Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development
Abstract
1. Introduction
2. Results
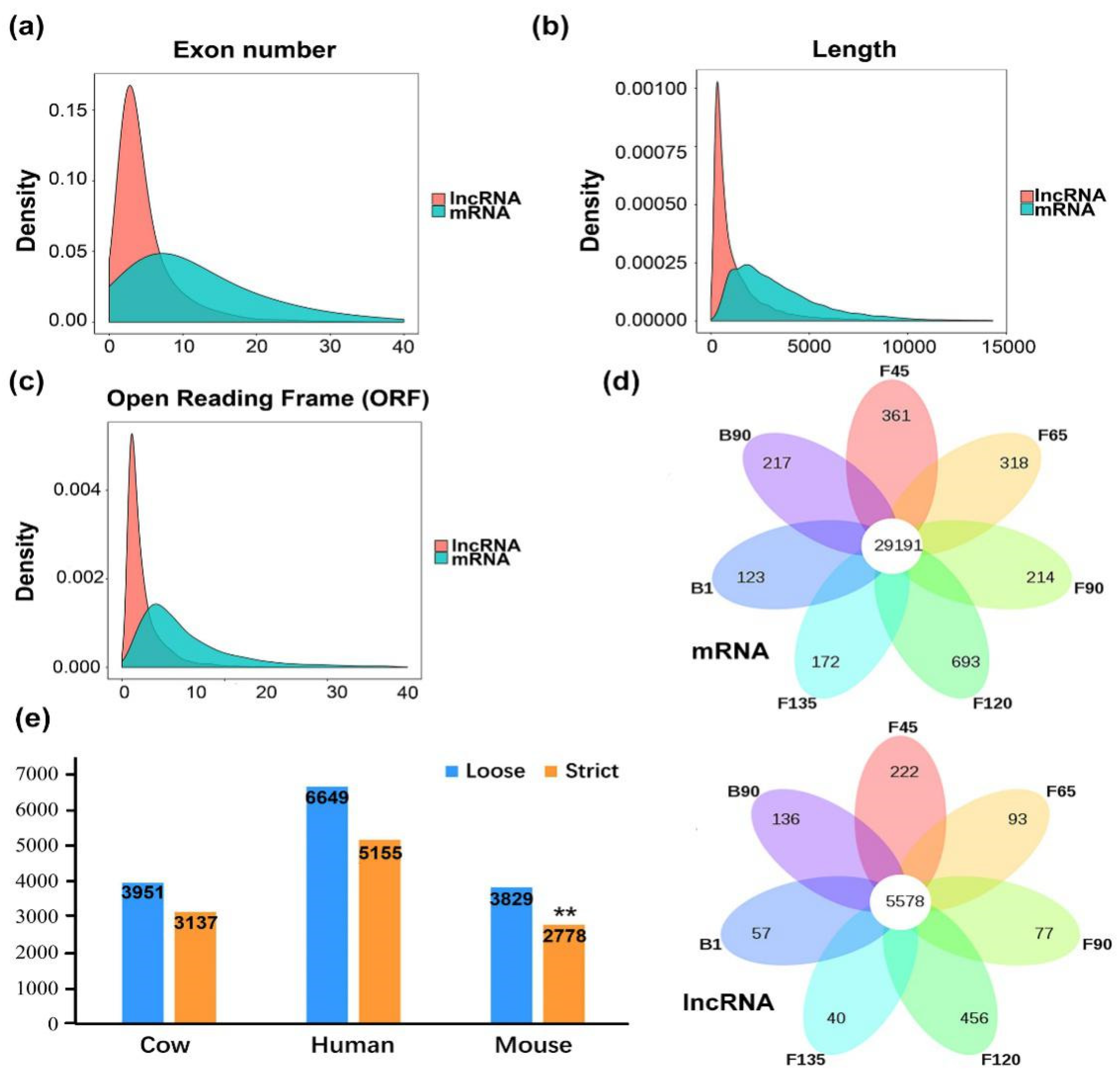
2.1. Identification and Characteristics of the Goat Skeletal Muscle Transcriptome
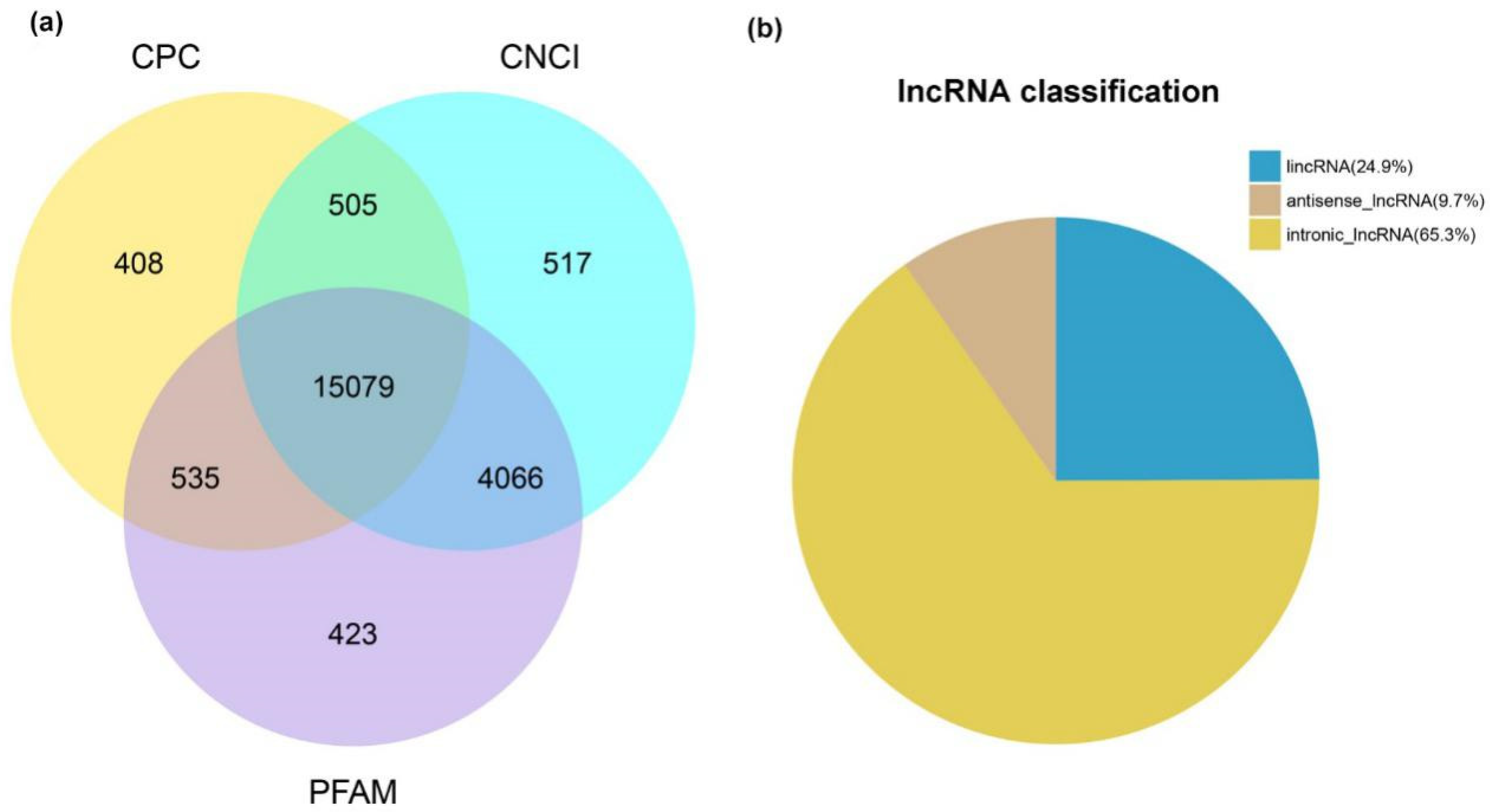
2.2. Characteristic and Conservation Analysis of Goat LncRNA
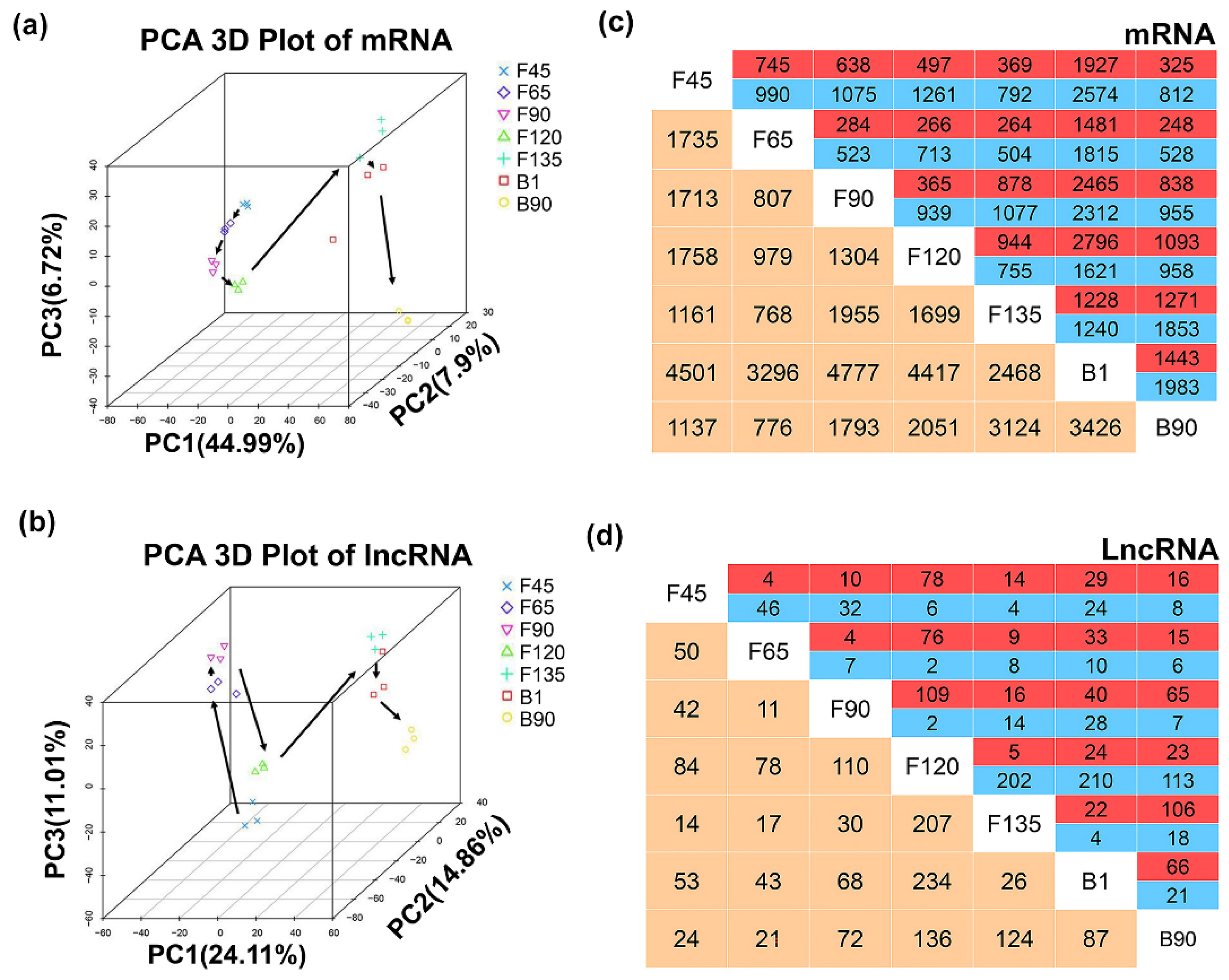
2.3. Dynamic Expression of LncRNA and mRNA
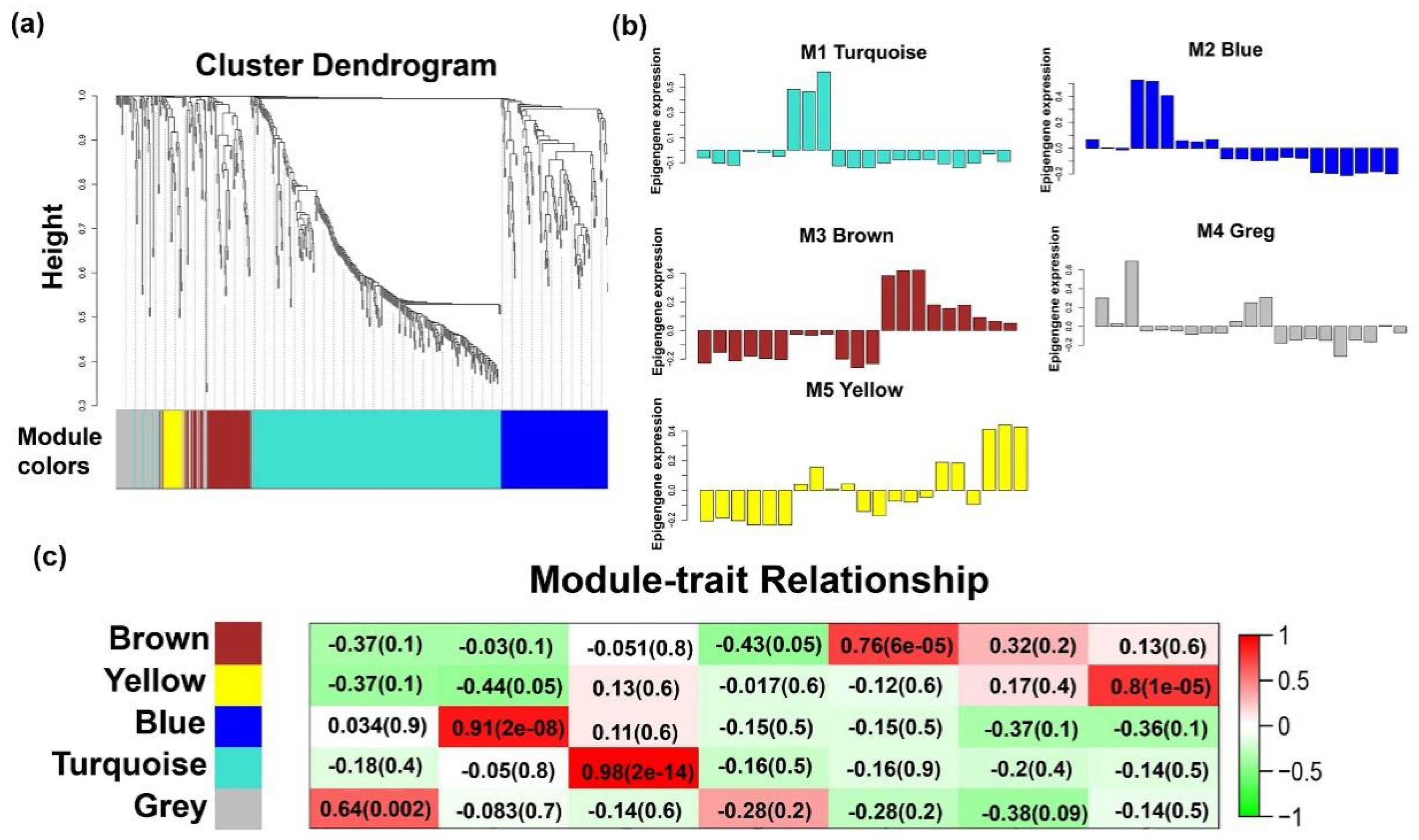
2.4. Temporal Expression Patterns of LncRNAs
2.4.1. LncRNAs Regulate Muscle Formation During Early Development
2.4.2. LncRNAs in Late Modules Are Associated With Muscle Differentiation and Hypertrophy
2.4.3. Functions of Individual-Stage-Specific LncRNAs
2.5. RT-qPCR Validation of DElncRNAs
3. Discussion
4. Materials and Methods
4.1. Animal Care and Tissue Collection
4.2. RNA Isolation, Lncrna and Small RNA Library Preparation and Sequencin
4.3. Data Analysis
4.4. Conservation Analysis
4.5. Quantification of Gene Expression Level and Differential Expression Analysis
4.6. Quantitative RT-PCR Validation
4.7. Target Gene Prediction
4.8. Functional and Pathway Enrichment Analysis
4.9. WGCNA Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
Abbreviations
| LncRNA | Long non-coding RNA |
| F45 | Fetus 45 days |
| F65 | Fetus 65 days |
| F90 | Fetus 90 days |
| F120 | Fetus 120 days |
| F135 | Fetus 135 days |
| B1 | Born 1 day |
| B90 | Born 90 days |
| AWG | Anhui white goats |
| FPKM | Fragment Per Kilobase of exon per Million fragments mapped |
| WGCNA | Weighted gene co-expression network analysis |
| PCA | Component analysis |
| DElncRNAs | Differentially expressed lncRNAs |
| DEmRNAs | Differentially expressed mRNAs |
References
