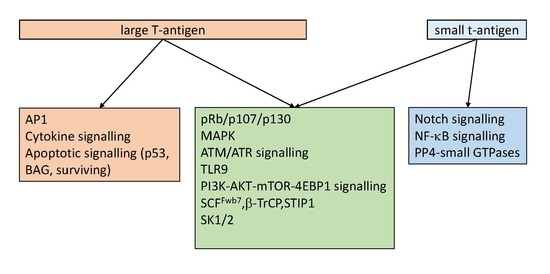
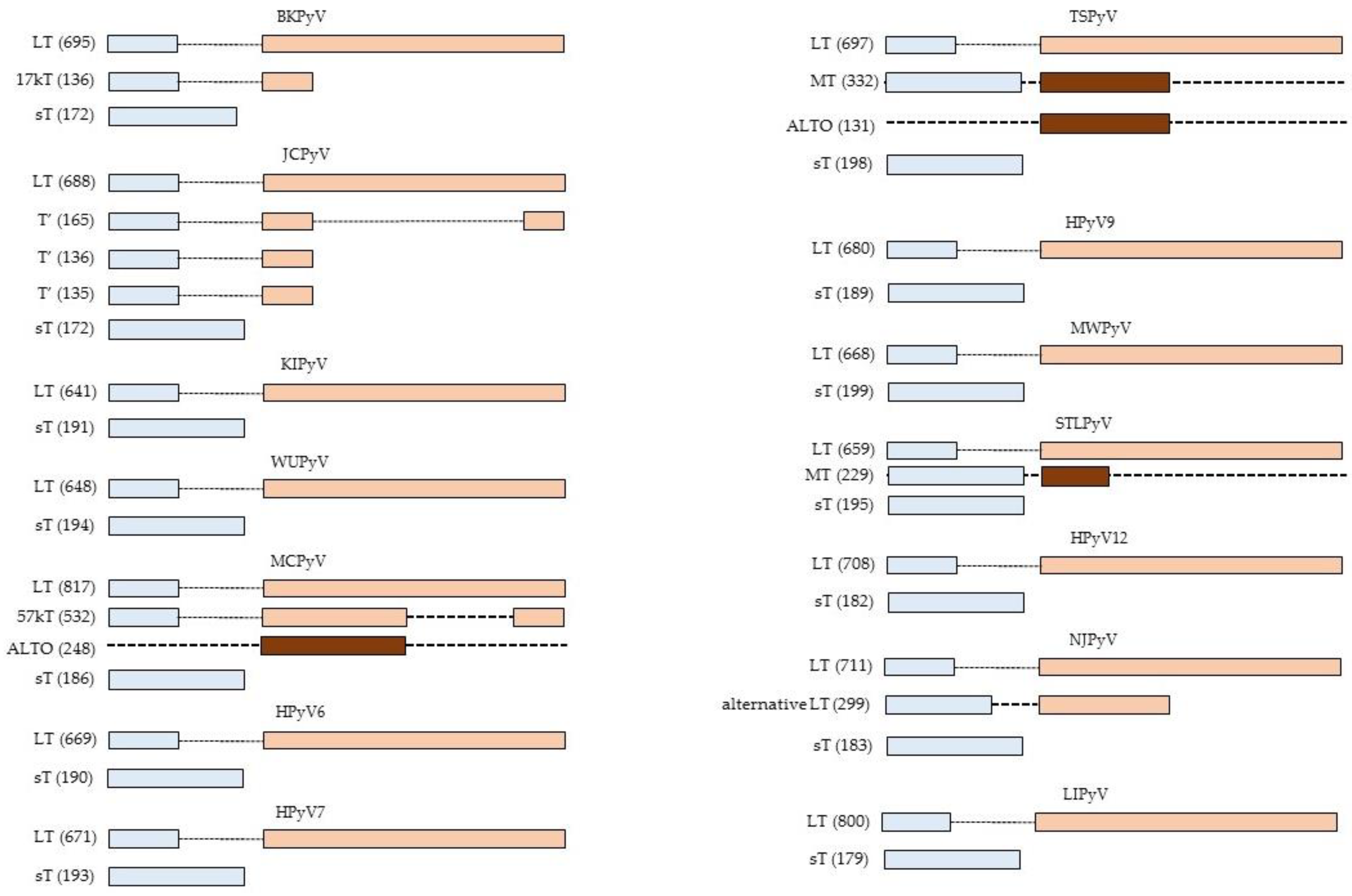
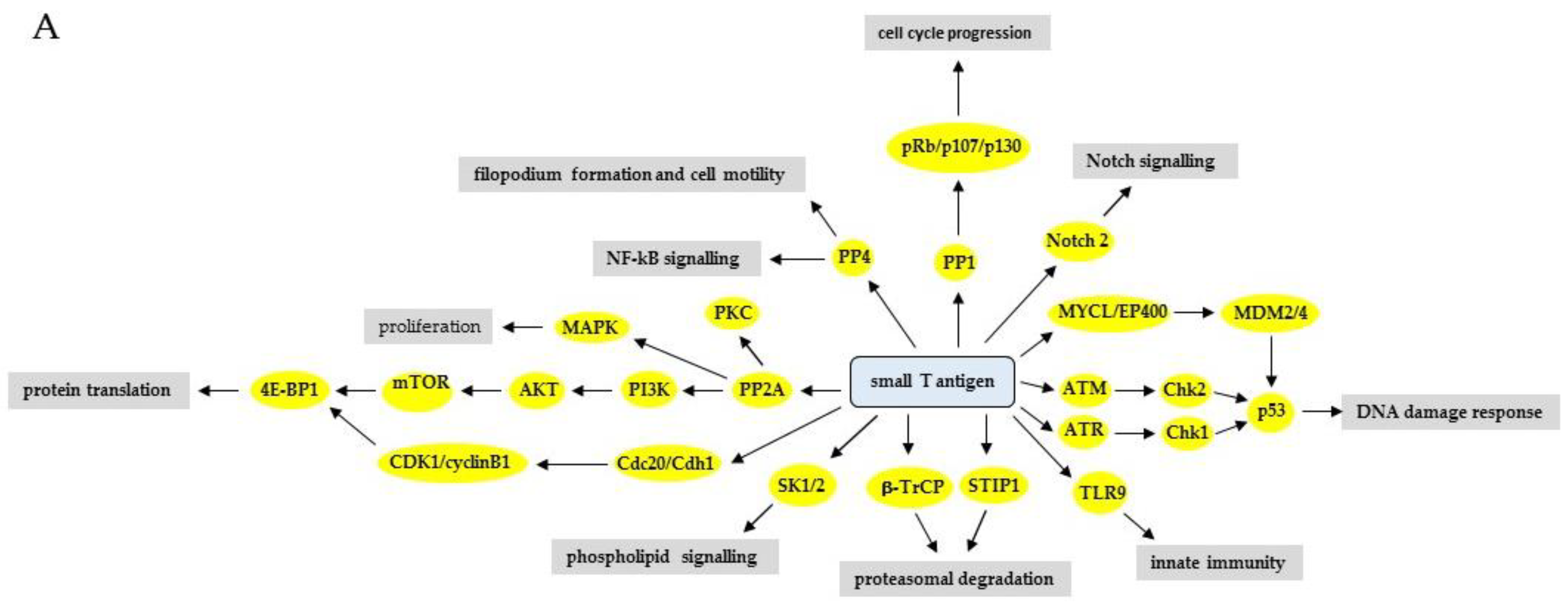
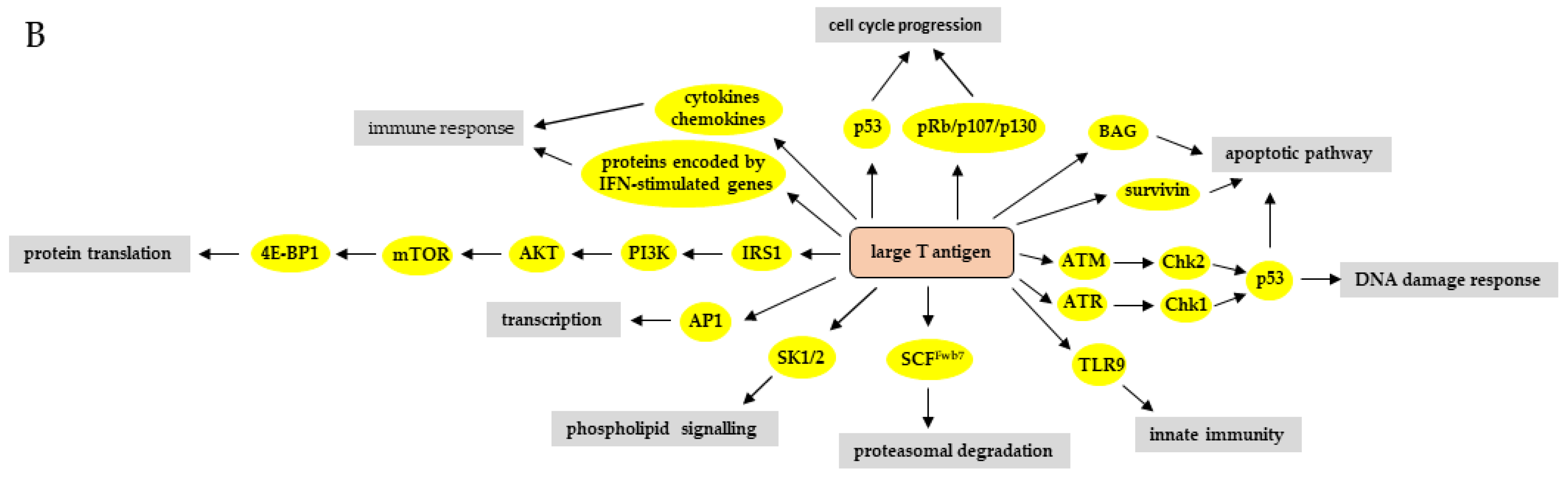
Effect of the Large and Small T-Antigens of Human Polyomaviruses on Signaling Pathways
Abstract
:1. Introduction
2. Interaction Partners of HPyV LT and sT
2.1. Protein Phosphatase 1 (PP1)
2.2. Protein Phosphatase 2A (PP2A)
2.3. Protein Phosphatase 4 (PP4)
3. The Effect of HPyV LT and sT on Signaling Pathways
3.1. Phosphatidyl-3-kinase/AKT/Mammalian Target of Rapamycin Pathway
3.2. Wnt Signalling
3.3. Protein Kinase C Pathway (PKC)
3.4. The Mitogen-Activated Protein Kinase (MAPK) Pathways
3.5. Notch Signaling Pathway
3.6. Hedgehog Signaling
3.7. DNA Damage Response Pathways
3.8. Retinoblastoma-E2F Pathway
3.9. p53 Pathway
3.10. Apoptotic Pathways
3.11. Ubiquitination-Proteasomal Degradation Pathway
3.12. Immune Response Pathways
3.12.1. NFκB Signaling Pathway
3.12.2. Innate Immune System
3.12.3. Interferon Signaling Pathway
3.12.4. Cytokines/Chemokines
3.13. Nuclear Receptor Signaling Pathway
3.14. Phospholipid Signaling Pathways
3.15. Metabolic Pathways
4. Conclusions and Future Research Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | 5′-adenosine monophosphate activated protein kinase |
| ATM | ataxia telangiectasia mutated |
| ATR | ATM- and Rad3-related |
| BAG | Bcl-2 associated athanogene protein |
| βTrCP | β-transducin-repeat containing protein |
| CCL17/TARC | chemokine (C-C motif) ligand 17/thymus and activation-regulated |
| Cdc | Cell division cycle protein |
