Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment
Abstract
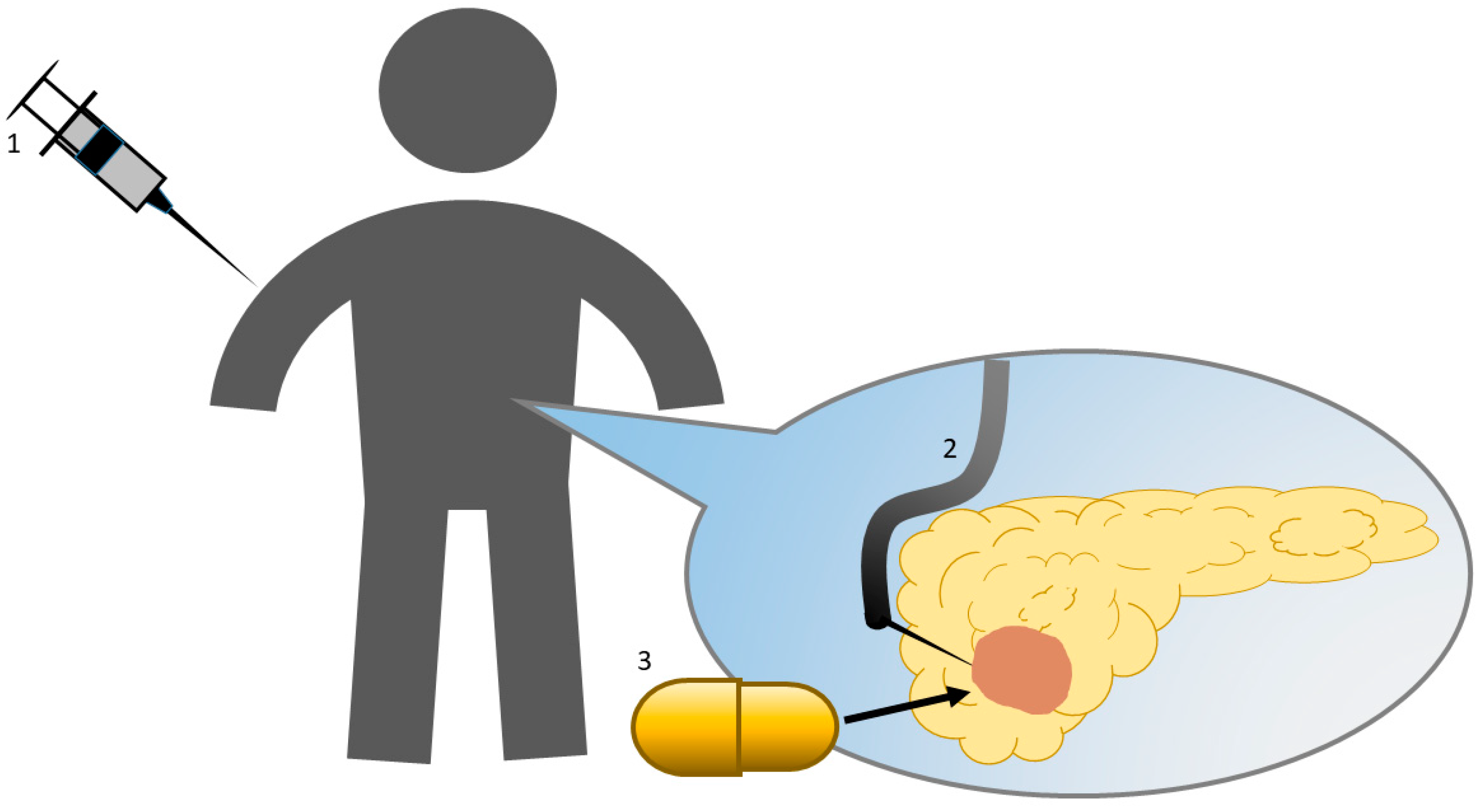
1. Introduction
2. Functions
2.1. Antisense Oligonucleotides
2.2. siRNAs
2.3. Aptamers
2.4. Decoys
2.5. CpG Oligos
3. Modifications of Nucleic Acid Drugs
3.1. Structural Modifications of Nucleic Acid Drugs
3.2. Conjugation of Ligand or Cell-Penetrating Peptides
4. Aptamers
5. Drug Delivery Systems of Nucleic Acid Drugs
6. Clinical Trials
6.1. Antisense Oligonucleotide
6.2. Clinical Trials for siRNAs
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Torres, C.; Grippo, P.J. Pancreatic cancer subtypes: A roadmap for precision medicine. Ann. Med. 2018, 50, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, F.C.; Kerr, S.E.; Kipp, B.R.; Voss, J.S.; Minot, D.M.; Tu, Z.J.; Henry, M.R.; Graham, R.P.; Vasmatzis, G.; Cheville, J.C.; et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016, 7, 54526–54536. [Google Scholar] [CrossRef] [PubMed]
- Barata, P.; Sood, A.K.; Hong, D.S. RNA-targeted therapeutics in cancer clinical trials: Current status and future directions. Cancer Treat. Rev. 2016, 50, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Jansen, B.; Zangemeister-Wittke, U. Antisense therapy for cancer—The time of truth. Lancet Oncol. 2002, 3, 672–683. [Google Scholar] [CrossRef]
- Moreno, P.M.; Pego, A.P. Therapeutic antisense oligonucleotides against cancer: Hurdling to the clinic. Front. Chem. 2014, 2, 87. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers (Basel) 2018, 10, 9. [Google Scholar] [CrossRef]
- Dausse, E.; Da Rocha Gomes, S.; Toulme, J.J. Aptamers: A new class of oligonucleotides in the drug discovery pipeline? Curr. Opin. Pharmacol. 2009, 9, 602–607. [Google Scholar] [CrossRef]
- Hecker, M.; Wagner, A.H. Transcription factor decoy technology: A therapeutic update. Biochem. Pharmacol. 2017, 144, 29–34. [Google Scholar] [CrossRef]
- Vollmer, J.; Weeratna, R.D.; Jurk, M.; Davis, H.L.; Schetter, C.; Wullner, M.; Wader, T.; Liu, M.; Kritzler, A.; Krieg, A.M. Impact of modifications of heterocyclic bases in CpG dinucleotides on their immune-modulatory activity. J. Leukoc. Biol. 2004, 76, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Yang, X.; Gharpure, K.M.; Hatakeyama, H.; Egli, M.; McGuire, M.H.; Nagaraja, A.S.; Miyake, T.M.; Rupaimoole, R.; Pecot, C.V.; et al. 2’-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat. Commun. 2014, 5, 3459. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.Z.; Grundy, J.S.; Geary, R.S. Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin. Drug Metab. Toxicol. 2013, 9, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Amarzguioui, M.; Holen, T.; Babaie, E.; Prydz, H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003, 31, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Pan, T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011, 44, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Fattal, E.; Barratt, G. Nanotechnologies and controlled release systems for the delivery of antisense oligonucleotides and small interfering RNA. Br. J. Pharmacol. 2009, 157, 179–194. [Google Scholar] [CrossRef]