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Agliano, F.; Rathinam, V.A.; Medvedev, A.E.; Vanaja, S.K.; Vella, A.T. Long Noncoding RNAs in Host-Pathogen Interactions. Trends Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.S.; Shin, S.B.; Kim, E.J.; Koo, J.H.; Yim, H.; Kim, S.G. Nrf2-lncRNA controls cell fate by modulating p53-dependent Nrf2 activation as an miRNA sponge for Plk2 and p21(cip1). Faseb J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Delas, M.J.; Jackson, B.T.; Kovacevic, T.; Vangelisti, S.; Munera Maravilla, E.; Wild, S.A.; Stork, E.M.; Erard, N.; Knott, S.R.V.; Hannon, G.J. lncRNA Spehd Regulates Hematopoietic Stem and Progenitor Cells and Is Required for Multilineage Differentiation. Cell Rep. 2019, 27, 719–729.e716. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, M.; Liu, F.; Zhang, Y.H.; Li, R.B.; Zhai, M.; Liu, F.; Zhou, L.Y.; Liu, C.Y.; Yan, K.W.; Dong, Y.H.; et al. The Long Non-Coding RNA CPR Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 2019. [Google Scholar] [CrossRef]
- Sun, M.; Gadad, S.S.; Kim, D.S.; Kraus, W.L. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol. Cell 2015, 59, 698–711. [Google Scholar] [CrossRef]
- Zhao, X.; Su, L.; He, X.; Zhao, B.; Miao, J. Long noncoding RNA CA7–4 promotes autophagy and apoptosis via sponging MIR877–3 P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 2019, 1–16. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Zhan, H.; Dai, J.; Chang, Y.; Wu, F.; Liu, T.; Liu, Z.; Gao, C.; Li, L.; et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- White, R.B.; Bierinx, A.S.; Gnocchi, V.F.; Zammit, P.S. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 2010, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Noulet, F.; Edom-Vovard, F.; Tozer, S.; Le Grand, F.; Duprez, D. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev. Cell 2010, 18, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cai, B.; Abdalla, B.A.; Zhu, X.; Zheng, M.; Han, P.; Nie, Q.; Zhang, X. LncIRS1 controls muscle atrophy via sponging miR−15 family to activate IGF1-PI3 K/AKT pathway. J. Cachexia Sarcopenia Muscle 2019, 10, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Parisi, A.; Lacour, F.; Giordani, L.; Colnot, S.; Maire, P.; Le Grand, F. APC is required for muscle stem cell proliferation and skeletal muscle tissue repair. J. Cell Biol. 2015, 210, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, W.R.; Wang, Y.M.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. MicroRNA−128 regulates the proliferation and differentiation of bovine skeletal muscle satellite cells by repressing Sp1. Mol. Cell. Biochem. 2016, 414, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Jin, J.P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene 2016, 582, 1–13. [Google Scholar] [CrossRef]
- Toyoda, T.; An, D.; Witczak, C.A.; Koh, H.J.; Hirshman, M.F.; Fujii, N.; Goodyear, L.J. Myo1 c regulates glucose uptake in mouse skeletal muscle. J. Biol. Chem. 2011, 286, 4133–4140. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Legendre, N.P.; Biswas, A.A.; Lawton, A.; Yamamoto, S.; Tajbakhsh, S.; Kardon, G.; Goldhamer, D.J. Loss of MyoD and Myf5 in Skeletal Muscle Stem Cells Results in Altered Myogenic Programming and Failed Regeneration. Stem. Cell Rep. 2018, 10, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.N.; Lee, I.J.; Park, M.C.; Baik, E.J. Role of JAK3 in myogenic differentiation. Cell. Signal. 2012, 24, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Nelson, B.R.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Requirement of MEF2 A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci.USA. 2014, 111, 4109–4114. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S. Muscle fiber type diversity revealed by anti-myosin heavy chain antibodies. Febs J. 2018, 285, 3688–3694. [Google Scholar] [CrossRef] [PubMed]
