, 42, 35–56. [Google Scholar] [CrossRef]
- Henderson, R.G.; Durando, J.; Oller, A.R.; Merkel, D.J.; Marone, P.A.; Bates, H.K. Acute oral toxicity of nickel compounds. Regul. Toxicol. Pharmacol. 2012, 62, 425–432. [Google Scholar] [CrossRef]
- Coogan, T.P.; Latta, D.M.; Snow, E.T.; Costa, M. Toxicity and carcinogenicity of nickel compounds. Crit. Rev. Toxicol. 1989, 19, 341–384. [Google Scholar] [CrossRef] [PubMed]
- Sunderman, F.W., Jr. Carcinogenicity of nickel compounds in animals. IARC Sci. Publ. 1984, 53, 127–142. [Google Scholar]
- Costa, M.; Davidson, T.L.; Chen, H.; Ke, Q.; Zhang, P.; Yan, Y.; Huang, C.; Kluz, T. Nickel carcinogenesis: Epigenetics and hypoxia signaling. Mutat. Res. 2005, 592, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, H. Biochemical and clinical aspects of nickel toxicity. Rev. Environ. Health 1996, 11, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, K.; Blasiak, J. Free radicals-mediated induction of oxidized DNA bases and DNA-protein cross-links by nickel chloride. Mutat. Res. 2002, 514, 233–243. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Zhou, B.B.S.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Chen, J. Chronic nickel-induced DNA damage and cell death: The protection role of ascorbic acid. Environ. Toxicol. 2008, 23, 401–406. [Google Scholar] [CrossRef]
- Magaye, R.; Zhao, J.S. Recent progress in studies of metallic nickel and nickel-based nanoparticles’ genotoxicity and carcinogenicity. Environ. Toxicol. Pharmacol. 2012, 34, 644–650. [Google Scholar] [CrossRef]
- Joyner, J.C.; Reichfield, J.; Cowan, J.A. Factors influencing the DNA nuclease activity of iron, cobalt, nickel, and copper chelates. J. Am. Chem. Soc. 2011, 133, 15613–15626. [Google Scholar] [CrossRef] [PubMed]
- Dumala, N.; Mangalampalli, B.; Chinde, S.; Kumari, S.I.; Mahoob, M.; Rahman, M.F.; Grover, P. Genotoxicity study of nickel oxide nanoparticles in female Wistar rats after acute oral exposure. Mutagenesis 2017, 32, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Doreswamy, K.; Shrilatha, B.; Rajeshkumar, T. Nickel-induced oxidative stress in testis of mice: Evidence of DNA damage and genotoxic effects. J. Androl. 2004, 25, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Guo, H.; Deng, J.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Wang, X.; Wu, B.; Li, J.; et al. Inhibitive effects of nickel chloride (NiCl(2)) on thymocytes. Biol. Trace Elem. Res. 2015, 164, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Wang, X.; Wu, B.; Guo, H. Toxic effect of NiCl2 on development of the bursa of Fabricius in broiler chickens. Oncotarget 2016, 7, 125–139. [Google Scholar] [PubMed]
- Guo, H.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Wang, X.; Wu, B.; Chen, K.; Deng, J. Dietary NiCl2 causes G2/M cell cycle arrest in the broiler’s kidney. Oncotarget 2015, 6, 35964–35977. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.R.; Cui, H.M.; Fang, J.; Zuo, Z.C.; Deng, J.L.; Wang, X.; Zhao, L.; Chen, K.J.; Deng, J. Nickel chloride (NiCl2) in hepatic toxicity: Apoptosis, G2/M cell cycle arrest and inflammatory response. Aging 2016, 8, 3009–3027. [Google Scholar] [CrossRef]
- Hosoya, N.; Miyagawa, K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014, 105, 370–388. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. The DNA damage response and cancer therapy. Nature 2012, 481, 287–294. [Google Scholar] [CrossRef]
- Jackson, S.P. The DNA-damage response: New molecular insights and new approaches to cancer therapy. Biochem. Soc. Trans. 2009, 37, 483–494. [Google Scholar] [CrossRef]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H.J. The key role of DNA damage on cancer, aging and longevity. Environ. Mol. Mutagenesis 2012, 53, S13. [Google Scholar]
- Schiewer, M.J.; Knudsen, K.E. DNA damage response in prostate cancer. CSH Perspect. Med. 2019, 9, a030486. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K.; Nohmi, T. Chemically-Induced DNA Damage, Mutagenesis, and Cancer. Int. J. Mol. Sci. 2018, 19, 1767. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H.J. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1914. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Bai, Y.N.; Pu, H.Q.; He, J.; Zheng, T.Z.; Li, H.Y.; Dai, M.; Cheng, N. Dynamic changes in DNA damage and repair biomarkers with employment length among nickel smelting workers. Biomed. Environ. Sci. 2015, 28, 679–682. [Google Scholar] [PubMed]
