Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat
Abstract
1. Introduction
2. Results
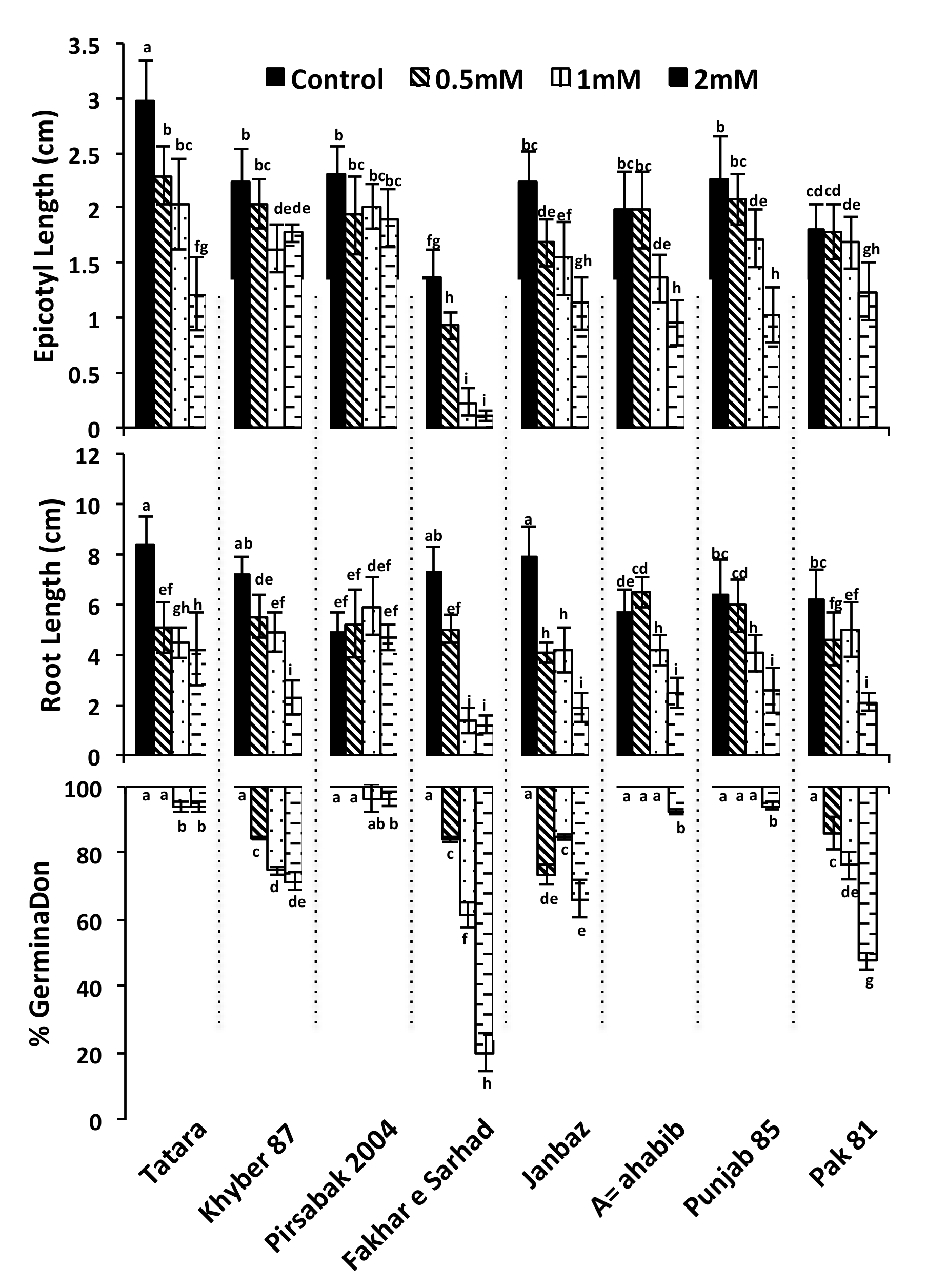
2.1. Genetic Diversity of Wheat Varieties Against Pb Toxicity
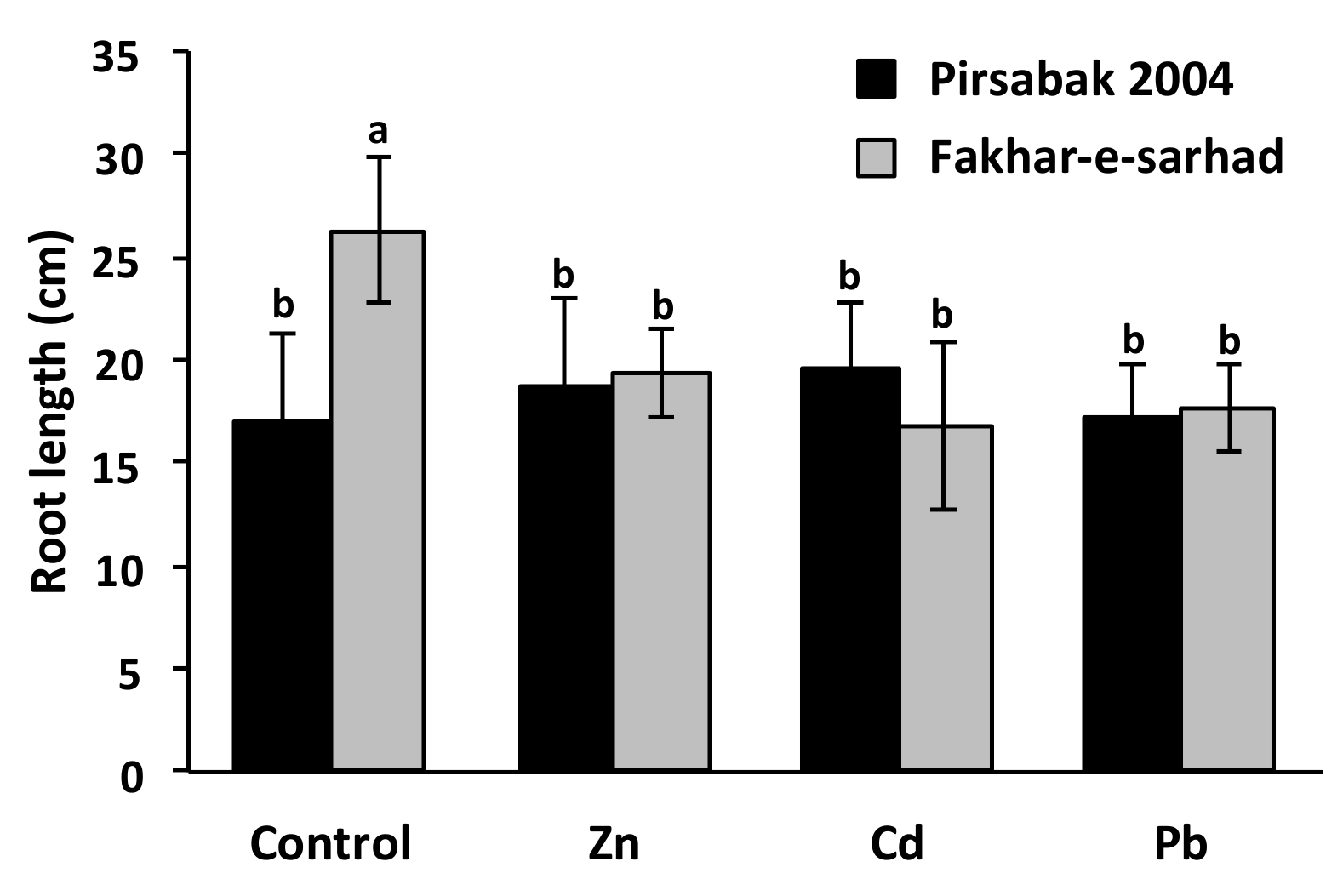
2.2. Pirsabak 2004 and Fakhar-e-sarhad Sensitivity to Pb, Cd and Zn
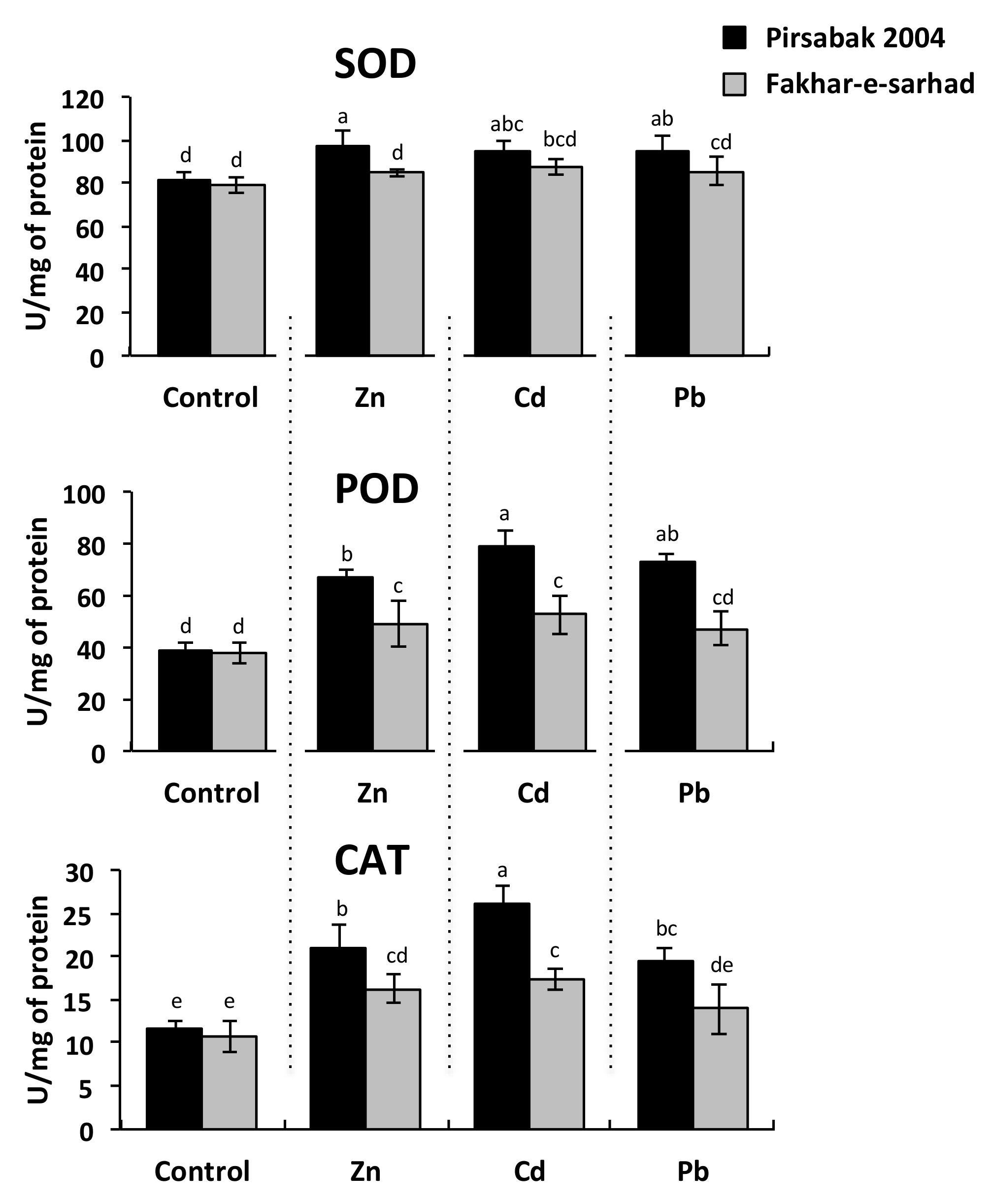
2.3. Antioxidant Activity in Pirsabak 2004 and Fakhar-e-sarhad in Response to Metal Stress
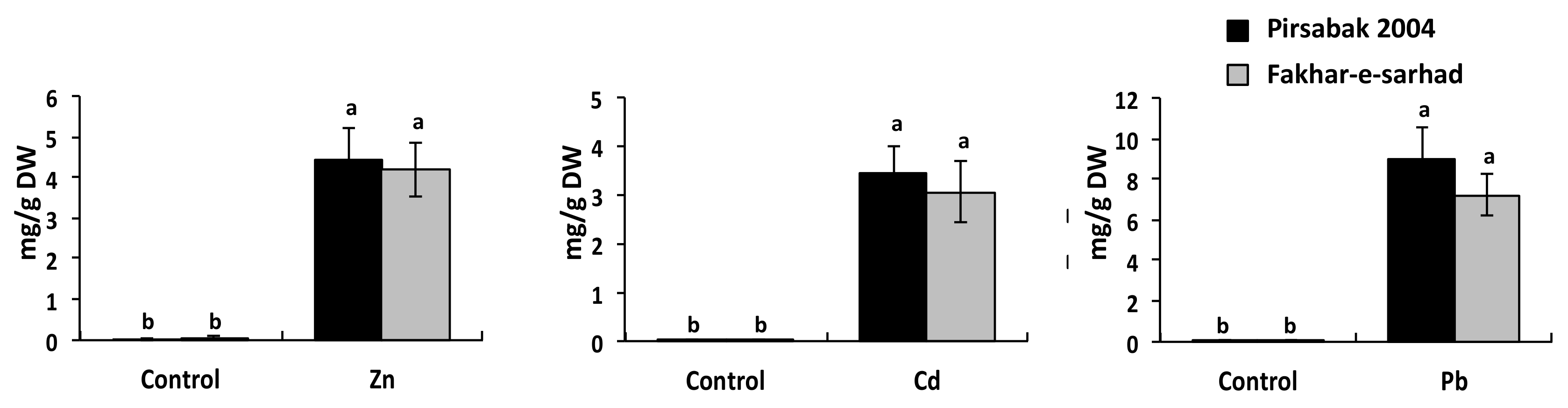
2.4. Accumulation of Pb, Cd and Zn in Pirsabak 2004 and Fakhar-e-Sarhad
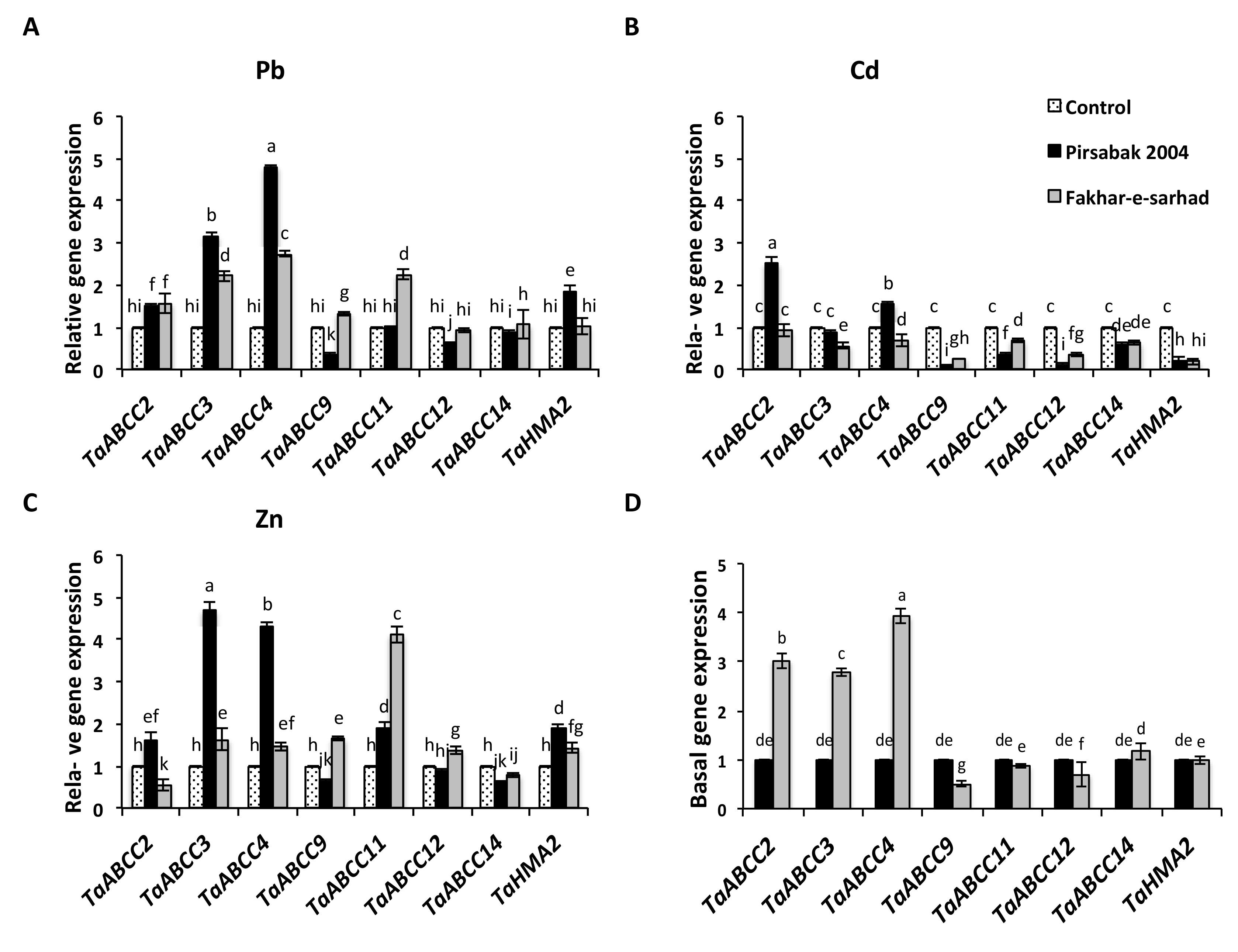
2.5. Expression of Root Expressed TaABCCs and TaHMA2 transporters in Response to Pb, Cd and Zn Metal Stresses
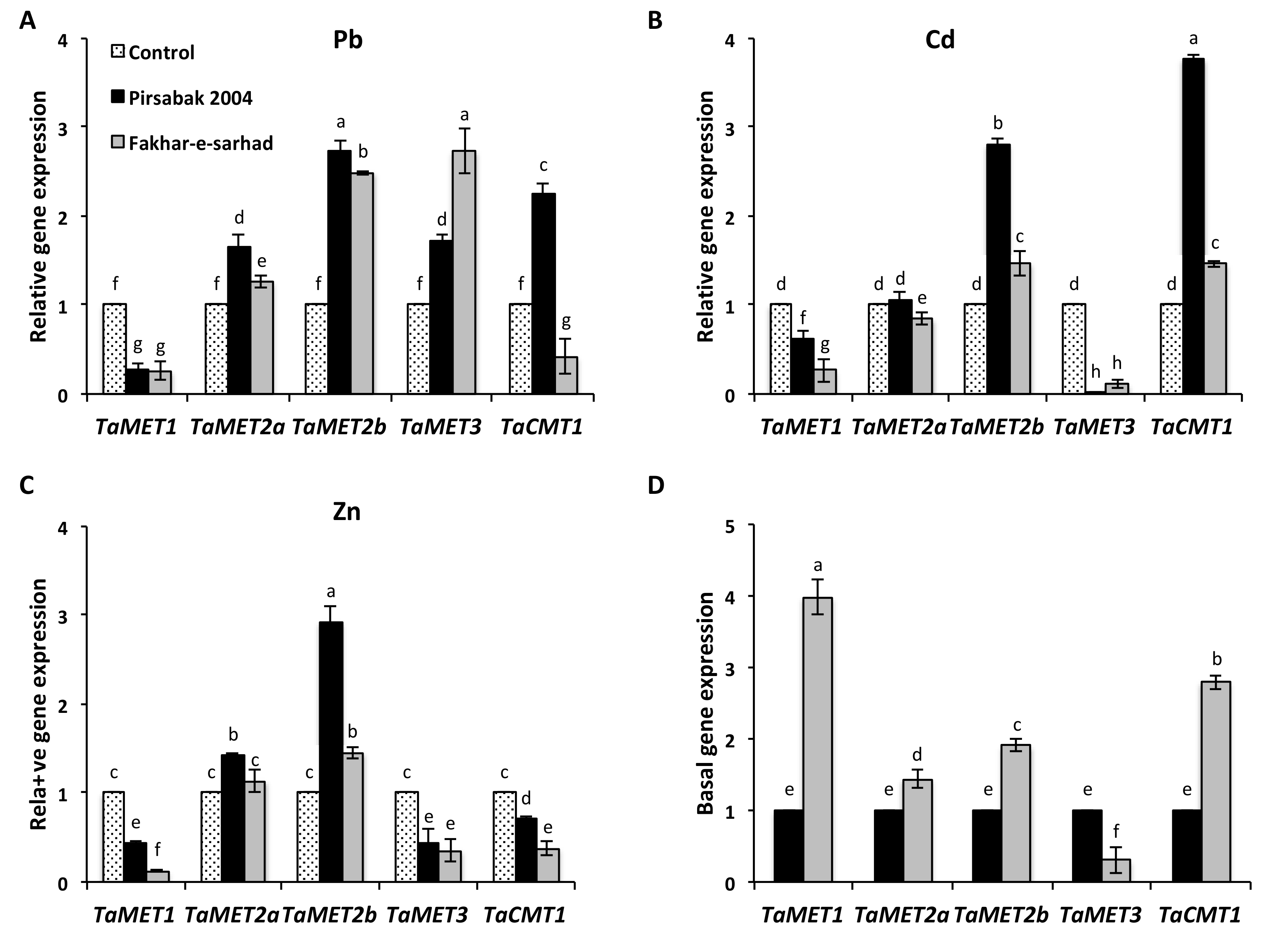
2.6. DNA Methyltransferase Expression in Response to Pb, Cd and Zn Metal Stresses
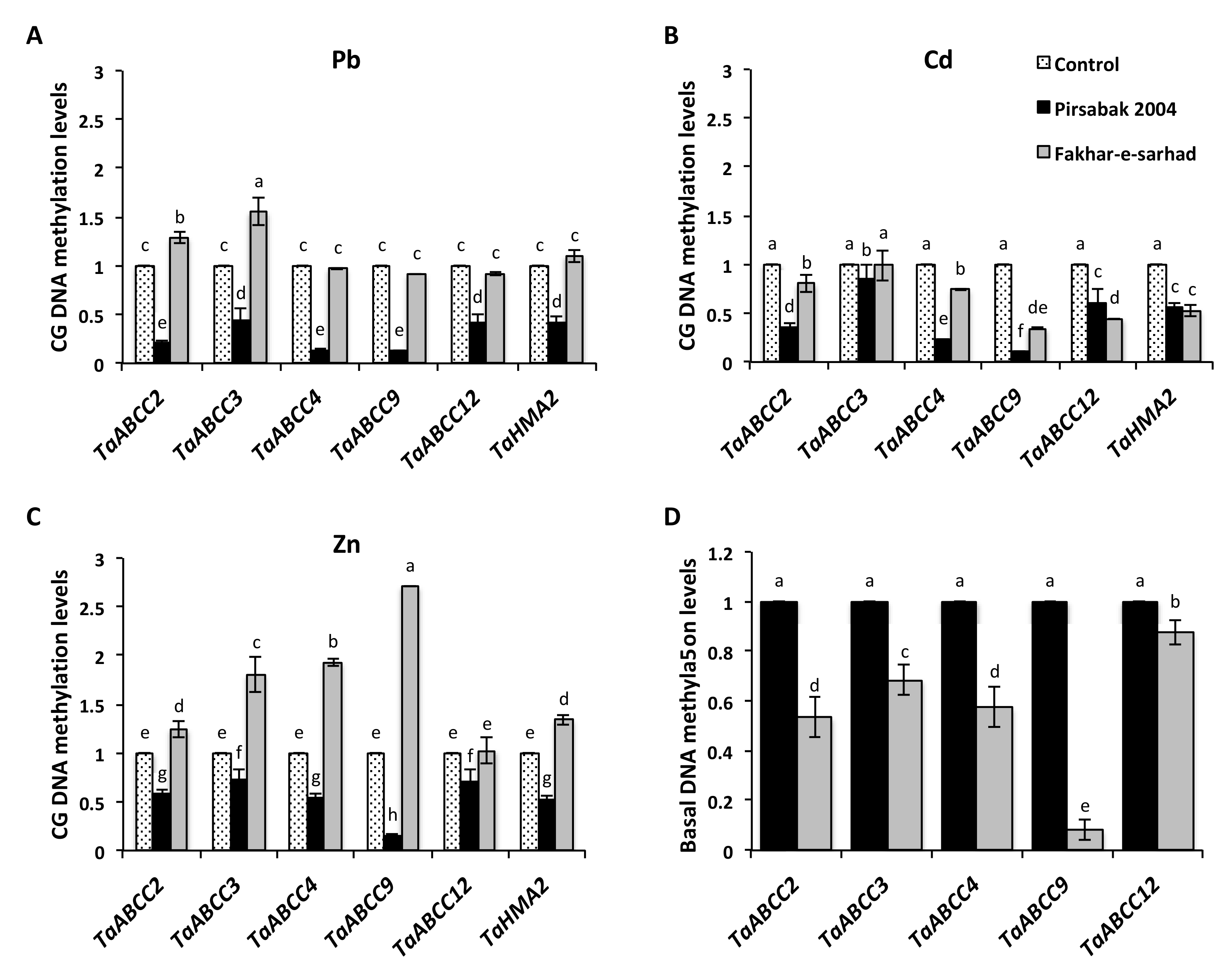
2.7. DNA Hypomethylation of Pirsabak 2004 in Response to Pb, Cd and Zn Metal Stresses
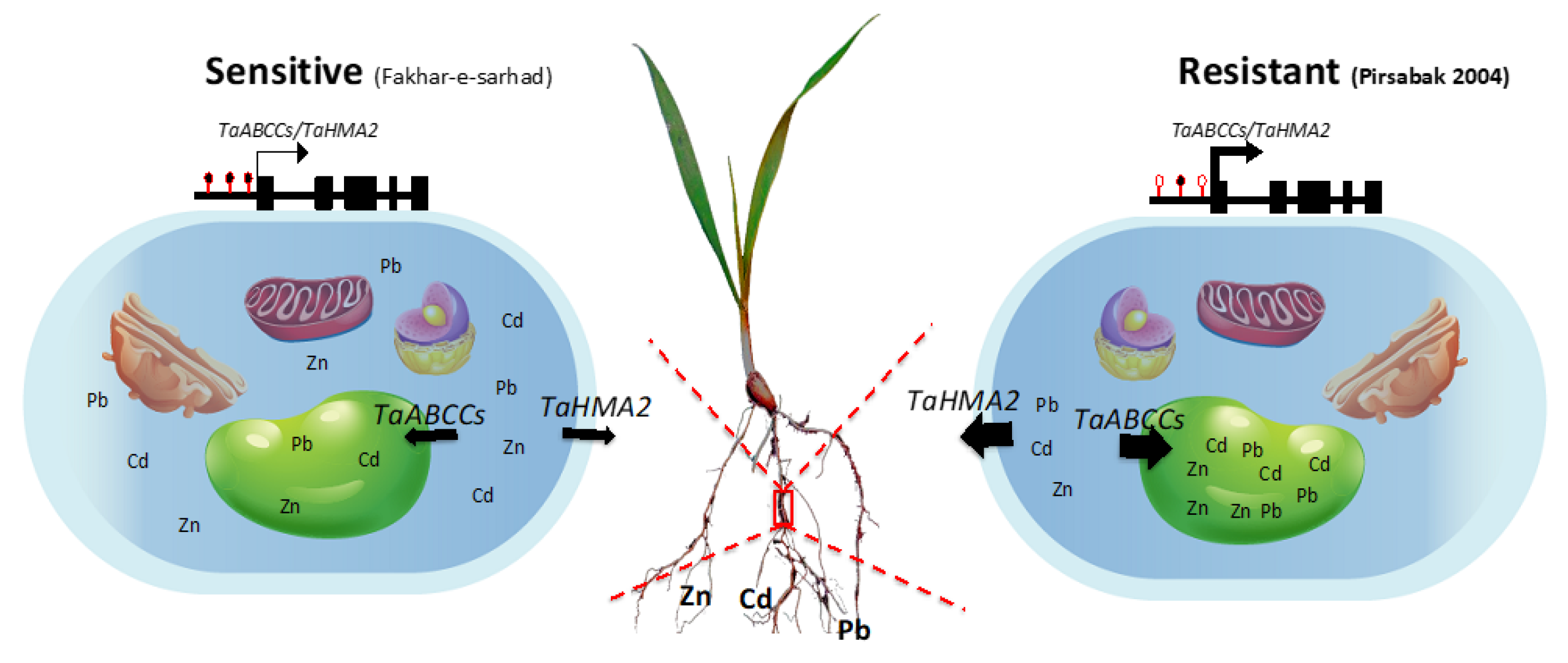
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Sowing and Growth Conditions for MS Media
4.3. Sowing and Growth Conditions for Hydroponics
4.4. Atomic Absorption Analysis
4.5. Extraction and Measurement of Antioxidant Enzymes
4.6. Gene Expression Analysis
4.7. DNA Methylation Chop-Quantitative PCR (Chop-qPCR)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Babst-Kostecka, A.A.; Waldmann, P.; Frérot, H.; Vollenweider, P. Plant adaptation to metal polluted environments—Physiological, morphological, and evolutionary insights from Biscutella laevigata. Environ. Exp. Bot. 2016, 127, 1–13. [Google Scholar] [CrossRef]