- Simeoni, F.; Morris, M.C.; Heitz, F.; Divita, G. Insight into the mechanism of the peptide-based gene delivery system MPG: Implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003, 31, 2717–2724. [Google Scholar] [CrossRef]
- Xie, X.; Yang, Y.; Lin, W.; Liu, H.; Liu, H.; Yang, Y.; Chen, Y.; Fu, X.; Deng, J. Cell-penetrating peptide-siRNA conjugate loaded YSA-modified nanobubbles for ultrasound triggered siRNA delivery. Colloids Surf. B Biointerfaces. 2015, 136, 641–650. [Google Scholar] [CrossRef]
- Arami, S.; Mahdavi, M.; Rashidi, M.R.; Yekta, R.; Rahnamay, M.; Molavi, L.; Hejazi, M.S.; Samadi, N. Apoptosis induction activity and molecular docking studies of survivin siRNA carried by Fe3O4-PEG-LAC-chitosan-PEI nanoparticles in MCF-7 human breast cancer cells. J. Pharm. Biomed. Anal. 2017, 142, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, M.M.; Meraviglia-Crivelli de Caso, D.; Menon, A.P.; Pastor, F. Aptamer-iRNAs as Therapeutics for Cancer Treatment. Pharmaceuticals (Basel) 2018, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Ramot, Y.; Rotkopf, S.; Gabai, R.M.; Zorde Khvalevsky, E.; Muravnik, S.; Marzoli, G.A.; Domb, A.J.; Shemi, A.; Nyska, A. Preclinical Safety Evaluation in Rats of a Polymeric Matrix Containing an siRNA Drug Used as a Local and Prolonged Delivery System for Pancreatic Cancer Therapy. Toxicol. Pathol. 2016, 44, 856–865. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef]
- Jing, H.; Cheng, W.; Li, S.; Wu, B.; Leng, X.; Xu, S.; Tian, J. Novel cell-penetrating peptide-loaded nanobubbles synergized with ultrasound irradiation enhance EGFR siRNA delivery for triple negative Breast cancer therapy. Colloids Surf. B Biointerfaces 2016, 146, 387–395. [Google Scholar] [CrossRef]
- Rabbani, P.S.; Zhou, A.; Borab, Z.M.; Frezzo, J.A.; Srivastava, N.; More, H.T.; Rifkin, W.J.; David, J.A.; Berens, S.J.; Chen, R.; et al. Novel lipoproteoplex delivers Keap1 siRNA based gene therapy to accelerate diabetic wound healing. Biomaterials 2017, 132, 1–15. [Google Scholar] [CrossRef]
- Wang, Y.; Miao, L.; Satterlee, A.; Huang, L. Delivery of oligonucleotides with lipid nanoparticles. Adv. Drug Deliv. Rev. 2015, 87, 68–80. [Google Scholar] [CrossRef]
- Amjadi, M.; Mostaghaci, B.; Sitti, M. Recent Advances in Skin Penetration Enhancers for Transdermal Gene and Drug Delivery. Curr. Gene Ther. 2017, 17, 139–146. [Google Scholar] [CrossRef]