| CK1α | Casein kinase 1α |
| DAMP | Danger-associated molecular pattern |
| DDR | DNA damage response |
| DHH | Desert hedgehog |
| DNA-PK | DNA protein kinase |
| DNMT1 | DNA methyltransferase 1 |
| Dvl | Dishevelled |
| ERK | Extracellular signal-regulated kinase |
| GSK3α | Glycogen synthase kinase 3α |
| HEK | Human embryonal kidney |
| HPyV | Human polyomavirus |
| IFN | Interferon |
| IGF | Insulin-like growth factor |
| IHH | Indian hedgehog |
| IKK | Inhibitor of NFκB kinase |
| IRF-1 | Interferon regulatory protein 1 |
| IRS-1 | Insulin receptor substrate 1 |
| JNK | c-Jun N-terminal kinase |
| LT | Large T-antigen |
| LEF | Lymphoid enhancer factor |
| MAPK | Mitogen-activated protein kinase |
| MCC | Merkel cell carcinoma |
| MCPyV | Merkel cell polyomavirus |
| MEK | Mitogen-activated kinase/ERK kinase |
| mT | Middle T-antigen |
| mTOR | Mammalian target of rapamycin |
| NEMO | NFκB essential modulator |
| NER | Nucleotide excision repair |
| PAMP | Pathogen-associated molecular pattern |
| p.i. | Post-infection |
| PI3K | Phosphatidyl-3-kinase |
| PP | Protein phosphatase |
| PRR | Pattern recognition receptor |
| PTCH | Patched |
| Rb | Retinoblastoma |
| RFC | Replication factor C |
| RPTE | Primary human renal proximal tubule epithelial |
| SHH | Sonic hedgehog |
| SK | Spingosine kinase |
| Smo | Smoothened |
| sT | Small t-antigen |
| STAT | Signal transducer and activator of transcription |
| STUB1 | STIP1 homology and U-box containing protein |
| TCF | T-cell factor |