- Saquib, Q.; Siddiqui, M.A.; Ahmad, J.; Ansari, S.M.; Faisal, M.; Wahab, R.; Alatar, A.A.; Al-Khedhairy, A.A.; Musarrat, J. Nickel oxide nanoparticles induced transcriptomic alterations in HEPG2 cells. Adv. Exp. Med. Biol. 2018, 1048, 163–174. [Google Scholar] [PubMed]
- Guillamet, E.; Creus, A.; Farina, M.; Sabbioni, E.; Fortaner, S.; Marcos, R. DNA-damage induction by eight metal compounds in TK6 human lymphoblastoid cells: Results obtained with the alkaline Comet assay. Mutat. Res. 2008, 654, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Latvala, S.; Vare, D.; Karlsson, H.L.; Elihn, K. In vitro genotoxicity of airborne Ni-NP in air-liquid interface. J. Appl. Toxicol. 2017, 37, 1420–1427. [Google Scholar] [CrossRef]
- Latvala, S.; Hedberg, J.; Di Bucchianico, S.; Moller, L.; Wallinder, I.O.; Elihn, K.; Karlsson, H.L. Nickel release, ROS generation and toxicity of Ni and NiO micro- and nanoparticles. PLoS ONE 2016, 11, e0159684. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Ma, L.Y. Quantification of metal ion induced DNA damage with single cell array based assay. Analyst 2013, 138, 5713–5718. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Jin, L.; Wu, N.; Tan, Y.; Song, Y.; Gao, M.; Liu, K.; Zhang, X.; He, J. DNA damage and oxidative stress in human B lymphoblastoid cells after combined exposure to hexavalent chromium and nickel compounds. Food Chem. Toxicol. 2013, 55, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.L.; Seo, Y.R. Molecular and genomic approach for understanding the gene-environment interaction between Nrf2 deficiency and carcinogenic nickel-induced DNA damage. Oncol. Rep. 2012, 28, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Saplakoglu, U.; Iscan, M.; Iscan, M. DNA single-strand breakage in rat lung, liver and kidney after single and combined treatments of nickel and cadmium. Mutat. Res. 1997, 394, 133–140. [Google Scholar] [CrossRef]
- Guo, H.; Wu, B.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Wang, X.; Deng, J.; Yin, S.; et al. NiCl2-down-regulated antioxidant enzyme mRNA expression causes oxidative damage in the broiler(’)s kidney. Biol. Trace Elem. Res. 2014, 162, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Guo, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Wang, X.; Zhao, L. Oxidative stress and inflammatory responses involved in dietary nickel chloride (NiCl2)-induced pulmonary toxicity in broiler chickens. Toxicol. Res. 2016, 5, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.C.; Shen, D.S.; Shentu, J.L.; Wang, M.Z.; Wan, M.Y. Could humic acid relieve the biochemical toxicities and DNA damage caused by nickel and deltamethrin in earthworms (Eisenia foetida)? Environ. Sci. Process. Impacts 2015, 17, 2074–2081. [Google Scholar] [CrossRef]
- Huffnagle, I.M.; Joyner, A.; Rumble, B.; Hysa, S.; Rudel, D.; Hvastkovs, E.G. Dual electrochemical and physiological apoptosis assay detection of in vivo generated nickel chloride induced DNA damage in Caenorhabditis elegans. Anal. Chem. 2014, 86, 8418–8424. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Costa, M. Pathway of nickel uptake influences its interaction with heterochromatic DNA. Toxicol. Appl. Pharmacol. 1986, 84, 278–285. [Google Scholar] [CrossRef]
- Ono, H.; Wada, O.; Ono, T. Distribution of trace metals in nuclei and nucleoli of normal and regenerating rat liver with special reference to the different behavior of nickel and chromium. J. Toxicol. Environ. Health 1981, 8, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, G.G.; Rossetto, F.E.; Turnbull, J.D.; Nieboer, E. Toxicity, uptake, and mutagenicity of particulate and soluble nickel compounds. Environ. Health Perspect. 1994, 102, 69–79. [Google Scholar] [PubMed]