- Yin, T.; Wang, L.; Yin, L.; Zhou, J.; Huo, M. Co-delivery of hydrophobic paclitaxel and hydrophilic AURKA specific siRNA by redox-sensitive micelles for effective treatment of breast cancer. Biomaterials 2015, 61, 10–25. [Google Scholar] [CrossRef]
- Shen, J.; Wang, Q.; Hu, Q.; Li, Y.; Tang, G.; Chu, P.K. Restoration of chemosensitivity by multifunctional micelles mediated by P-gp siRNA to reverse MDR. Biomaterials 2014, 35, 8621–8634. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Xu, J.; Zhang, B.; Yao, W.; Xu, W.; Wu, W.; Xu, Y.; Wang, H.; Ni, Q.; Hou, H.; et al. RGD-conjugated albumin nanoparticles as a novel delivery vehicle in pancreatic cancer therapy. Cancer Biol. Ther. 2012, 13, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Kahalekar, V.; Gupta, D.T.; Bhatt, P.; Shukla, A.; Bhatia, S. Fully covered self-expanding metallic stent placement for benign refractory esophageal strictures. Indian J. Gastroenterol. 2017, 36, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Matsukawa, M.; Fujii, Y.; Matsuda, Y.; Arai, T.; Ochiai, Y.; Itoi, T.; Yahagi, N. Effects of EUS-guided intratumoral injection of oligonucleotide STNM01 on tumor growth, histology, and overall survival in patients with unresectable pancreatic cancer. Gastrointest Endosc. 2018, 87, 1126–1131. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; David, E.B.; Raskin, S.; et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015, 6, 24560–24570. [Google Scholar] [CrossRef] [PubMed]
- Shemi, A.; Khvalevsky, E.Z.; Gabai, R.M.; Domb, A.; Barenholz, Y. Multistep, effective drug distribution within solid tumors. Oncotarget 2015, 6, 39564–39577. [Google Scholar] [CrossRef]
- Yamahatsu, K.; Matsuda, Y.; Ishiwata, T.; Uchida, E.; Naito, Z. Nestin as a novel therapeutic target for pancreatic cancer via tumor angiogenesis. Int. J. Oncol. 2012, 40, 1345–1357. [Google Scholar]
- Matsuda, Y.; Ishiwata, T.; Yoshimura, H.; Yamashita, S.; Ushijima, T.; Arai, T. Systemic Administration of Small Interfering RNA Targeting Human Nestin Inhibits Pancreatic Cancer Cell Proliferation and Metastasis. Pancreas 2016, 45, 93–100. [Google Scholar] [CrossRef]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whalley, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Mahadevan, D.; Chalasani, P.; Rensvold, D.; Kurtin, S.; Pretzinger, C.; Jolivet, J.; Ramanathan, R.K.; Von Hoff, D.D.; Weiss, G.J. Phase I trial of AEG35156 an antisense oligonucleotide to XIAP plus gemcitabine in patients with metastatic pancreatic ductal adenocarcinoma. Am. J. Clin. Oncol. 2013, 36, 239–243. [Google Scholar] [CrossRef]