| TEFb | Transcription elongation factor b |
| TLR | Toll-like receptor |
| TNFα | Tumour necrosis factor α |
References
- Buck, C.B.; Van Doorslaer, K.; Peretti, A.; Geoghegan, E.M.; Tisza, M.J.; An, P.; Katz, J.P.; Pipas, J.M.; McBride, A.A.; Camus, A.C.; et al. The Ancient Evolutionary History of Polyomaviruses. PLoS Pathog. 2016, 12, e1005574. [Google Scholar] [CrossRef]
- Moens, U.; Krumbholz, A.; Ehlers, B.; Zell, R.; Johne, R.; Calvignac-Spencer, S.; Lauber, C. Biology, evolution, and medical importance of polyomaviruses: An update. Infect Genet. Evol. 2017, 54, 18–38. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef]
- Allander, T.; Andreasson, K.; Gupta, S.; Bjerkner, A.; Bogdanovic, G.; Persson, M.A.; Dalianis, T.; Ramqvist, T.; Andersson, B. Identification of a third human polyomavirus. J. Virol. 2007, 81, 4130–4136. [Google Scholar] [CrossRef]
- Gaynor, A.M.; Nissen, M.D.; Whiley, D.M.; Mackay, I.M.; Lambert, S.B.; Wu, G.; Brennan, D.C.; Storch, G.A.; Sloots, T.P.; Wang, D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007, 3, e64. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Schowalter, R.M.; Pastrana, D.V.; Pumphrey, K.A.; Moyer, A.L.; Buck, C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 2010, 7, 509–515. [Google Scholar] [CrossRef]
- van der Meijden, E.; Janssens, R.W.; Lauber, C.; Bouwes Bavinck, J.N.; Gorbalenya, A.E.; Feltkamp, M.C. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010, 6, e1001024. [Google Scholar] [CrossRef]