- Sengar, R.S.; Gautam, M.; Sengar, R.S.; Garg, S.K.; Sengar, K.; Chaudhary, R. Lead stress effects on physiobiochemical activities of higher plants. Rev. Env. Contam. Toxicol. 2008, 196, 73–93. [Google Scholar]
- Hartwig, A. Cadmium and cancer. Met. Ions. Life Sci. 2013, 11, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Pourrut, B.; Shahid, M.; Dumat, C.; Winterton, P.; Pinelli, E. Lead Uptake, Toxicity, and Detoxification in Plants. In Reviews of Environmental Contamination and Toxicology; Whitacre, D.M., Ed.; Springer: New York, NY, USA, 2011; Volume 213, pp. 113–136. [Google Scholar]
- Gill, S.S.; Tuteja, N. Cadmium stress tolerance in crop plants. Plant. Signal. Behav. 2011, 6, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Andresen, E.; Küpper, H. Cadmium Toxicity in Plants. In Cadmium: From Toxicity to Essentiality; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 395–413. [Google Scholar]
- Kobayashi, T.; Nishizawa, N.K. Iron Uptake, Translocation, and Regulation in Higher Plants. Annu. Rev. Plant. Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Sadeghzadeh, B.; Rengel, Z. Zinc in Soils and Crop Nutrition. In The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops; Wiley-Blackwell: Brussels, Belgium, 2011; pp. 335–375. [Google Scholar]
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in plants. New Phytol. 2007, 173, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, A.; Rani, R.; Kumar, S.; Gautam, A. Heavy metal detoxification and tolerance mechanisms in plants: Implications for phytoremediation. Environ. Rev. 2015, 24, 39–51. [Google Scholar] [CrossRef]