- Reilich, P.; Krause, S.; Schramm, N.; Klutzny, U.; Bulst, S.; Zehetmayer, B.; Schneiderat, P.; Walter, M.C.; Schoser, B.; Lochmuller, H. A novel mutation in the myotilin gene (MYOT) causes a severe form of limb girdle muscular dystrophy 1 A (LGMD1 A). J. Neurol. 2011, 258, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.J.; Hansen, J.M.; Merrell, A.J.; Murphy, M.M.; Lawson, J.A.; Hutcheson, D.A.; Hansen, M.S.; Angus-Hill, M.; Kardon, G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Dev. (Camb. Engl.) 2011, 138, 371–384. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, J.; Ryu, J.Y.; Lee, S.; Yang, D.K.; Jeong, D.; Kim, J.; Lee, S.H.; Kim, J.M.; Hajjar, R.J.; et al. Transcription coactivator Eya2 is a critical regulator of physiological hypertrophy. J. Mol. Cell. Cardiol. 2012, 52, 718–726. [Google Scholar] [CrossRef]
- Nguyen, P.D.; Gurevich, D.B.; Sonntag, C.; Hersey, L.; Alaei, S.; Nim, H.T.; Siegel, A.; Hall, T.E.; Rossello, F.J.; Boyd, S.E.; et al. Muscle Stem Cells Undergo Extensive Clonal Drift during Tissue Growth via Meox1-Mediated Induction of G2 Cell-Cycle Arrest. Cell Stem Cell 2017, 21, 107–119.e106. [Google Scholar] [CrossRef]
- Ojima, K. Myosin: Formation and maintenance of thick filaments. J. Anim. Sci. 2019, 90, 801–807. [Google Scholar] [CrossRef]
- Dumont, N.A.; Wang, Y.X.; Von Maltzahn, J.; Pasut, A.; Bentzinger, C.F.; Brun, C.E.; Rudnicki, M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015, 21, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Aradhya, R.; Zmojdzian, M.; Da Ponte, J.P.; Jagla, K. Muscle niche-driven Insulin-Notch-Myc cascade reactivates dormant Adult Muscle Precursors in Drosophila. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.C.; Gidlund, E.K.; Norrbom, J.; Valencia, A.P.; Thomson, D.M.; Schuh, R.A.; Neufer, P.D.; Spangenburg, E.E. BRCA1 is a novel regulator of metabolic function in skeletal muscle. J. Lipid Res. 2014, 55, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ectors, F.; Davis, E.E.; Pirottin, D.; Cheng, H.; Farnir, F.; Hadfield, T.; Cockett, N.; Charlier, C.; Georges, M.; et al. Ectopic Expression of Retrotransposon-Derived PEG11/RTL1 Contributes to the Callipyge Muscular Hypertrophy. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Ma, K. Circadian clock regulation of skeletal muscle growth and repair. F1000 Res. 2016, 5, 1549. [Google Scholar] [CrossRef] [PubMed]
