Reaction Kinetic Models of Antibiotic Heteroresistance
Abstract
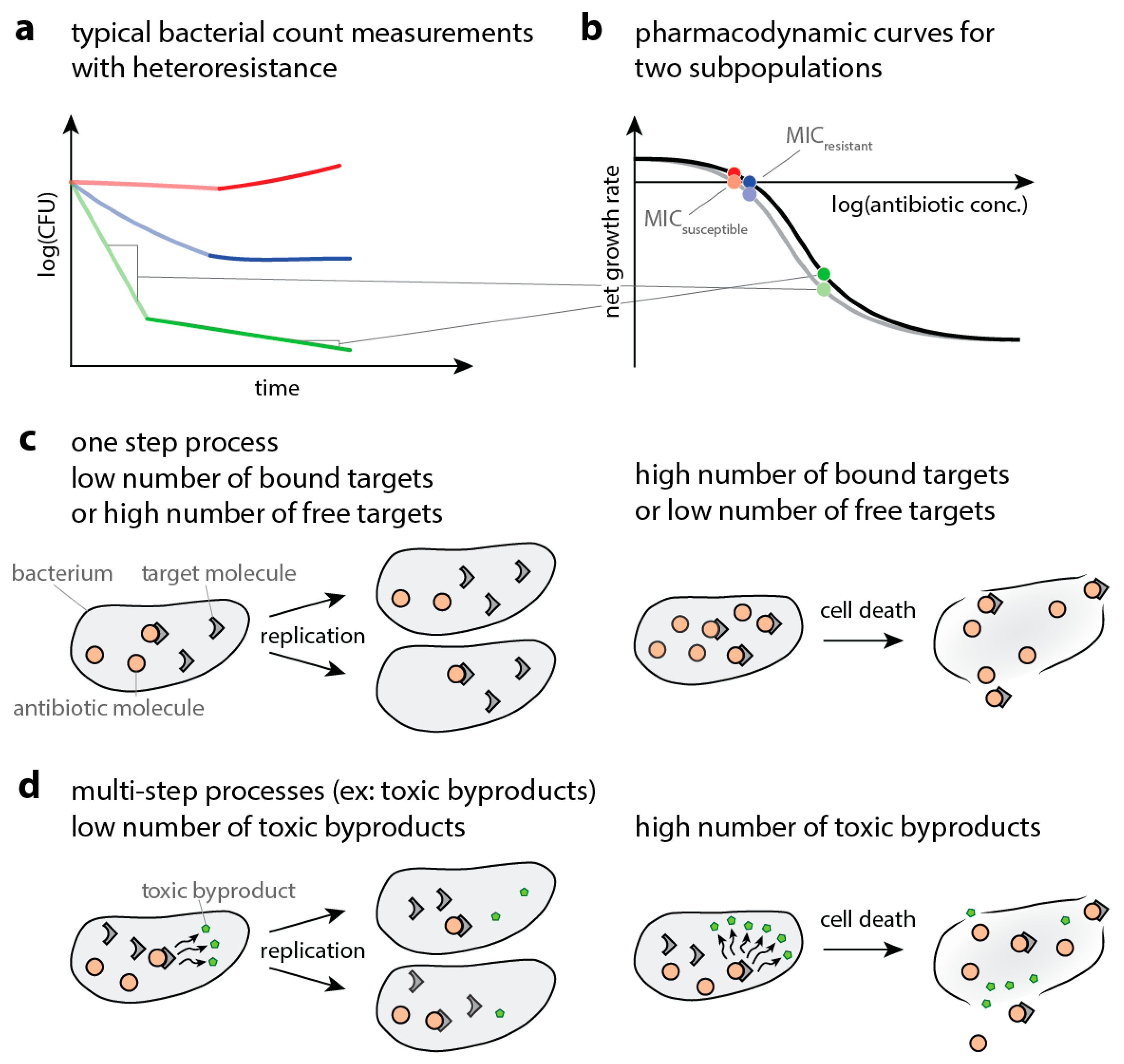
1. Introduction
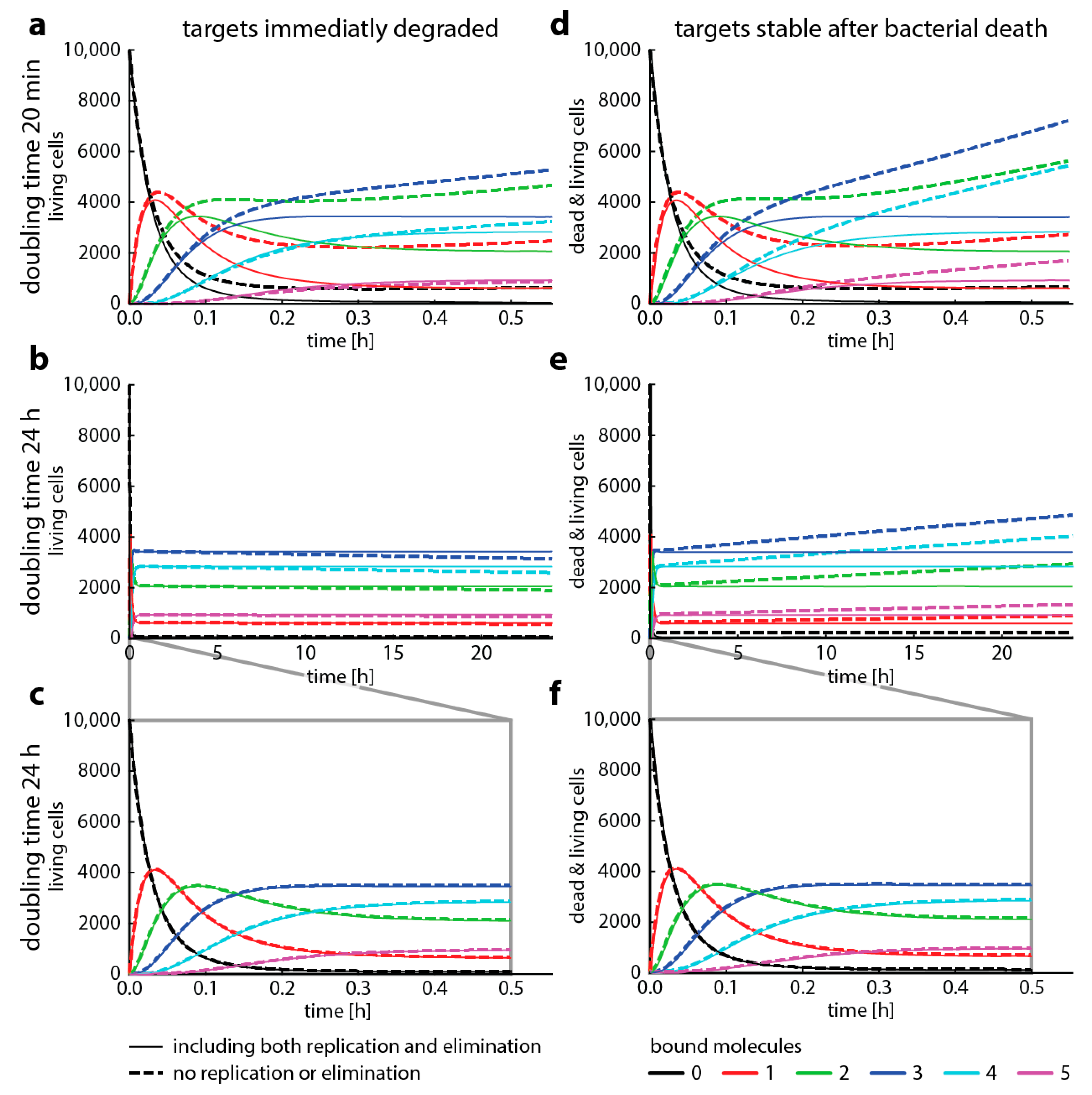
2. Results
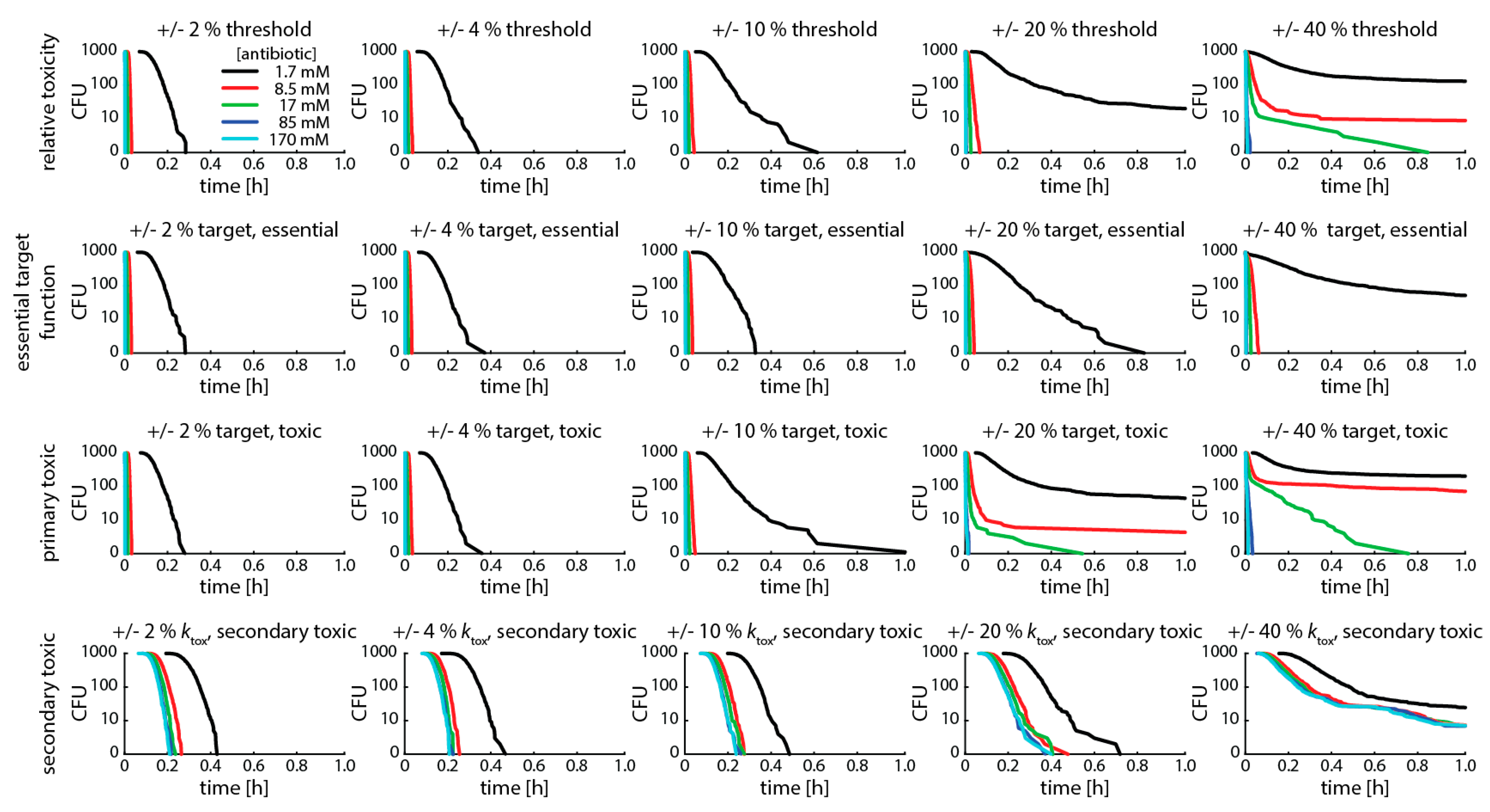
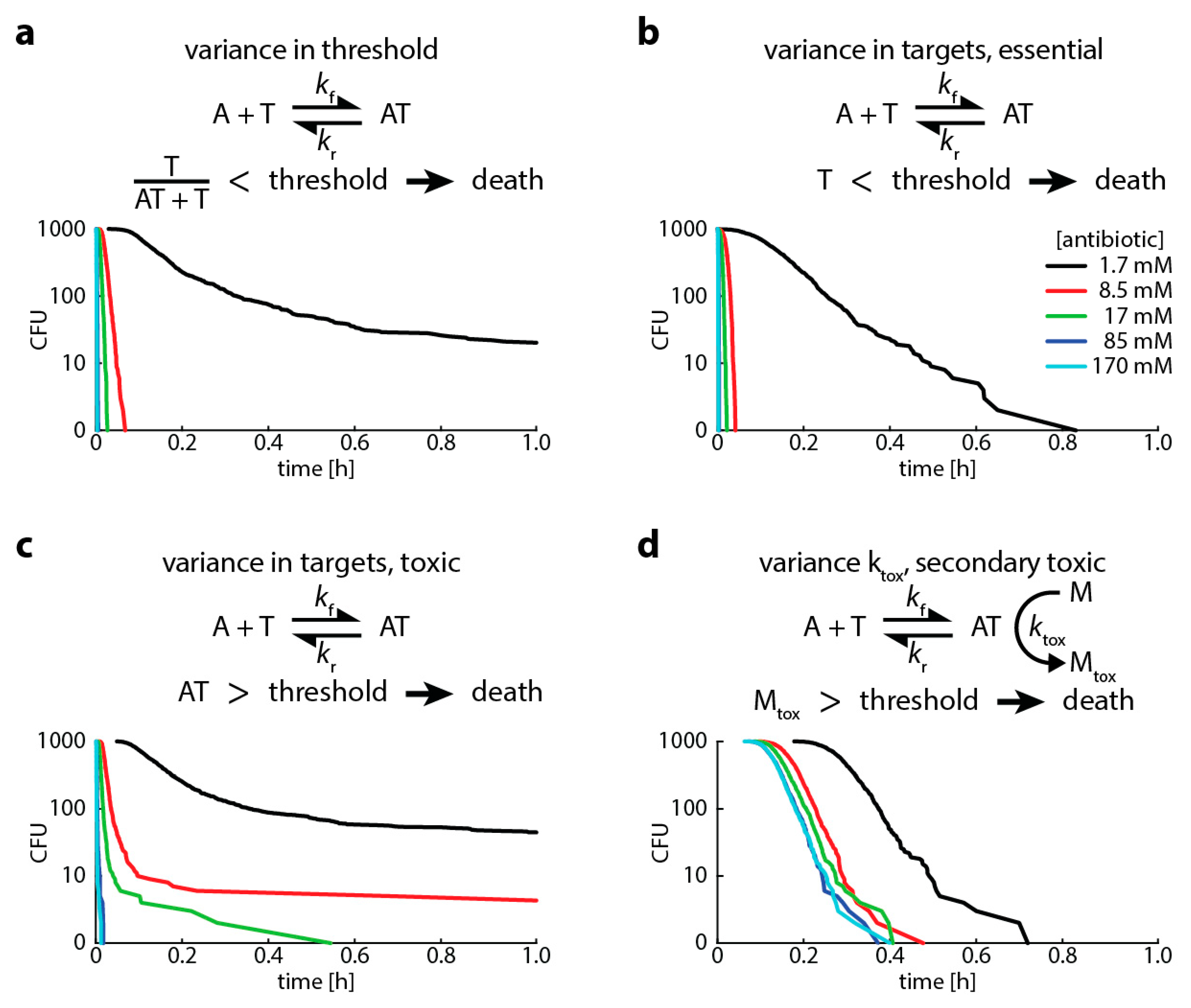
- A high proportion of bound versus unbound target is toxic (i.e., “relative toxicity”). In this case, we assume a bacterium is dead once the ratio of bound target and total target molecules exceeds a threshold. Here, we introduce heterogeneity by assuming that the number of target molecules is normally distributed around its mean.
- The target has an “essential” function (i.e., the cell dies when less than a certain amount of target is free). In this case, we assume a bacterium is dead once the number of free target falls below a threshold. Here, we introduce heterogeneity by assuming that the threshold for killing is normally distributed around its mean.
- The antibiotic-target complex is “primary toxic” (i.e., the cell is immediately damaged by bound targets). In this case, we assume a bacterium is dead once the number of bound targets exceeds a threshold. Here, we introduce heterogeneity by assuming that the number of target molecules is normally distributed around its mean.
- The antibiotic-target complex is “secondary toxic” (i.e., triggers the accumulation of toxic metabolites that ultimately kill the cell). In this case, we assume a bacterium is dead once the number of toxic metabolites exceeds a threshold. Here, we introduce heterogeneity by assuming that the rate of toxic metabolite production is normally distributed around its mean.