- Oruambo, I.F.; Kachikwu, S.; Idabor, L. Dose-related increased binding of nickel to chromatin proteins; and changes to DNA concentration in the liver of guinea pigs treated with Nigerian light crude oil. Int. J. Environ. Res. Public Health 2007, 4, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Swartz, J.R. A filter microplate assay for quantitative analysis of DNA binding proteins using fluorescent DNA. Anal. Biochem. 2011, 415, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Bal, W.; Liang, R.; Lukszo, J.; Lee, S.H.; Dizdaroglu, M.; Kasprzak, K.S. Ni(II) specifically cleaves the C-terminal tail of the major variant of histone H2A and forms an oxidative damage-mediating complex with the cleaved-off octapeptide. Chem. Res. Toxicol. 2000, 13, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Bal, W.; Karantza, V.; Moudrianakis, E.N.; Kasprzak, K.S. Interaction of Nickel(II) with histones: In vitro binding of nickel(II) to the core histone tetramer. Arch. Biochem. Biophys. 1999, 364, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Karaczyn, A.A.; Wojciech, B.; North, S.L.; Bare, R.M.; Hoang, V.M.; Fisher, R.J.; Kasprzak, K.S. The octapeptidic end of the C-terminal tail of histone H2A is cleaved off in cells exposed to carcinogenic nickel(II). Chem. Res. Toxicol. 2003, 16, 1555–1559. [Google Scholar] [CrossRef]
- Oliveira, S.C.; Corduneanu, O.; Oliveira-Brett, A.M. In situ evaluation of heavy metal-DNA interactions using an electrochemical DNA biosensor. Bioelectrochemistry 2008, 72, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Ciccarelli, R.B.; Wetterhahn, K.E. Nickel distribution and DNA lesions induced in rat tissues by the carcinogen nickel carbonate. Cancer Res. 1982, 42, 3544–3549. [Google Scholar]
- Bonsignore, R.; Terenzi, A.; Spinello, A.; Martorana, A.; Lauria, A.; Almerico, A.M.; Keppler, B.K.; Barone, G. G-quadruplex vs. duplex-DNA binding of nickel(II) and zinc(II) Schiff base complexes. J. Inorg. Biochem. 2016, 161, 115–121. [Google Scholar] [CrossRef]
- Polo-Ceron, D. Cu(II) and Ni(II) Complexes with New Tridentate NNS Thiosemicarbazones: Synthesis, Characterisation, DNA Interaction, and Antibacterial Activity. Bioinorg. Chem. Appl. 2019, 2019, 3520837. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett. 2017, 387, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Akhtar, M.J.; Alhadlaq, H.A.; Khan, M.A.M.; Alrokayan, S.A. Comparative cytotoxic response of nickel ferrite nanoparticles in human liver HepG2 and breast MFC-7 cancer cells. Chemosphere 2015, 135, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Zou, L.; Gan, X.; Li, Y.; Liu, F.; Chang, X.; Zhang, X.; Tian, M.; Li, S.; Su, L. ROS generation and MAPKs activation contribute to the Ni-induced testosterone synthesis disturbance in rat Leydig cells. Toxicol. Lett. 2018, 290, 36–45. [Google Scholar] [CrossRef]
- Kong, L.; Hu, W.C.; Lu, C.C.; Cheng, K.P.; Tang, M. Mechanisms underlying nickel nanoparticle induced reproductive toxicity and chemo-protective effects of vitamin C in male rats. Chemosphere 2019, 218, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Terpilowska, S.; Siwicki, A.K. Pro- and antioxidant activity of chromium(III), iron(III), molybdenum(III) or nickel(II) and their mixtures. Chem. Biol. Interact. 2019, 298, 43–51. [Google Scholar] [CrossRef]
- Sousa, C.A.; Soares, H.M.V.M.; Soares, E.V. Toxic effects of nickel oxide (NiO) nanoparticles on the freshwater alga Pseudokirchneriella subcapitata. Aquat. Toxicol. 2018, 204, 80–90. [Google Scholar] [CrossRef]
- Sousa, C.A.; Soares, H.M.V.M.; Soares, E.V. Nickel Oxide (NiO) Nanoparticles Induce Loss of Cell Viability in Yeast Mediated by Oxidative Stress. Chem. Res. Toxicol. 2018, 31, 658–665. [Google Scholar] [CrossRef]
- Das, D.; Das, P.; Moniruzzaman, M.; Sarkar, M.P.; Mukherjee, J.; Chakraborty, S.B. Consequences of oxidative damage and mitochondrial dysfunction on the fatty acid profile of muscle of indian major carps considering metal toxicity. Chemosphere 2018, 207, 385–396. [Google Scholar] [CrossRef]
- Gupta, V.; Jatav, P.K.; Verma, R.; Kothari, S.L.; Kachhwaha, S. Nickel accumulation and its effect on growth, physiological and biochemical parameters in millets and oats. Environ. Sci. Pollut. Res. 2017, 24, 23915–23925. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Gao, X.J.; Zhu, J.Q.; Cheng, K.P.; Tang, M. Mechanisms Involved in Reproductive Toxicity Caused by Nickel Nanoparticle in Female Rats. Environ. Toxicol. 2016, 31, 1674–1683. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Lim, S.S.; Baek, B.J.; An, J.M.; Nam, H.S.; Woo, K.M.; Cho, M.K.; Kim, S.H.; Lee, S.H. Nickel(II)-induced nasal epithelial toxicity and oxidative mitochondrial damage. Environ. Toxicol. Pharmacol. 2016, 42, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Wang, Y.F.; Lin, Y.H.; Yen, S.F. Nickel-induced oxidative stress and effect of antioxidants in human lymphocytes. Arch. Toxicol. 2003, 77, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Salnikow, K.; Sutherland, J.E.; Broday, L.; Peng, W.; Zhang, Q.; Kluz, T. The role of oxidative stress in nickel and chromate genotoxicity. Mol. Cell. Biochem. 2002, 234–235, 265–275. [Google Scholar] [CrossRef]