- Elabd, C.; Cousin, W.; Upadhyayula, P.; Chen, R.Y.; Chooljian, M.S.; Li, J.; Kung, S.; Jiang, K.P.; Conboy, I.M. Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 2014, 5, 4082. [Google Scholar] [CrossRef] [PubMed]
- Terry, E.E.; Zhang, X.; Hoffmann, C.; Hughes, L.D.; Lewis, S.A.; Li, J.; Wallace, M.J.; Riley, L.A.; Douglas, C.M.; Gutierrez-Monreal, M.A.; et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. eLife 2018, 7. [Google Scholar] [CrossRef]
- Ren, C.; Deng, M.; Fan, Y.; Yang, H.; Zhang, G.; Feng, X.; Li, F.; Wang, D.; Wang, F.; Zhang, Y. Genome-Wide Analysis Reveals Extensive Changes in LncRNAs during Skeletal Muscle Development in Hu Sheep. Genes 2017, 8, 191. [Google Scholar] [CrossRef]
- Zhan, S.; Dong, Y.; Zhao, W.; Guo, J.; Zhong, T.; Wang, L.; Li, L.; Zhang, H. Genome-wide identification and characterization of long non-coding RNAs in developmental skeletal muscle of fetal goat. BMC Genom. 2016, 17, 666. [Google Scholar] [CrossRef]
- Singh, M. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 2012, 3, 326. [Google Scholar] [CrossRef] [PubMed]
- Fico, A.; Fiorenzano, A.; Pascale, E.; Patriarca, E.J.; Minchiotti, G. Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cell. Mol. Life Sci. 2019, 76, 1459–1471. [Google Scholar] [CrossRef] [PubMed]
- Karlic, R.; Ganesh, S.; Franke, V.; Svobodova, E.; Urbanova, J.; Suzuki, Y.; Aoki, F.; Vlahovicek, K.; Svoboda, P. Long non-coding RNA exchange during the oocyte-to-embryo transition in mice. Dna Res. 2017, 24, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.L.; Van der Linde, S.M.; Vogel, I.S.P.; Haroon, M.; Offringa, C.; De Wit, G.M.J.; Koolwijk, P.; Van der Laarse, W.J.; Jaspers, R.T. IGF−1 Attenuates Hypoxia-Induced Atrophy but Inhibits Myoglobin Expression in C2 C12 Skeletal Muscle Myotubes. Int. J. Mol. Sci. 2017, 18, 1889. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- Bennett, R.D.; Caride, A.J.; Mauer, A.S.; Strehler, E.E. Interaction with the IQ3 motif of myosin−10 is required for calmodulin-like protein-dependent filopodial extension. Febs Lett. 2008, 582, 2377–2381. [Google Scholar] [CrossRef]
- Pisconti, A.; Banks, G.B.; Babaeijandaghi, F.; Betta, N.D.; Rossi, F.M.; Chamberlain, J.S.; Olwin, B.B. Loss of niche-satellite cell interactions in syndecan−3 null mice alters muscle progenitor cell homeostasis improving muscle regeneration. Skelet. Muscle 2016, 6, 34. [Google Scholar] [CrossRef]
- Schneider, M.; Kostin, S.; Strom, C.C.; Aplin, M.; Lyngbaek, S.; Theilade, J.; Grigorian, M.; Andersen, C.B.; Lukanidin, E.; Lerche Hansen, J.; et al. S100 A4 is upregulated in injured myocardium and promotes growth and survival of cardiac myocytes. Cardiovasc. Res. 2007, 75, 40–50. [Google Scholar] [CrossRef]
- Fleming-Waddell, J.N.; Olbricht, G.R.; Taxis, T.M.; White, J.D.; Vuocolo, T.; Craig, B.A.; Tellam, R.L.; Neary, M.K.; Cockett, N.E.; Bidwell, C.A. Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PLoS ONE 2009, 4, e7399. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinform. (Oxf. Engl.) 2008, 24, 719–720. [Google Scholar] [CrossRef]
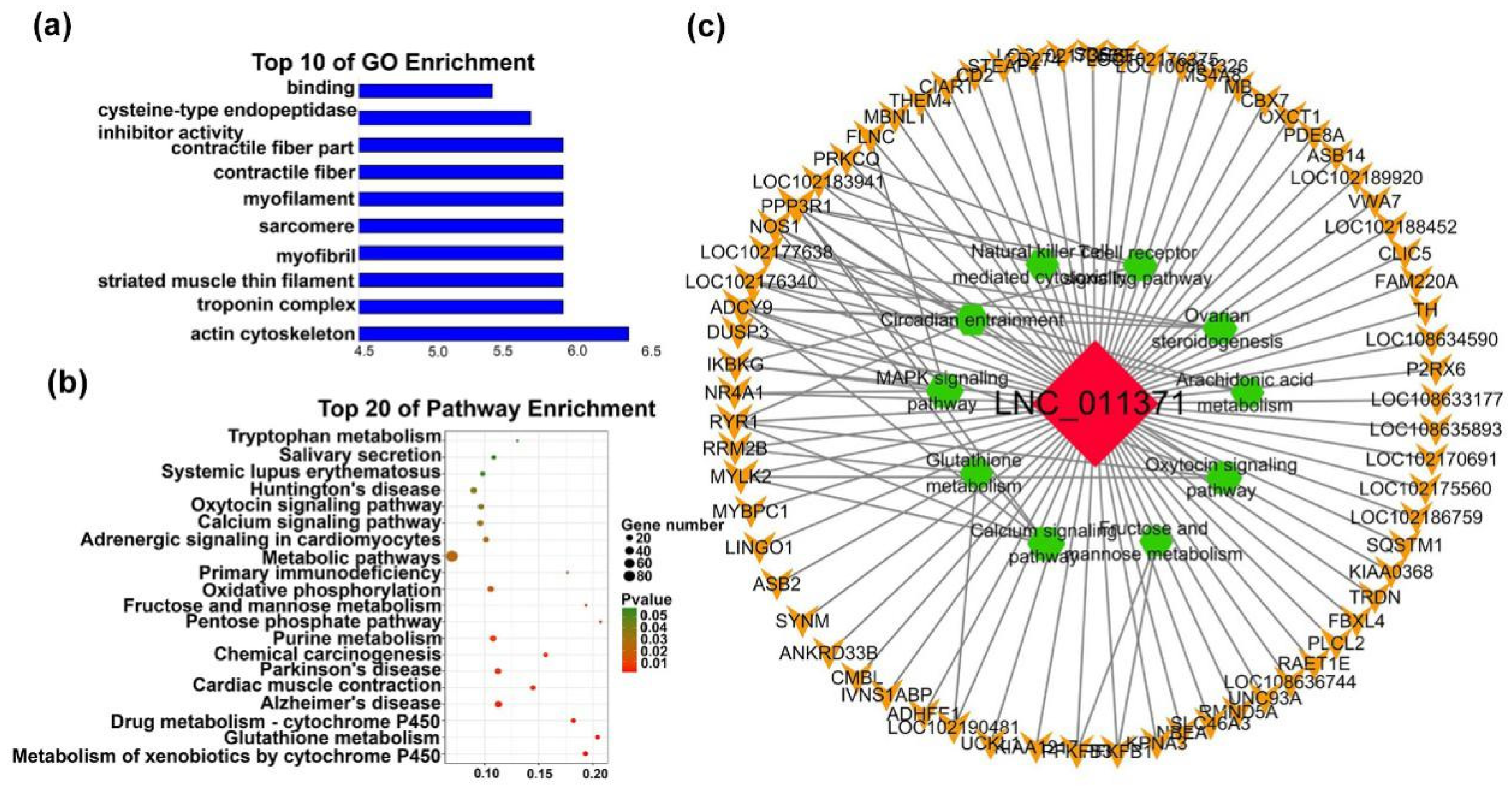
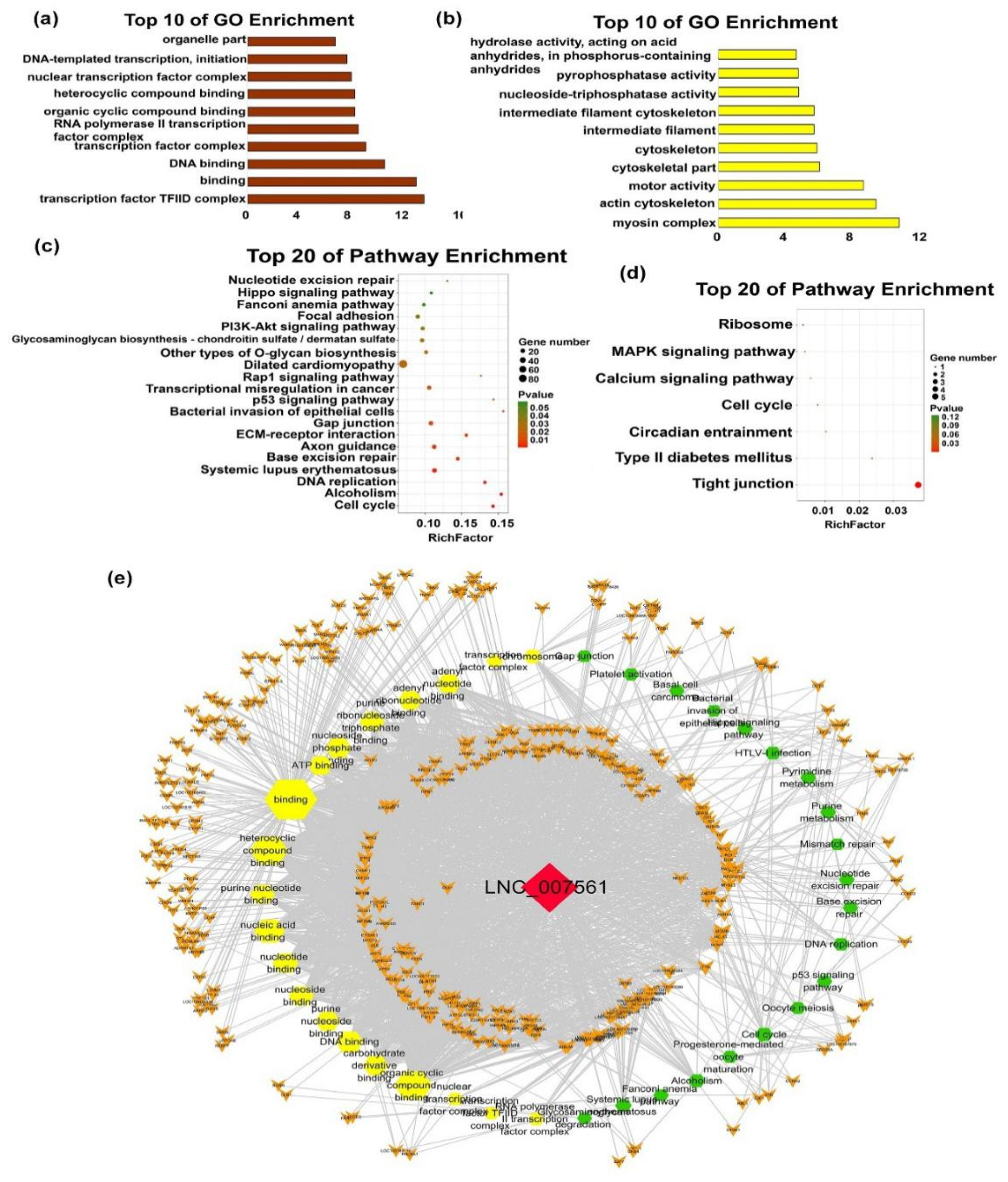
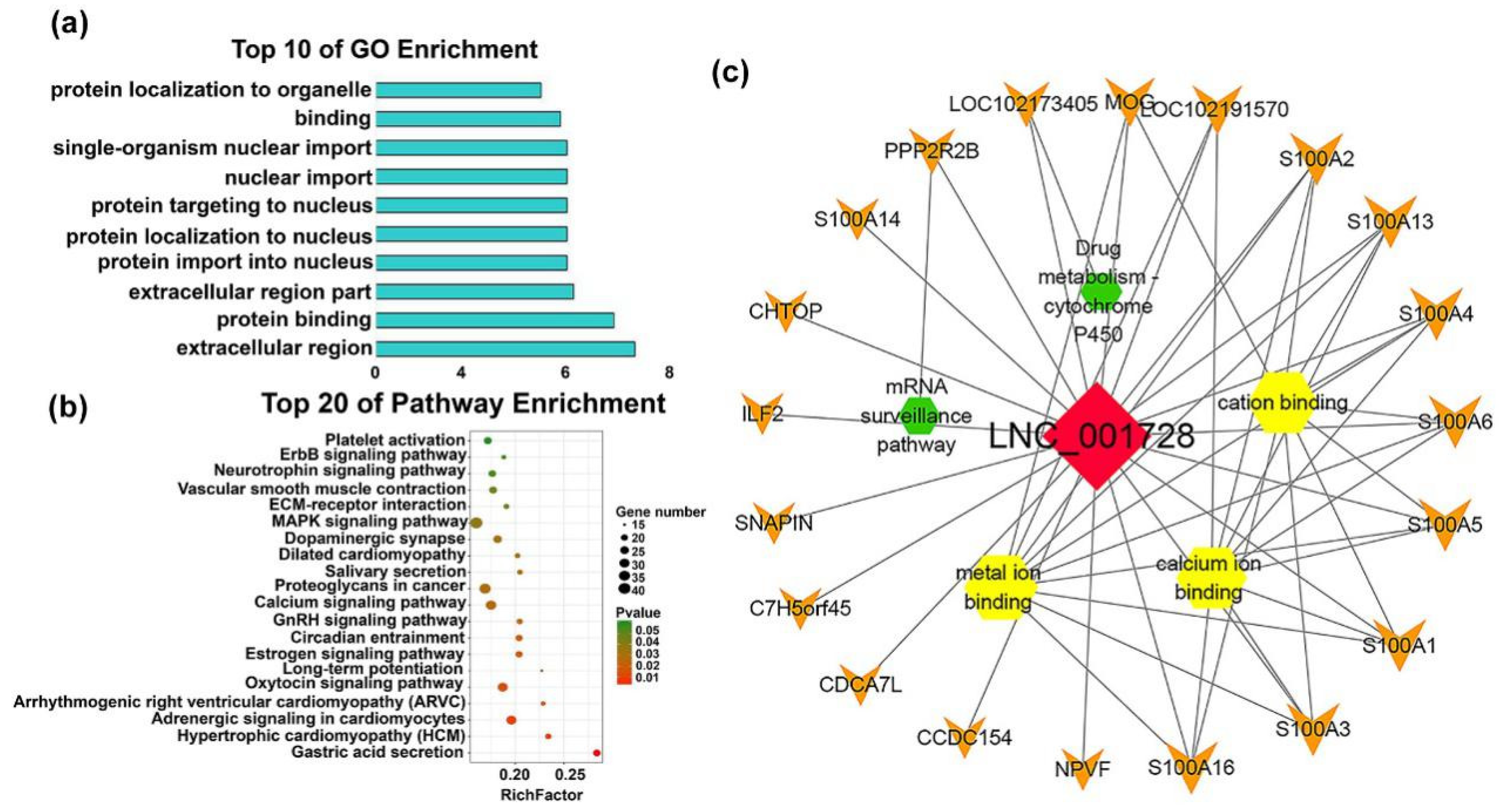
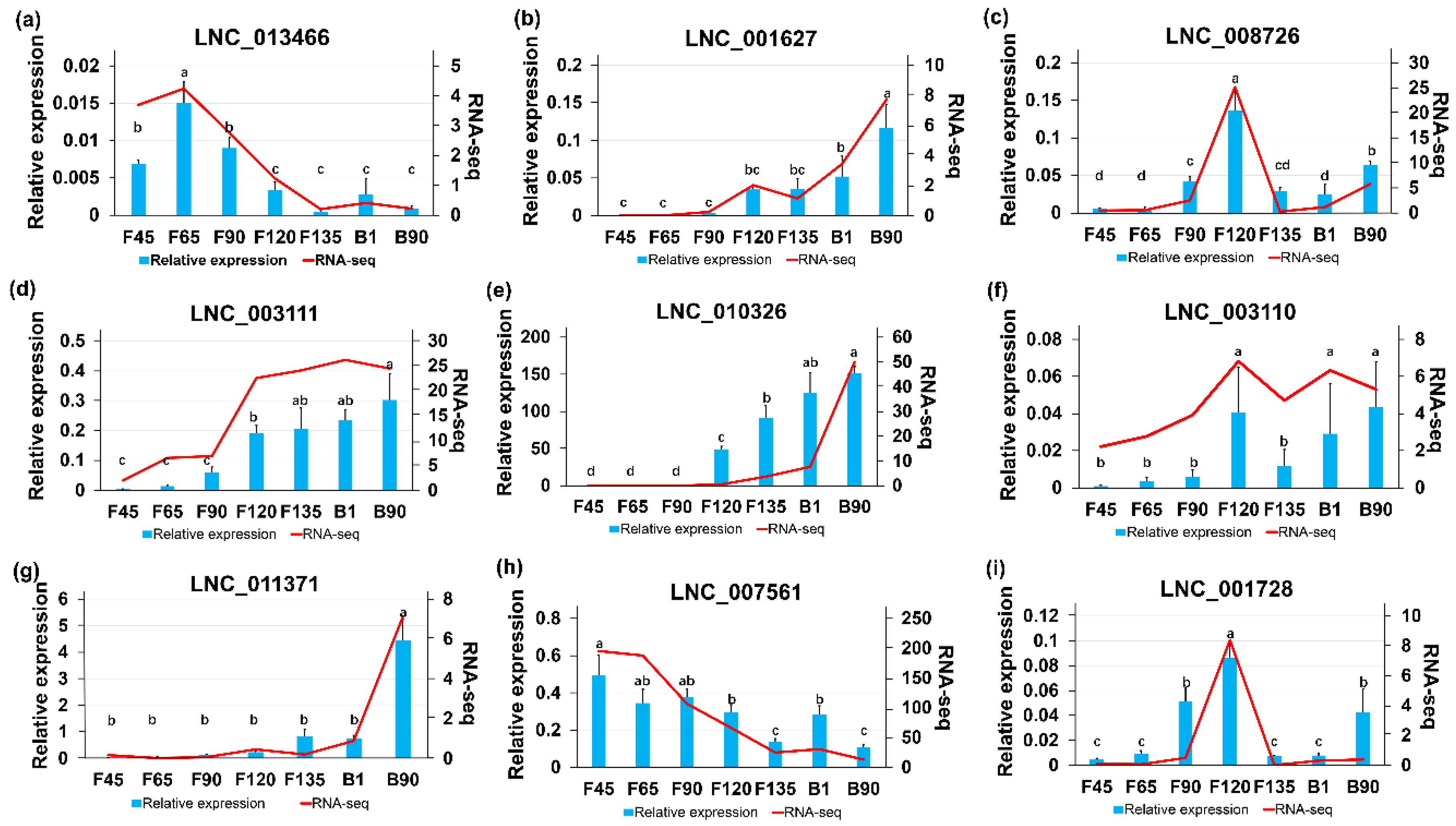
| mRNAs | Transcript Source | Function of mRNA | Associated lncRNAs |
|---|---|---|---|
| APC | Module1 | APC is required for muscle stem cell proliferation and skeletal muscle tissue repair [18]. | LNC_006306; LNC_003007 |
| SP1 | Module1 | Sp1 is an activator of MyoD and a suppressor of CDKN1 A that plays an important role in bovine muscle cell proliferation and differentiation [19]. | LNC_002771; LNC_002368; LNC_009507; LNC_011751 |