3. Discussion
4. Materials and Methods
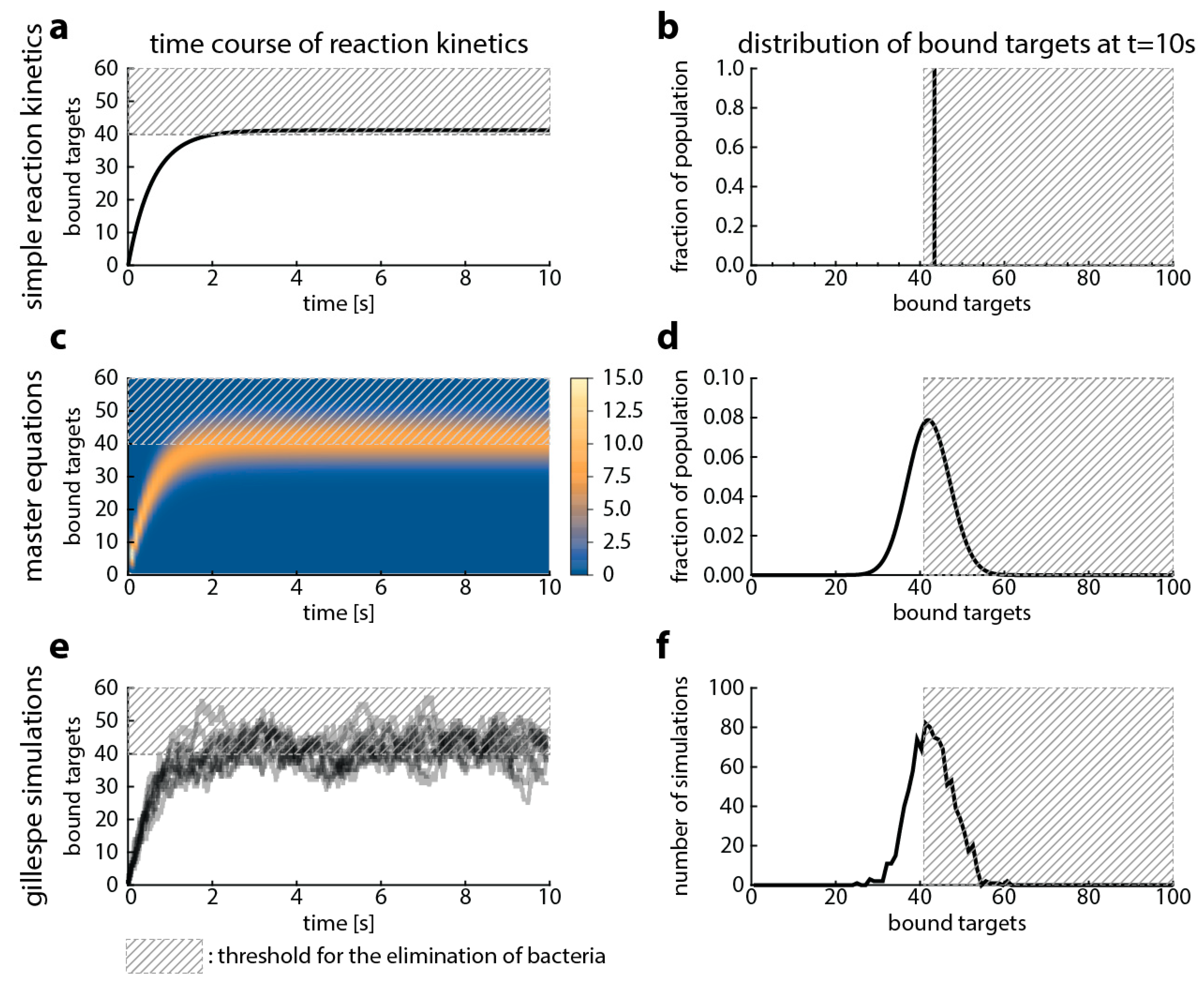
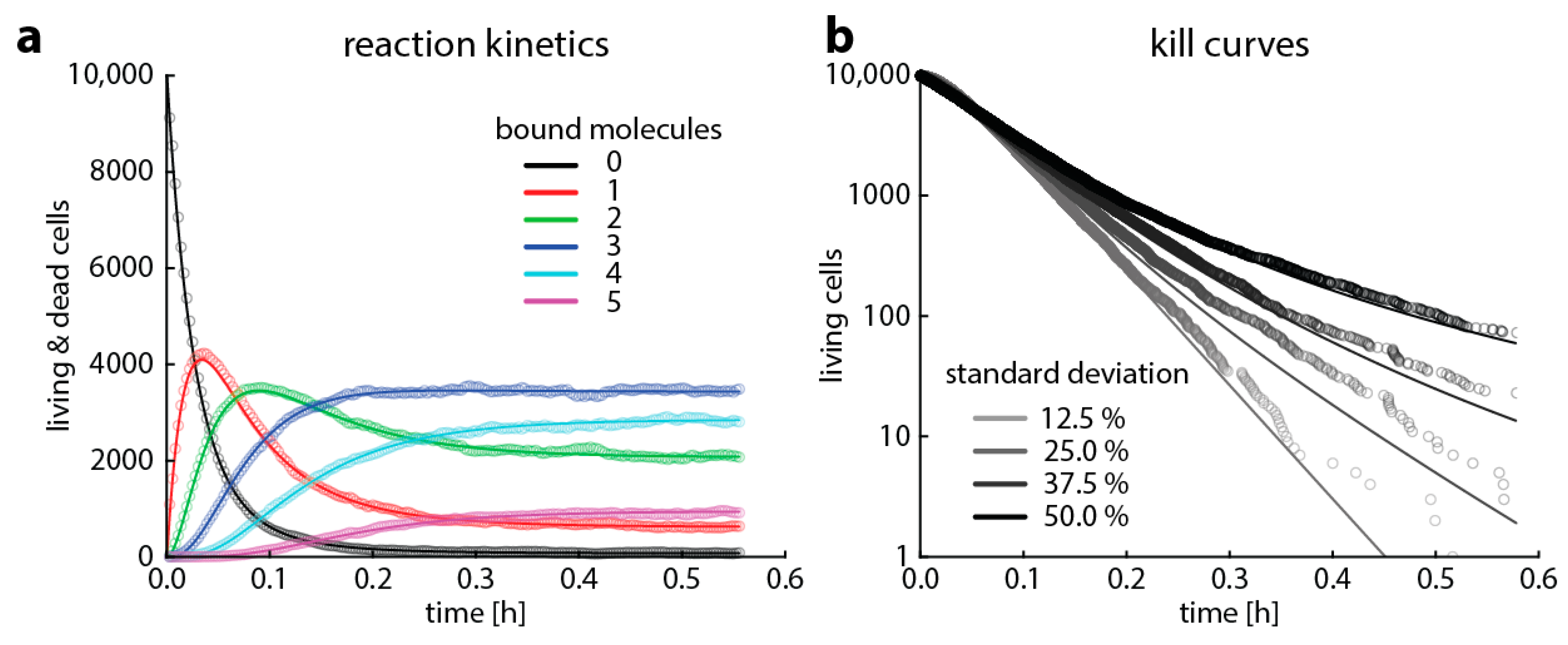
4.1. Simple Reaction Kinetics (Excluding Bacterial Replication and Death)
- is the adjusted forward reaction rate to accommodate working with the number molecules inside the cells instead of concentrations. Here, is the forward reaction rate, is the average cell volume, and is the Avogadro number.
- is the reverse reaction rate.
- is the number of antibiotic molecules inside the cell.
- is the number of free target molecules inside the cell.
- is the number of bound target molecules inside the cell.
4.2. Master Equations (Including Bacterial Replication and Death)
- is the number of bound targets,
- is the number of targets,
- is the number of bacteria with bound targets,
- is the elimination rate of bacteria at bound targets,
- —describes the inheritance of bound targets from the mother cells (“inflow”) using the hypergeometric distribution for the random distribution of targets.
- is the replication rate of cells at bound targets.