- Huang, J.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Wu, B. The association between splenocyte apoptosis and alterations of Bax, Bcl-2 and caspase-3 mRNA expression, and oxidative stress induced by dietary nickel chloride in broilers. Int. J. Environ. Res. Public Health 2013, 10, 7310–7326. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Huang, J. Dietary nickel chloride induces oxidative intestinal damage in broilers. Int. J. Environ. Res. Public Health 2013, 10, 2109–2119. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Huang, J. Dietary nickel chloride induces oxidative stress, apoptosis and alters Bax/Bcl-2 and caspase-3 mRNA expression in the cecal tonsil of broilers. Food Chem. Toxicol. 2014, 63, 18–29. [Google Scholar] [CrossRef]
- Tang, K.; Li, J.; Yin, S.; Guo, H.; Deng, J.; Cui, H. Effects of nickel chloride on histopathological lesions and oxidative damage in the thymus. Health 2014, 6, 2875–2882. [Google Scholar] [CrossRef]
- Yin, S.; Guo, H.; Cui, H.; Peng, X.; Fang, J.; Zuo, Z.; Deng, J.; Wang, X.; Tang, K.; Li, J. Nickel Chloride (NiCl2) Induces Histopathological Lesions via Oxidative Damage in the Broiler’s Bursa of Fabricius. Biol. Trace Elem. Res. 2016, 171, 214–223. [Google Scholar] [CrossRef]
- Ahamed, M.; Ali, D.; Alhadlaq, H.A.; Akhtar, M.J. Nickel oxide nanoparticles exert cytotoxicity via oxidative stress and induce apoptotic response in human liver cells (HepG2). Chemosphere 2013, 93, 2514–2522. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.Y.; Su, L.; Sun, Y.F.; Han, A.J.; Chang, X.H.; Zhu, A.; Liu, F.F.; Li, J.; Sun, Y. Nickel sulfate induced apoptosis via activating ROS-dependent mitochondria and endoplasmic reticulum stress pathways in rat Leydig cells. Environ. Toxicol. 2017, 32, 1918–1926. [Google Scholar] [CrossRef] [PubMed]
- Di Bucchianico, S.; Gliga, A.R.; Akerlund, E.; Skoglund, S.; Wallinder, I.O.; Fadeel, B.; Karlsson, H.L. Calcium-dependent cyto- and genotoxicity of nickel metal and nickel oxide nanoparticles in human lung cells. Part. Fibre Toxicol. 2018, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Patel, E.; Lynch, C.; Ruff, V.; Reynolds, M. Co-exposure to nickel and cobalt chloride enhances cytotoxicity and oxidative stress in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2012, 258, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.J.; Chang, Q.S.; Wang, X.; Son, Y.; Zhang, Z.; Chen, G.; Luo, J.; Bi, Y.Y.; Chen, F.; Shi, X.L. Reactive Oxygen Species-Activated Akt/ASK1/p38 Signaling Pathway in Nickel Compound-Induced Apoptosis in BEAS 2B Cells. Chem. Res. Toxicol. 2010, 23, 568–577. [Google Scholar] [CrossRef]
- Henkler, F.; Brinkmann, J.; Luch, A. The role of oxidative stress in carcinogenesis induced by metals and xenobiotics. Cancers 2010, 2, 376–396. [Google Scholar] [CrossRef]
- Bal, W.; Kozlowski, H.; Kasprzak, K.S. Molecular models in nickel carcinogenesis. J. Inorg. Biochem. 2000, 79, 213–218. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.; Vellayappan, B.A.; Jeyasekharan, A.D. Ros and the DNA damage response in cancer. Redox Biol. 2018. [Google Scholar] [CrossRef]
- Wu, Q.; Ni, X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 2015, 16, 13–19. [Google Scholar] [CrossRef]
- Scott, T.L.; Rangaswamy, S.; Wicker, C.A.; Izumi, T. Repair of oxidative DNA damage and cancer: Recent progress in DNA base excision repair. Antioxid. Redox Signal. 2014, 20, 708–726. [Google Scholar] [CrossRef]