- Goel, S.; Desai, K.; Macapinlac, M.; Wadler, S.; Goldberg, G.; Fields, A.; Einstein, M.; Volterra, F.; Wong, B.; Martin, R.; et al. A phase I safety and dose escalation trial of docetaxel combined with GEM231, a second generation antisense oligonucleotide targeting protein kinase A R1alpha in patients with advanced solid cancers. Investig. New Drugs 2006, 24, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Alberts, S.R.; Schroeder, M.; Erlichman, C.; Steen, P.D.; Foster, N.R.; Moore, D.F., Jr.; Rowland, K.M., Jr.; Nair, S.; Tschetter, L.K.; Fitch, T.R. Gemcitabine and ISIS-2503 for patients with locally advanced or metastatic pancreatic adenocarcinoma: A North Central Cancer Treatment Group phase II trial. J. Clin. Oncol. 2004, 22, 4944–4950. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.H.; Murphy, P.B.; Peyton, J.D.; Shipley, D.L.; Al-Hazzouri, A.; Rodriguez, F.A.; Womack, M.S.; Xiong, H.Q.; Waterhouse, D.M.; Tempero, M.A.; et al. A Randomized, Double-Blinded, Phase II Trial of Gemcitabine and Nab-Paclitaxel Plus Apatorsen or Placebo in Patients with Metastatic Pancreatic Cancer: The RAINIER Trial. Oncologist 2017, 22, 1427–e129. [Google Scholar] [CrossRef] [PubMed]
- Jaschinski, F.; Rothhammer, T.; Jachimczak, P.; Seitz, C.; Schneider, A.; Schlingensiepen, K.H. The antisense oligonucleotide trabedersen (AP 12009) for the targeted inhibition of TGF-beta2. Curr. Pharm. Biotechnol. 2011, 12, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Vitravene Study Group. A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. Am. J. Ophthalmol. 2002, 133, 467–474. [Google Scholar]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Raal, F.J.; Santos, R.D.; Blom, D.J.; Marais, A.D.; Charng, M.J.; Cromwell, W.C.; Lachmann, R.H.; Gaudet, D.; Tan, J.L.; Chasan-Taber, S.; et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 375, 998–1006. [Google Scholar] [CrossRef]
- Stein, C.A. Eteplirsen Approved for Duchenne Muscular Dystrophy: The FDA Faces a Difficult Choice. Mol. Ther. 2016, 24, 1884–1885. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010, 24, 1634–1644. [Google Scholar] [CrossRef]
- Splawn, L.M.; Bailey, C.A.; Medina, J.P.; Cho, J.C. Heplisav-B vaccination for the prevention of hepatitis B virus infection in adults in the United States. Drugs Today (Barc.) 2018, 54, 399–405. [Google Scholar] [CrossRef]
- Gales, L. Tegsedi (Inotersen): An Antisense Oligonucleotide Approved for the Treatment of Adult Patients with Hereditary Transthyretin Amyloidosis. Pharmaceuticals (Basel) 2019, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2019, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.J.; Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yokoyama, J.; Kawauchi, Y.; Honda, Y.; Sato, H.; Aoyagi, Y.; Terai, S.; Okazaki, K.; Suzuki, Y.; Sameshima, Y.; et al. Phase 1 Clinical Study of siRNA Targeting