- Ji, H.; Peng, Y.; Meckes, N.; Allen, S.; Stewart, C.N., Jr.; Traw, M.B. ATP-dependent binding cassette transporter G family member 16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant. Physiol. 2014, 166, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Sanchez-Fernandez, R.; Li, Z.S.; Rea, P.A. Enhanced multispecificity of arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 2001, 276, 8648–8656. [Google Scholar] [CrossRef] [PubMed]
- Nagy, R.; Grob, H.; Weder, B.; Green, P.; Klein, M.; Frelet-Barrand, A.; Schjoerring, J.K.; Brearley, C.; Martinoia, E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622. [Google Scholar] [CrossRef]
- Park, J.; Song, W.-Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant. J. 2012, 69, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-S.; Szczypka, M.; Lu, Y.-P.; Thiele, D.J.; Rea, P.A. The Yeast Cadmium Factor Protein (YCF1) Is a Vacuolar Glutathione S-Conjugate Pump. J. Biol. Chem. 1996, 271, 6509–6517. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.G.; Mason, D.L.; Liu, G.; Rea, P.A.; Bachhawat, A.K.; Michaelis, S. Localization, Regulation, and Substrate Transport Properties of Bpt1p, a Saccharomyces cerevisiae MRP-Type ABC Transporter. Eukaryot. Cell 2002, 1, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Bhati, K.K.; Sharma, S.; Aggarwal, S.; Kaur, M.; Shukla, V.; Kaur, J.; Mantri, S.; Pandey, A.K. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front. Plant. Sci. 2015, 6, 488. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Clark, I.M.; He, X.L.; Pallett, K.E.; Cole, D.J.; Hallahan, D.L. Co-induction of glutathione-S-transferases and multidrug resistance associated protein by xenobiotics in wheat. Pest. Manag. Sci. 2003, 59, 202–214. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P(1B)-ATPases--an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Arguello, J.M. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 2003, 195, 93–108. [Google Scholar] [CrossRef]
- Axelsen, K.B.; Palmgren, M.G. Evolution of Substrate Specificities in the P-Type ATPase Superfamily. J. Mol. Evol. 1998, 46, 84–101. [Google Scholar] [CrossRef]
- Tan, J.; Wang, J.; Chai, T.; Zhang, Y.; Feng, S.; Li, Y.; Zhao, H.; Liu, H.; Chai, X. Functional analyses of TaHMA2, a P1B-type ATPase in wheat. Plant Biotechnol. J. 2013, 11, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Berr, A.; Shafiq, S.; Shen, W.H. Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 2011, 1809, 567–576. [Google Scholar] [CrossRef]
- Zhang, M.; Kimatu, J.N.; Xu, K.; Liu, B. DNA cytosine methylation in plant development. J. Genet. Genom. 2010, 37, 1–12. [Google Scholar] [CrossRef]
- Shafiq, S.; Khan, A.R. Plant Epigenetics and Crop Improvement. In PlantOmics: The Omics of Plant Science; Barh, D., Khan, M.S., Davies, E., Eds.; Springer: New Delhi, India, 2015; pp. 157–179. [Google Scholar]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Kankel, M.W.; Ramsey, D.E.; Stokes, T.L.; Flowers, S.K.; Haag, J.R.; Jeddeloh, J.A.; Riddle, N.C.; Verbsky, M.L.; Richards, E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003, 163, 1109. [Google Scholar] [PubMed]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jacobsen, S.E. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef]
- Dai, Y.; Ni, Z.; Dai, J.; Zhao, T.; Sun, Q. Isolation and expression analysis of genes encoding DNA methyltransferase in wheat (Triticum aestivum L.). Biochim. Biophys. Acta 2005, 1729, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, J. Plant genomic DNA methylation in response to stresses: Potential applications and challenges in plant breeding. Prog. Nat. Sci. 2009, 19, 1037–1045. [Google Scholar] [CrossRef]
- Dowen, H.R.; Pelizzola, M.; Schmitz, R.; Lister, R.; Dowen, J.; Nery, J.; Dixon, J.; R Ecker, J. Widespread dynamic DNA methylation in response to biotic stress. P. Nat. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Vilahur, N.; Vahter, M.; Broberg, K. The Epigenetic Effects of Prenatal Cadmium Exposure. Curr. Environ. Health. Rep. 2015, 2, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Arita, A.; Costa, M. Epigenetics in metal carcinogenesis: Nickel, Arsenic, Chromium and Cadmium. Metallomics 2009, 1, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Aina, R.; Sgorbati, S.; Santagostino, A.; Labra, M.; Ghiani, A.; Citterio, S. Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plant 2004, 121, 472–480. [Google Scholar] [CrossRef]
- Filek, M.; Keskinen, R.; Hartikainen, H.; Szarejko, I.; Janiak, A.; Miszalski, Z.; Golda, A. The protective role of selenium in rape seedlings subjected to cadmium stress. J. Plant Physiol. 2008, 165, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Liu, L.W.; Gong, Y.Q.; Huang, D.Q.; Wang, F.; He, L.L. Analysis of genomic DNA methylation level in radish under cadmium stress by methylation-sensitive amplified polymorphism technique. J. Plant Physiol. Mol. Biol. 2007, 33, 219–226. [Google Scholar]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, G.; Shi, G.; Pan, F. Toxicity of Cu, Pb, and Zn on Seed Germination and Young Seedlings of Wheat (Triticum aestivum L.). In International Conference on Computer and Computing Technologies in Agriculture; Springer: Heidelberg/Berlin, Germany, 2010; pp. 231–240. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Elstner, E.F. Oxygen Activation and Oxygen Toxicity. Annu. Rev. Plant. Physiol. 1982, 33, 73–96. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Vitte, C. Transposable elements, a treasure trove to decipher epigenetic variation: Insights from Arabidopsis and crop epigenomes. J. Exp. Bot. 2014, 65, 2801–2812. [Google Scholar] [CrossRef] [PubMed]
- Taspinar, M.S.; Agar, G.; Alpsoy, L.; Yildirim, N.; Bozari, S.; Sevsay, S. The protective role of zinc and calcium in Vicia faba seedlings subjected to cadmium stress. Toxicol. Ind. Health 2010, 27, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, X.; Li, S.; Wang, Z. Effect of nickel chloride on Arabidopsis genomic DNA and methylation of 18S rDNA. Electron. J. Biotechnol. 2014, 283, 51–57. [Google Scholar] [CrossRef][Green Version]