- Li, R.; Zhao, L.; Zhang, L.; Chen, M.; Dong, C.; Cai, Z. DNA damage and repair, oxidative stress and metabolism biomarker responses in lungs of rats exposed to ambient atmospheric 1-nitropyrene. Environ. Toxicol. Pharmacol. 2017, 54, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Barzilai, A.; Yamamoto, K. DNA damage responses to oxidative stress. DNA Repair 2004, 3, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.N.; Liu, B.D.; Yin, J.F.; Xu, T.; Zhao, S.L.; Xu, Q.; Chen, X.; Wang, H.L. Detection of 8-hydroxydeoxyguanosine (8-OHdG) as a biomarker of oxidative damage in peripheral leukocyte DNA by UHPLC-MS/MS. J. Chromatogr. B 2017, 1064, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.M.; Williams, T.D.; Hodges, N.J.; Waring, R.H. Reactive oxygen species and oxidative DNA damage mediate the cytotoxicity of tungsten-nickel-cobalt alloys in vitro. Toxicol. Appl. Pharm. 2011, 250, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, S.Y.; Jia, L.; Zhang, L.; Ba, J.C.; Han, D.; Yu, C.P.; Wu, Y.H. Nickel-Refining Fumes Induced DNA Damage and Apoptosis of NIH/3T3 Cells via Oxidative Stress. Int. J. Environ. Res. Public Health 2016, 13, 629. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.Q.; Huang, Y.; Wang, X.L.; Zhang, J.W.; Wu, K.S. Associations of neonatal lead, cadmium, chromium and nickel co-exposure with DNA oxidative damage in an electronic waste recycling town. Sci. Total Environ. 2014, 472, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Zheng, G.H.; Ming, Q.L.; Chao, C.; Sun, J.M. Sesamin protects mouse liver against nickel-induced oxidative DNA damage and apoptosis by the PI3K-Akt pathway. J. Agric. Food Chem. 2013, 61, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, A.; Baluce, B.; Visalli, G.; La Maestra, S.; Micale, R.; Izzotti, A. Ex vivo study for the assessment of behavioral factor and gene polymorphisms in individual susceptibility to oxidative DNA damage metals-induced. Int. J. Hyg. Environ. Health 2011, 214, 210–218. [Google Scholar] [CrossRef]
- Kelly, M.C.; Whitaker, G.; White, B.; Smyth, M.R. Nickel(II)-catalysed oxidative guanine and DNA damage beyond 8-oxoguanine. Free Radic. Biol. Med. 2007, 42, 1680–1689. [Google Scholar] [CrossRef]
- Misra, M.; Olinski, R.; Dizdaroglu, M.; Kasprzak, K.S. Enhancement by L-histidine of nickel(II)-induced DNA-protein cross-linking and oxidative DNA base damage in the rat kidney. Chem. Res. Toxicol. 1993, 6, 33–37. [Google Scholar] [CrossRef]
- Kawanishi, S.; Oikawa, S.; Inoue, S.; Nishino, K. Distinct mechanisms of oxidative DNA damage induced by carcinogenic nickel subsulfide and nickel oxides. Environ. Health Perspect. 2002, 110, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.C.; He, M.D.; Lu, Y.H.; Li, L.; Zhong, M.; Zhang, Y.W.; Wang, Y.; Yu, Z.P.; Zhou, Z. Nickel exposure induces oxidative damage to mitochondrial DNA in Neuro2a cells: The neuroprotective roles of melatonin. J. Pineal Res. 2011, 51, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.Q.; Jiang, M.Z.; Zhang, Y.; Wan, R.; Li, J.; Zhong, C.J.; Li, H.Y.; Tang, S.C.; Zhang, Q.W. Comparative mouse lung injury by nickel nanoparticles with differential surface modification. J. Nanobiotechnol. 2019, 17, 2. [Google Scholar] [CrossRef] [PubMed]
- Abudayyak, M.; Guzel, E.; Ozhan, G. Nickel oxide nanoparticles are highly toxic to SH-SY5Y neuronal cells. Neurochem. Int. 2017, 108, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Abudayyak, M.; Guzel, E.; Ozhan, G. Nickel oxide nanoparticles induce oxidative DNA damage and apoptosis in kidney cell line (NRK-52E). Biol. Trace Elem. Res. 2017, 178, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Dogra, V.; Kaur, G.; Kaur, A.; Kumar, R.; Kumar, S. In vitro assessment of antimicrobial and genotoxic effect of metallosurfactant based nickel hydroxide nanoparticles against Escherichia coli and its genomic DNA. Colloid Surf. B 2018, 170, 99–108. [Google Scholar] [CrossRef]
- Boran, H.; Saffak, S. Comparison of Dissolved Nickel and Nickel Nanoparticles Toxicity in Larval Zebrafish in Terms of Gene Expression and DNA Damage. Arch. Environ. Contam. Toxicol. 2018, 74, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Magaye, R.; Gu, Y.L.; Wang, Y.F.; Su, H.; Zhou, Q.; Mao, G.C.; Shi, H.B.; Yue, X.; Zou, B.B.; Xu, J.; et al. In vitro and in vivo evaluation of the toxicities induced by metallic nickel nano and fine particles. J. Mol. Histol. 2016, 47, 273–286. [Google Scholar] [CrossRef]
- Akerlund, E.; Cappellini, F.; Di Bucchianico, S.; Islam, S.; Skoglund, S.; Derr, R.; Wallinder, I.O.; Hendriks, G.; Karlsson, H.L. Genotoxic and mutagenic properties of Ni and NiO nanoparticles investigated by comet assay,-H2AX staining, Hprt mutation assay and ToxTracker reporter cell lines. Environ. Mol. Mutagenesis 2018, 59, 211–222. [Google Scholar] [CrossRef]
- Alarifi, S.; Ali, D.; Alakhtani, S.; Al Suhaibani, E.S.; Al-Qahtani, A.A. Reactive oxygen species-mediated DNA damage and apoptosis in human skin epidermal cells after exposure to nickel nanoparticles. Biol. Trace Elem. Res. 2014, 157, 84–93. [Google Scholar] [CrossRef]