Carbohydrate Sulphotransferase 15 in Crohn’s Disease Patients with Active Mucosal Lesions. J. Crohns Colitis 2017, 11, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Schultheis, B.; Strumberg, D.; Kuhlmann, J.; Wolf, M.; Link, K.; Seufferlein, T.; Kaufmann, J.; Gebhardt, F.; Bruyniks, N.; Pelzer, U. A phase Ib/IIa study of combination therapy with gemcitabine and Atu027 in patients with locally advanced or metastatic pancreatic adenocarcinoma. J. Clin. Oncol. 2016, 34, 385. [Google Scholar] [CrossRef]
- Demeure, M.J.; Armaghany, T.; Ejadi, S.; Ramanathan, R.K.; Elfiky, A.; Strosberg, J.R.; Smith, D.C.; Whitsett, T.; Liang, W.S.; Sekar, S.; et al. A phase I/II study of TKM-080301, a PLK1-targeted RNAi in patients with adrenocortical cancer (ACC). J. Clin. Oncol. 2016, 34, 2547. [Google Scholar] [CrossRef]
| Antisense Oligonucleotides | siRNAs | Antisense miRNAs | miRNA Mimics | Decoys | Aptamers | CpG Oligodeoxynucleotides | |
|---|---|---|---|---|---|---|---|
| Structure | Single strand DNA/RNA | Double strand RNA | Single strand DNA/RNA | Double strand RNA | Double strand DNA | Single strand DNA/RNA | Single strand DNA |
| Length (base pairs) | 12–21 20–30 | 20–25 | 12–16 | 20–25 | 20 | 26–45 | 20 |
| Site | Intracellular (nucleus, cytoplasm) | Intracellular (cytoplasm) | Intracellular (cytoplasm) | Intracellular (cytoplasm) | Intracellular (nucleus) | Extracellular | Extracellular (endosome) |
| Target | mRNA pre-mRNA miRNA | mRNA | miRNA | mRNA | Protein (transcription factor) | Protein | Protein (TLR9) |
| Function | mRNA degradation Translational inhibition miRNA inhibition Splicing inhibition | mRNA degradation | miRNA degradation | mRNA degradation Translational inhibition | Transcriptional inhibition | Inhibition of protein function | Activation of natural immunity via TLR9 |
| Drug delivery system | Modified or unnecessary | Necessary | Necessary | Necessary | Necessary | PEGylation | Antigen |
| Structural Modifications | Contents | Stability | Cellular Uptake | Gene Silencing Effect | Cytotoxicity | Binding Affinity |
|---|---|---|---|---|---|---|
| Diester modification | Phosphorothioate | superior | superior | inferior | superior | |
| Ribose modification | 2’-O-Me, 2’-O-A, 2’-F | superior | inferior | |||
| Base modification | Adenine methylation and deamination, cytosine methylation, hydroxy methylation and carboxy substitution, Guanine oxidation | superior | ||||
| Oligonucleotide analogues replacement | Peptide nucleic acid, locked nucleic acid, morpholino phosphamide | superior | superior | inferior | ||
| Conjugation to cell-penetrating peptides | Cysteine, transactivator of transcription peptide, gelatin | superior | superior | inferior | ||
| Aptamer | 20–100 nucleotides | superior | superior |