- Scuda, N.; Hofmann, J.; Calvignac-Spencer, S.; Ruprecht, K.; Liman, P.; Kuhn, J.; Hengel, H.; Ehlers, B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J. Virol. 2011, 85, 4586–4590. [Google Scholar] [CrossRef]
- Siebrasse, E.A.; Reyes, A.; Lim, E.S.; Zhao, G.; Mkakosya, R.S.; Manary, M.J.; Gordon, J.I.; Wang, D. Identification of MW polyomavirus, a novel polyomavirus in human stool. J. Virol. 2012, 86, 10321–10326. [Google Scholar] [CrossRef]
- Yu, G.; Greninger, A.L.; Isa, P.; Phan, T.G.; Martinez, M.A.; de la Luz Sanchez, M.; Contreras, J.F.; Santos-Preciado, J.I.; Parsonnet, J.; Miller, S.; et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS ONE 2012, 7, e49449. [Google Scholar] [CrossRef]
- Lim, E.S.; Reyes, A.; Antonio, M.; Saha, D.; Ikumapayi, U.N.; Adeyemi, M.; Stine, O.C.; Skelton, R.; Brennan, D.C.; Mkakosya, R.S.; et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 2013, 436, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Korup, S.; Rietscher, J.; Calvignac-Spencer, S.; Trusch, F.; Hofmann, J.; Moens, U.; Sauer, I.; Voigt, S.; Schmuck, R.; Ehlers, B. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS ONE 2013, 8, e58021. [Google Scholar] [CrossRef]
- Mishra, N.; Pereira, M.; Rhodes, R.H.; An, P.; Pipas, J.M.; Jain, K.; Kapoor, A.; Briese, T.; Faust, P.L.; Lipkin, W.I. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J. Infect. Dis. 2014, 210, 1595–1599. [Google Scholar] [CrossRef]
- Gheit, T.; Dutta, S.; Oliver, J.; Robitaille, A.; Hampras, S.; Combes, J.D.; McKay-Chopin, S.; Le Calvez-Kelm, F.; Fenske, N.; Cherpelis, B.; et al. Isolation and characterization of a novel putative human polyomavirus. Virology 2017, 506, 45–54. [Google Scholar] [CrossRef]