- Kumar, V.; Mishra, R.K.; Kaur, G.; Dutta, D. Cobalt and nickel impair DNA metabolism by the oxidative stress independent pathway. Metallomics 2017, 9, 1596–1609. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagenesis 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Rivera, B.; Polak, P.; Foulkes, W.D. Monogenic diseases of DNA repair. N. Engl. J. Med. 2018, 378, 491. [Google Scholar] [PubMed]
- Keijzers, G.; Bakula, D.; Scheibye-Knudsen, M. Monogenic Diseases of DNA Repair. N. Engl. J. Med. 2017, 377, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.P.; Hu, Z.H.; Zhang, Y.; Gou, X.J.; Mu, Y.; Wang, L.R.; Xie, X.Q. Metal binding mediated conformational change of XPA protein: A potential cytotoxic mechanism of nickel in the nucleotide excision repair. J. Mol. Model. 2016, 22, 156. [Google Scholar] [CrossRef] [PubMed]
- Krueger, I.; Mullenders, L.H.; Hartwig, A. Nickel(II) increases the sensitivity of V79 Chinese hamster cells towards cisplatin and transplatin by interference with distinct steps of DNA repair. Carcinogenesis 1999, 20, 1177–1184. [Google Scholar] [CrossRef]
- Hartwig, A.; Mullenders, L.H.F.; Schlepegrell, R.; Kasten, U.; Beyersmann, D. Nickel(II) interferes with the tncision step in nucleotide excision repair in mammalian cells. Cancer Res. 1994, 54, 4045–4051. [Google Scholar]
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Bürkle, A. Interference by Toxic Metal Ions with DNA Repair Processes and Cell Cycle Control: Molecular Mechanisms. Environ. Health Perspect. 2002, 110, 797–799. [Google Scholar] [CrossRef]
- Scanlon, S.E.; Scanlon, C.D.; Hegan, D.C.; Sulkowski, P.L.; Glazer, P.M. Nickel induces transcriptional down-regulation of DNA repair pathways in tumorigenic and non-tumorigenic lung cells. Carcinogenesis 2017, 38, 627–637. [Google Scholar] [CrossRef]
- Arita, A.; Munoz, A.; Chervona, Y.; Niu, J.; Qu, Q.; Zhao, N.; Ruan, Y.; Kiok, K.; Kluz, T.; Sun, H.; et al. Gene expression profiles in peripheral blood mononuclear cells of Chinese nickel refinery workers with high exposures to nickel and control subjects. Cancer Epidemiol. Prev. Biomark. 2013, 22, 261–269. [Google Scholar] [CrossRef]
- Hartwig, A. Carcinogenicity of metal compounds: Possible role of DNA repair inhibition. Toxicol. Lett. 1998, 102–103, 235–239. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef]
- Van Gent, D.C.; Kanaar, R. Exploiting DNA repair defects for novel cancer therapies. Mol. Biol. Cell 2016, 27, 2145–2148. [Google Scholar] [CrossRef]
- Jackson, S.P.; Helleday, T. Drugging DNA repair. Science 2016, 352, 1178–1179. [Google Scholar] [CrossRef]
- Saez, G.T. DNA injury and repair systems. Int. J. Mol. Sci. 2018, 19, 1902. [Google Scholar] [CrossRef]
- Lin, W.W.; Yuan, N.; Wang, Z.; Cao, Y.; Fang, Y.X.; Li, X.; Xu, F.; Song, L.; Wang, J.; Zhang, H.; et al. Autophagy confers DNA damage repair pathways to protect the hematopoietic system from nuclear radiation injury. Sci. Rep. 2015, 5, 12362. [Google Scholar] [CrossRef]
- Ji, W.; Yang, L.; Yu, L.; Yuan, J.; Hu, D.; Zhang, W.; Yang, J.; Pang, Y.; Li, W.; Lu, J.; et al. Epigenetic silencing of O6-methylguanine DNA methyltransferase gene in NiS-transformed cells. Carcinogenesis 2008, 29, 1267–1275. [Google Scholar] [CrossRef]
- Iwitzki, F.; Schlepegrell, R.; Eichhorn, U.; Kaina, B.; Beyersmann, D.; Hartwig, A. Nickel(II) inhibits the repair of O6-methylguanine in mammalian cells. Arch. Toxicol. 1998, 72, 681–689. [Google Scholar] [CrossRef]
- Chen, H.B.; Giri, N.C.; Zhang, R.H.; Yamane, K.; Zhang, Y.; Maroney, M.; Costa, M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous tron in the catalytic centers. J. Biol. Chem. 2010, 285, 7374–7383. [Google Scholar] [CrossRef]
- Chervona, Y.; Arita, A.; Costa, M. Carcinogenic metals and the epigenome: Understanding the effect of nickel, arsenic, and chromium. Metallomics 2012, 4, 619–627. [Google Scholar] [CrossRef]
- Wozniak, K.; Blasiak, J. Nickel impairs the repair of UV- and MNNG-damaged DNA. Cell. Mol. Biol. Lett. 2004, 9, 83–94. [Google Scholar]
- Hu, W.; Feng, Z.; Tang, M. Nickel (II) enhances benzo[a]pyrene diol epoxide-induced mutagenesis through inhibition of nucleotide excision repair in human cells: A possible mechanism for nickel (II)-induced carcinogenesis. Carcinogenesis 2004, 25, 455–462. [Google Scholar] [CrossRef][Green Version]