| TNNT2 | Module1 | TNNT2 is a muscle type-specific TNT for cardiac muscle [20]. | LNC_009262; LNC_009263 |
| MYO1 C | Module1 | Myo1 c regulates glucose uptake in mouse skeletal muscle [21]. | LNC_010262; LNC_010263 |
| MYF5 | Module1 | Myf5 in muscle regeneration indicates that their expression after injury stabilizes myogenic identity and confers the capacity for muscle differentiation [22]. | LNC_003108; LNC_003109; LNC_003110; LNC_003111; LNC_003112; LNC_003113; |
| JAK3 | Module1,2 | JAK3 inhibition potently facilitates myoblast differentiation through antagonistic STAT1/STAT3 activities [23]. | LNC_003114 |
| MEF2 A | Module2 | Requirement of MEF2 A, C, and D for skeletal muscle regeneration and differentiate [24]. | LNC_004659; LNC_004660 |
| MYH13 | Module2,5 | MYH13 associates with extraocular muscles [25]. | LNC_010326; LNC_010329 |
| TNNT1 | Module1,2 | TNNT1 is a muscle type-specific TNT for slow skeletal muscle [20]. | LNC_010184; LNC_010185 |
| MYOT | Module3 | Mutation in the myotilin gene (MYOT) causes a severe form of limb girdle muscular dystrophy 1 A [26]. | LNC_004077 |
| TCF4 | Module3 | TCF4 regulates myogenesis [27]. | LNC_000994; LNC_006624; LNC_007561 |
| EYA2 | Module3 | Eya2 is a critical regulator of physiological hypertrophy [28]. | LNC_000994; LNC_001379; LNC_006624 |
| MEOX1 | Module3 | Meox1 initiates G2 cell-cycle arrest within muscle stem cells [29]. | LNC_004123 |
| BAMBI | Module3 | BAMBI promotes C2 C12 myogenic differentiation by enhancing Wnt/β-catenin signaling [29]. | LNC_014703 |
| MYO10 | Module3 | Myosin is a component of myofibrils [30] | LNC_005183; LNC_007561; LNC_013466 |
| MARK2 | Module4 | Mark2 (also known as Par1 b) is an important regulator of cell polarity [31]. | LNC_014128 |