4.3. Gillespie Simulations (Excluding Bacterial Replication And Including Bacterial Death)
4.4. Implementation
Author Contributions
Funding
Conflicts of Interest
Appendix A

References
- Band, V.I.; Weiss, D.S. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLOS Pathog. 2019, 15, e1007726. [Google Scholar] [CrossRef] [PubMed]
- Band, V.I.; Crispell, E.K.; Napier, B.A.; Herrera, C.M.; Tharp, G.K.; Vavikolanu, K.; Pohl, J.; Read, T.D.; Bosinger, S.E.; Trent, M.S.; et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 2016, 1, 16053. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef] [PubMed]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial Heteroresistance: An Emerging Field in Need for Clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Abel zur Wiesch, P.; Abel, S.; Gkotzis, S.; Ocampo, P.; Engelstädter, J.; Hinkley, T.; Magnus, C.; Waldor, M.K.; Udekwu, K.; Cohen, T. Classic reaction kinetics can explain complex patterns of antibiotic action. Sci. Transl. Med. 2015, 7, 287ra73. [Google Scholar] [CrossRef] [PubMed]
- Martinecz, A.; Abel zur Wiesch, P. Estimating treatment prolongation for persistent infections. Pathog. Dis. 2018, 76, fty065. [Google Scholar] [CrossRef]
- Rego, E.H.; Audette, R.E.; Rubin, E.J. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 2017, 546, 153–157. [Google Scholar] [CrossRef]
- Bergmiller, T.; Andersson, A.M.C.; Tomasek, K.; Balleza, E.; Kiviet, D.J.; Hauschild, R.; Tkačik, G.; Guet, C.C. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science 2017, 356, 311–315. [Google Scholar] [CrossRef]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.B.; et al. Enhanced Efflux Activity Facilitates Drug Tolerance in Dormant Bacterial Cells. Mol. Cell 2016, 62, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Tonge, P.J. Drug-Target Kinetics in Drug Discovery. ACS Chem. Neurosci. 2018, 9, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Walkup, G.K.; You, Z.; Ross, P.L.; Allen, E.K.H.; Daryaee, F.; Hale, M.R.; O’Donnell, J.; Ehmann, D.E.; Schuck, V.J.A.; Buurman, E.T.; et al. Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat. Chem. Biol. 2015, 11, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Daryaee, F.; Tonge, P.J. Pharmacokinetic–pharmacodynamic models that incorporate drug–target binding kinetics. Curr. Opin. Chem. Biol. 2019, 50, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Malmström, J.; Beck, M.; Schmidt, A.; Lange, V.; Deutsch, E.W.; Aebersold, R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 2009, 460, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Schmidt, A.; Güell, M.; Kühner, S.; Gavin, A.-C.; Aebersold, R.; Serrano, L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011, 7, 511. [Google Scholar] [CrossRef]
- Belenky, P.; Ye, J.D.; Porter, C.B.M.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef]
- Gillespie, D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977, 81, 2340–2361. [Google Scholar] [CrossRef]
- Gillespie, D.T.; Hellander, A.; Petzold, L.R. Perspective: Stochastic algorithms for chemical kinetics. J. Chem. Phys. 2013, 138, 170901. [Google Scholar] [CrossRef]
- Gillespie, D.T. A rigorous derivation of the chemical master equation. Phys. Stat. Mech. Appl. 1992, 188, 404–425. [Google Scholar] [CrossRef]
- Spratt, B.G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA. 1975, 72, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Terrak, M.; Ghosh, T.K.; van Heijenoort, J.; Van Beeumen, J.; Lampilas, M.; Aszodi, J.; Ayala, J.A.; Ghuysen, J.M.; Nguyen-Distèche, M. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol. Microbiol. 1999, 34, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D.T. Stochastic Simulation of Chemical Kinetics. Annu. Rev. Phys. Chem. 2007, 58, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, Y.; Petzold, L.R.; Gillespie, D.T. Algorithms and Software for Stochastic Simulation of Biochemical Reacting Systems. Biotechnol. Prog. 2008, 24, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Wehrli, W. Kinetic Studies of the Interaction between Rifampicin and DNA-Dependent RNA Polymerase of Escherichia coli. Eur. J. Biochem. 1977, 80, 325–330. [Google Scholar] [CrossRef]