- Lee-Chen, S.F.; Wang, M.C.; Yu, C.T.; Wu, D.R.; Jan, K.Y. Nickel chloride inhibits the DNA repair of UV-treated but not methyl methanesulfonate-treated chinese hamster ovary cells. Biol. Trace Elem. Res. 1993, 37, 39–50. [Google Scholar] [CrossRef]
- Wozniak, K.; Czechowska, A.; Blasiak, J. Nickel(II) affects poly(ADP-ribose) polymerase-mediated DNA repair in normal and cancer cells. Z. Nat. C 2006, 61, 142–148. [Google Scholar] [CrossRef]
- Hartmann, M.; Hartwig, A. Disturbance of DNA damage recognition after UV-irradiation by nickel(II) and cadmium(II) in mammalian cells. Carcinogenesis 1998, 19, 617–621. [Google Scholar] [CrossRef]
- Kasprzak, K.S.; Waalkes, M.P.; Poirier, L.A. Antagonism by essential divalent metals and amino acids of nickel(II)-DNA binding in vitro. Toxicol. Appl. Pharmacol. 1986, 82, 336–343. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, Y.J.; Kim, H.J.; Kim, H.S.; Kang, M.S.; Lee, S.K.; Park, M.K.; Murata, K.; Kim, H.L.; Seo, Y.R. A molecular mechanism of nickel (II): Reduction of nucleotide excision repair activity by structural and functional disruption of p53. Carcinogenesis 2018, 39, 1157–1164. [Google Scholar] [CrossRef]
- Morales, M.E.; Derbes, R.S.; Ade, C.M.; Ortego, J.C.; Stark, J.; Deininger, P.L.; Roy-Engel, A.M. Heavy metal exposure influences double strand break DNA repair outcomes. PLoS ONE 2016, 11, e0151367. [Google Scholar] [CrossRef]
- Yi, C.; He, C. DNA repair by reversal of DNA damage. CSH Perspect. Biol. 2013, 5, a012575. [Google Scholar] [CrossRef]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 2012, 12, 104–120. [Google Scholar] [CrossRef]
- Gerson, S.L. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef]
- Loenarz, C.; Schofield, C.J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011, 36, 7–18. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate-and iron-dependent dioxygenases. Genome Biol. 2001, 2, research0007.1. [Google Scholar] [CrossRef]
- Permata, T.B.M.; Hagiwara, Y.; Sato, H.; Yasuhara, T.; Oike, T.; Gondhowiardjo, S.; Held, K.D.; Nakano, T.; Shibata, A. Base excision repair regulates PD-L1 expression in cancer cells. Oncogene 2019, 38, 4452–4466. [Google Scholar] [CrossRef]
- Krokan, H.E.; Magnar, B.R.S. Base excision repair. CSH Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Nimrat, C.; Yunfu, L.; Santillan, B.A.; Patricia, Y.; Wilson, J.H. Environmental stress induces trinucleotide repeat mutagenesis in human cells. Proc. Nat. Acad. Sci. USA 2015, 112, 3764–3769. [Google Scholar]
- Apostolou, Z.; Chatzinikolaou, G.; Stratigi, K.; Garinis, G.A. Nucleotide excision repair and transcription-associated genome instability. Bioessays 2019, 41, e1800201. [Google Scholar] [CrossRef]
- Zhang, G.H.; Ren, J.C.; Luo, M.; Cui, J.; Du, Y.; Yang, D.; Cui, S.; Wang, X.; Wu, W.; Cao, J.; et al. Association of BER and NER pathway polymorphism haplotypes and micronucleus frequencies with global DNA methylation in benzene-exposed workers of China: Effects of DNA repair genes polymorphisms on genetic damage. Mutat. Res. 2019, 839, 13–20. [Google Scholar] [CrossRef]
- Fagbemi, A.F.; Orelli, B.; Schärer, O.D. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair 2011, 10, 722–729. [Google Scholar] [CrossRef]
- Vincent, M.; Jean Philippe, L.; Thilo, R.; Zhou, Y.; Lee, M.Y.; Jean Marc, E. Sequential recruitment of the repair factors during NER: The role of XPG in initiating the resynthesis step. EMBO J. 2014, 27, 155–167. [Google Scholar]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Sugitani, N.; Sivley, R.M.; Perry, K.E.; Capra, J.A.; Chazin, W.J. XPA: A key scaffold for human nucleotide excision repair. DNA Repair 2016, 44, 123–135. [Google Scholar] [CrossRef]
- Friedberg, E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 2001, 1, 22–33. [Google Scholar] [CrossRef]
- Iyama, T.; Iii, D.M.W. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 2013, 12, 620–636. [Google Scholar] [CrossRef]
- Xing, J.L.; Dinney, C.P.; Shete, S.; Huang, M.S.; Hildebrandt, M.A.; Chen, Z.N.; Gu, J. Comprehensive pathway-based interrogation of genetic variations in the nucleotide excision DNA repair pathway and risk of bladder cancer. Cancer 2012, 118, 205–215. [Google Scholar] [CrossRef]