| MEF2 C | Module4 | Requirement of MEF2 A, C, and D for skeletal muscle regeneration and differentiation [24]. | LNC_004357 |
| MYC | Module4 | Myc is involved in regulating myoblast proliferation during muscle development and regeneration [32]. | LNC_008001 |
| BRCA1 | Module4 | BRCA1 is a regulator of metabolic function in skeletal muscle [33] | LNC_010762; LNC_010763; LNC_010766 |
| TNNT3 | Module4 | TNNT1 is a muscle type-specific TnT for fast skeletal muscle [20]. | LNC_014032 |
| LOC102180883 (myosin-4) | Module5 | Myosin is a component of myofibrils [30]. | LNC_010329 |
| LOC102181155 (myosin-2) | Module5 | Myosin II is the myosin type responsible for producing muscle contraction in muscle cells in most animal cell types [34]. | LNC_010329 |
| LOC102181426 (myosin-1) | Module5 | Myosin is a component of myofibrils [30]. | LNC_010329 |
| MYH8 | Module5 | MYH8 is associated with developing muscle [25]. | LNC_010329 |
| RTL1 | Module5 | Ectopic expression of PEG11/RTL1 contributes to the callipyge muscular hypertrophy [35]. | LNC_010329 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. https://doi.org/10.3390/ijms20163950
Ling Y, Zheng Q, Sui M, Zhu L, Xu L, Zhang Y, Liu Y, Fang F, Chu M, Ma Y, et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. International Journal of Molecular Sciences. 2019; 20(16):3950. https://doi.org/10.3390/ijms20163950
Chicago/Turabian StyleLing, Yinghui, Qi Zheng, Menghua Sui, Lu Zhu, Lina Xu, Yunhai Zhang, Ya Liu, Fugui Fang, Mingxing Chu, Yuehui Ma, and et al. 2019. "Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development" International Journal of Molecular Sciences 20, no. 16: 3950. https://doi.org/10.3390/ijms20163950
APA StyleLing, Y., Zheng, Q., Sui, M., Zhu, L., Xu, L., Zhang, Y., Liu, Y., Fang, F., Chu, M., Ma, Y., & Zhang, X. (2019). Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. International Journal of Molecular Sciences, 20(16), 3950. https://doi.org/10.3390/ijms20163950