- Szczypka, M.S.; Wemmie, J.A.; Moye-Rowley, W.S.; Thiele, D.J. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 1994, 269, 22853–22857. [Google Scholar] [PubMed]
- Ghosh, M.; Shen, J.; Rosen, B.P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1999, 96, 5001–5006. [Google Scholar] [CrossRef]
- Song, W.-Y.; Ju Sohn, E.; Martinoia, E.; Jik Lee, Y.; Yang, Y.-Y.; Jasinski, M.; Forestier, C.; Hwang, I.; Lee, Y. Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat. Biotech. 2003, 21, 914–919. [Google Scholar] [CrossRef]
- Shim, D.; Kim, S.; Choi, Y.I.; Song, W.Y.; Park, J.; Youk, E.S.; Jeong, S.C.; Martinoia, E.; Noh, E.W.; Lee, Y. Transgenic poplar trees expressing yeast cadmium factor 1 exhibit the characteristics necessary for the phytoremediation of mine tailing soil. Chemosphere 2013, 90, 1478–1486. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.U.; Min, S.R.; Jeong, W.J.; Sultana, S.; Choi, K.S.; Song, W.Y.; Lee, Y.; Lim, Y.P.; Liu, J.R. Overexpression of a yeast cadmium factor 1 (YCF1) enhances heavy metal tolerance and accumulation in Brassica juncea. Plant. Cell Tiss. Org. Cult. 2011, 105, 85–91. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Shahzad, M.; Witzel, K.; Zörb, C.; Mühling, K.H. Growth-Related Changes in Subcellular Ion Patterns in Maize Leaves (Zea mays L.) under Salt Stress. J. Agron. Crop. Sci. 2012, 198, 46–56. [Google Scholar] [CrossRef]