- Baeder, D.Y.; Yu, G.; Hozé, N.; Rolff, J.; Regoes, R.R. Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016, 371. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.C.; Solomkin, J.S.; Citron, D.M.; Alder, J.D. Clinical Efficacy and Correlation of Clinical Outcomes with In Vitro Susceptibility for Anaerobic Bacteria in Patients with Complicated Intra-abdominal Infections Treated With Moxifloxacin. Clin. Infect. Dis. 2011, 53, 1074–1080. [Google Scholar] [CrossRef]
- Colangeli, R.; Jedrey, H.; Kim, S.; Connell, R.; Ma, S.; Chippada Venkata, U.D.; Chakravorty, S.; Gupta, A.; Sizemore, E.E.; Diem, L.; et al. Bacterial Factors That Predict Relapse after Tuberculosis Therapy. N. Engl. J. Med. 2018, 379, 823–833. [Google Scholar] [CrossRef]
- Doern, G.V.; Brecher, S.M. The Clinical Predictive Value (or Lack Thereof) of the Results of In Vitro Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2011, 49, S11–S14. [Google Scholar] [CrossRef]
- Onufrak, N.J.; Forrest, A.; Gonzalez, D. Pharmacokinetic and Pharmacodynamic Principles of Anti-infective Dosing. Clin. Ther. 2016, 38, 1930–1947. [Google Scholar] [CrossRef]
- Hill, A.V. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 1910, 40, iv–vii. [Google Scholar]
- Holford, N. Pharmacodynamic principles and the time course of immediate drug effects. Transl. Clin. Pharm. 2017, 25, 157. [Google Scholar] [CrossRef]
- Kouyos, R.D.; Abel zur Wiesch, P.; Bonhoeffer, S. On Being the Right Size: The Impact of Population Size and Stochastic Effects on the Evolution of Drug Resistance in Hospitals and the Community. PLoS Pathog. 2011, 7, e1001334. [Google Scholar] [CrossRef] [PubMed]
- Avican, K.; Fahlgren, A.; Huss, M.; Heroven, A.K.; Beckstette, M.; Dersch, P.; Fällman, M. Reprogramming of Yersinia from Virulent to Persistent Mode Revealed by Complex In Vivo RNA-seq Analysis. PLOS Pathog. 2015, 11, e1004600. [Google Scholar] [CrossRef] [PubMed]
- Thorsted, A.; Thygesen, P.; Agersø, H.; Laursen, T.; Kreilgaard, M. Translational mixed-effects PKPD modelling of recombinant human growth hormone-from hypophysectomized rat to patients: Translational PKPD model of rhGH. Br. J. Pharm. 2016, 173, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Mouton, J.W.; Muller, A.E.; Canton, R.; Giske, C.G.; Kahlmeter, G.; Turnidge, J. MIC-based dose adjustment: Facts and fables. J. Antimicrob. Chemother. 2018, 73, 564–568. [Google Scholar] [CrossRef] [PubMed]
| Model | Reactions | Description | Rate |
|---|---|---|---|
| Single step model | Forward reaction | ||
| Backward reaction | |||
| Multi–step model | Forward reaction | ||
| Backward reaction | |||
| Toxic metabolite production |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinecz, A.; Clarelli, F.; Abel, S.; Abel zur Wiesch, P. Reaction Kinetic Models of Antibiotic Heteroresistance. Int. J. Mol. Sci. 2019, 20, 3965. https://doi.org/10.3390/ijms20163965
Martinecz A, Clarelli F, Abel S, Abel zur Wiesch P. Reaction Kinetic Models of Antibiotic Heteroresistance. International Journal of Molecular Sciences. 2019; 20(16):3965. https://doi.org/10.3390/ijms20163965
Chicago/Turabian StyleMartinecz, Antal, Fabrizio Clarelli, Sören Abel, and Pia Abel zur Wiesch. 2019. "Reaction Kinetic Models of Antibiotic Heteroresistance" International Journal of Molecular Sciences 20, no. 16: 3965. https://doi.org/10.3390/ijms20163965
APA StyleMartinecz, A., Clarelli, F., Abel, S., & Abel zur Wiesch, P. (2019). Reaction Kinetic Models of Antibiotic Heteroresistance. International Journal of Molecular Sciences, 20(16), 3965. https://doi.org/10.3390/ijms20163965