- Bal, W.; Schwerdtle, T.; Hartwig, A. Mechanism of nickel assault on the zinc finger of DNA repair protein XPA. Chem. Res. Toxicol. 2003, 16, 242–248. [Google Scholar] [CrossRef]
- Staresincic, L.; Fagbemi, A.F.; Enzlin, J.H.; Gourdin, A.M.; Wijgers, N.; Dunand-Sauthier, I.; Giglia-Mari, G.; Clarkson, S.G.; Vermeulen, W.; Scharer, O.D. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009, 28, 1111–1120. [Google Scholar] [CrossRef]
- Li, G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008, 18, 85–98. [Google Scholar] [CrossRef]
- Martin, S.A.; Lord, C.J.; Ashworth, A. Therapeutic targeting of the DNA mismatch repair pathway. Clin. Cancer Res. 2010, 16, 5107–5113. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef]
- Karamurzin, Y.; Rutgers, J.K. DNA mismatch repair deficiency in endometrial carcinoma. Int. J. Gynecol. Pathol. 2009, 28, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Felsberg, J.; Thon, N.; Eigenbrod, S.; Hentschel, B.; Sabel, M.C.; Westphal, M.; Schackert, G.; Kreth, F.W.; Pietsch, T.; Löffler, M. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 2011, 129, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Erie, D.A. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; McCabe, N.; Mullarkey, M.; Cummins, R.; Burgess, D.J.; Nakabeppu, Y.; Oka, S.; Kay, E.; Lord, C.J.; Ashworth, A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell 2010, 17, 235–248. [Google Scholar] [CrossRef]
- Fishel, R.; Lee, J.B. Mismatch repair. In DNA Replication, Recombination, and Repair; Springer: Berlin/Heidelberg, Germany, 2016; pp. 305–339. [Google Scholar]
- Massey, D.J.; Koren, A. Mismatch repair prefers exons. Nat. Genet. 2017, 49, 1673–1674. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Benson, F.E.; Baumann, P.; West, S.C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 1998, 391, 401–404. [Google Scholar] [CrossRef]
- Zhao, Z.; Oh, S.; Li, D.P.; Ni, D.J.; Pirooz, S.D.; Lee, J.H.; Yang, S.H.; Lee, J.Y.; Ghozalli, I.; Costanzo, V.; et al. A Dual Role for UVRAG in Maintaining Chromosomal Stability Independent of Autophagy. Dev. Cell 2012, 22, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003, 4, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Takeda, E.; Kubota, Y.; Okayasu, R. Inhibition of repair of radiation-induced DNA double-strand breaks by nickel and arsenite. Radiat. Res. 2000, 154, 686–691. [Google Scholar] [CrossRef]
- Janion, C. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 2008, 4, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Jiang, Q.; Woodgate, R.; Cox, M.M.; Goodman, M.F. A new model for SOS-induced mutagenesis: How RecA protein activates DNA polymerase V. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Žgur-Bertok, D. DNA damage repair and bacterial pathogens. PLoS Pathog. 2013, 9, e1003711. [Google Scholar] [CrossRef] [PubMed]

| Item | Suppression | Enhancement | No Alteration |
|---|---|---|---|
| Direct reversal | Ji et al. [137], Iwitzki et al. [138], Chen et al. [139], Chervona et al. [140] | ||
| BER | Wu et al. [47], Wozniak and Blasiak [141] | ||
| NER | Hartwig et al. [127], Hu et al. [142], Lee-Chen et al. [143], Wozniak et al. [144], [145], Hartmann and Hartwig [146], Hartwig et al. [128], Hu et al. [125], Wozniak and Blasiak [25], Kim et al. [147] | ||
| MMR | Scanlon et al. [129] | Ji et al. [137] | |
| HR | Scanlon et al. [129] | ||
| NHEJ | Morales et al. [148] | Scanlon et al. [129] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Liu, H.; Wu, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Nickel Carcinogenesis Mechanism: DNA Damage. Int. J. Mol. Sci. 2019, 20, 4690. https://doi.org/10.3390/ijms20194690
Guo H, Liu H, Wu H, Cui H, Fang J, Zuo Z, Deng J, Li Y, Wang X, Zhao L. Nickel Carcinogenesis Mechanism: DNA Damage. International Journal of Molecular Sciences. 2019; 20(19):4690. https://doi.org/10.3390/ijms20194690
Chicago/Turabian StyleGuo, Hongrui, Huan Liu, Hongbin Wu, Hengmin Cui, Jing Fang, Zhicai Zuo, Junliang Deng, Yinglun Li, Xun Wang, and Ling Zhao. 2019. "Nickel Carcinogenesis Mechanism: DNA Damage" International Journal of Molecular Sciences 20, no. 19: 4690. https://doi.org/10.3390/ijms20194690
APA StyleGuo, H., Liu, H., Wu, H., Cui, H., Fang, J., Zuo, Z., Deng, J., Li, Y., Wang, X., & Zhao, L. (2019). Nickel Carcinogenesis Mechanism: DNA Damage. International Journal of Molecular Sciences, 20(19), 4690. https://doi.org/10.3390/ijms20194690