- Csiszár, J.; Lantos, E.; Tari, I.; Emilian, M.; Wodala, B.; Vashegyi, A.; Horvath, F.; Pécsváradi, A.; Szabo, M.; Bartha-Dima, B.; et al. Antioxidant enzyme activities in Allium species and their cultivars under water stress. Plant. Soil Environ. 2007, 53, 517–523. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Shafiq, S.; Berr, A.F.; Shen, W.H. Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. 2014, 201, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tang, K.; Wang, B.; Duan, C.-G.; Lang, Z.; Zhu, J.-K. Protocol: A beginner’s guide to the analysis of RNA-directed DNA methylation in plants. Plant Methods 2014, 10, 18. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shafiq, S.; Zeb, Q.; Ali, A.; Sajjad, Y.; Nazir, R.; Widemann, E.; Liu, L. Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. Int. J. Mol. Sci. 2019, 20, 4676. https://doi.org/10.3390/ijms20194676
Shafiq S, Zeb Q, Ali A, Sajjad Y, Nazir R, Widemann E, Liu L. Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. International Journal of Molecular Sciences. 2019; 20(19):4676. https://doi.org/10.3390/ijms20194676
Chicago/Turabian StyleShafiq, Sarfraz, Qudsia Zeb, Asim Ali, Yasar Sajjad, Rashid Nazir, Emilie Widemann, and Liangyu Liu. 2019. "Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat" International Journal of Molecular Sciences 20, no. 19: 4676. https://doi.org/10.3390/ijms20194676
APA StyleShafiq, S., Zeb, Q., Ali, A., Sajjad, Y., Nazir, R., Widemann, E., & Liu, L. (2019). Lead, Cadmium and Zinc Phytotoxicity Alter DNA Methylation Levels to Confer Heavy Metal Tolerance in Wheat. International Journal of Molecular Sciences, 20(19), 4676. https://doi.org/10.3390/ijms20194676