| Materials | |
|---|---|
| Liposomes | |
| Cationic liposome | DOTAP, DODMA, DOGS, DC-Chol |
| Neutral liposome | PC, Chol, DOPE |
| Ionizable liposome | DODMA, DODAP |
| Micelles | |
| Polymeric micelles | Amphiphilic copolymer, PEG, polyamino acid, polylactic or glycolic acid, polycaprolactone, and short phospholipid chains |
| Cationic polymer micelles | PEG-PLL-PLLeu, PEI-CG-PEI, PgP |
| Nanoparticles | |
| Albumin-based | thiol, arginine-glycine-aspartic acid peptide |
| Metal-based | gold, silver, magnetic |
| Drug | Nucleic Acid | Disease | Modification | Administration | Company |
|---|---|---|---|---|---|
| Vitravene [45] | ASO | Cytomegalovirus retinitis | Phosphorothioated | Intravitreous | Isis Pharmaceuticals, Carlsbad, CA |
| Macugen [46] | Aptamer | Age-related macular degeneration | PEGylation 2’-F 2’-OMe | Intravitreous | Valeant Pharmaceuticals, Laval, Canada |
| Kynamro [47] | ASO | Homozygous familial hypercholesterolemia | Phosphorothioated 2’-MOE | Subcutaneous | Kastle Therapeutics, Chicago, IL |
| Exondys 51 [48] | ASO | Duchenne muscular dystrophy | Morpholino nucleic acid | Intravenous | Sarepta Therapeutics, Cambridge, MA |
| Spinraza [49] | ASO | Myelopathic muscular atrophy | Phosphorothioated 2’-MOE | Intraspinal | Biogen, Cambridge, MA |
| Heplisav-B [50] | CpG oligo | Hepatitis B | Phosphorothioated | Intramuscular | Dynavax Technologies, Berkeley, CA |
| Tegsedi [51] | ASO | Hereditary transthyretin-mediated amyloidosis | Phosphorothioated 2’-MOE | Subcutaneous | Akcea Therapeutics, Boston, MA |
| Onpattro [52] | siRNA | Hereditary transthyretin-mediated amyloidosis | 2’-MOE | Intravenous | Alnylam Pharmaceuticals, Cambridge, MA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamakawa, K.; Nakano-Narusawa, Y.; Hashimoto, N.; Yokohira, M.; Matsuda, Y. Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment. Int. J. Mol. Sci. 2019, 20, 4224. https://doi.org/10.3390/ijms20174224
Yamakawa K, Nakano-Narusawa Y, Hashimoto N, Yokohira M, Matsuda Y. Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment. International Journal of Molecular Sciences. 2019; 20(17):4224. https://doi.org/10.3390/ijms20174224
Chicago/Turabian StyleYamakawa, Keiko, Yuko Nakano-Narusawa, Nozomi Hashimoto, Masanao Yokohira, and Yoko Matsuda. 2019. "Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment" International Journal of Molecular Sciences 20, no. 17: 4224. https://doi.org/10.3390/ijms20174224
APA StyleYamakawa, K., Nakano-Narusawa, Y., Hashimoto, N., Yokohira, M., & Matsuda, Y. (2019). Development and Clinical Trials of Nucleic Acid Medicines for Pancreatic Cancer Treatment. International Journal of Molecular Sciences, 20(17), 4224. https://doi.org/10.3390/ijms20174224