- Gerits, N.; Moens, U. Agnoprotein of mammalian polyomaviruses. Virology 2012, 432, 316–326. [Google Scholar] [CrossRef] [Green Version]
- Schowalter, R.M.; Buck, C.B. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 2013, 9, e1003558. [Google Scholar] [CrossRef]
- Kamminga, S.; van der Meijden, E.; Feltkamp, M.C.W.; Zaaijer, H.L. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS ONE 2018, 13, e0206273. [Google Scholar] [CrossRef]
- Kean, J.M.; Rao, S.; Wang, M.; Garcea, R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009, 5, e1000363. [Google Scholar] [CrossRef]
- DeCaprio, J.A.; Garcea, R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013, 11, 264–276. [Google Scholar] [CrossRef]
- Dalianis, T.; Hirsch, H.H. Human polyomaviruses in disease and cancer. Virology 2013, 437, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Kazem, S.; van der Meijden, E.; Feltkamp, M.C. The trichodysplasia spinulosa-associated polyomavirus: Virological background and clinical implications. Apmis 2013, 121, 770–782. [Google Scholar] [CrossRef]
- Haley, C.T.; Mui, U.N.; Vangipuram, R.; Rady, P.L.; Tyring, S.K. Human Oncoviruses: Mucocutaneous Manifestations, Pathogenesis, Therapeutics, and Prevention: Papillomaviruses and Merkel cell polyomavirus. J. Am. Acad. Dermatol. 2019, 81, 23–41. [Google Scholar] [CrossRef]
- Anzivino, E.; Rodio, D.M.; Mischitelli, M.; Bellizzi, A.; Sciarra, A.; Salciccia, S.; Gentile, V.; Pietropaolo, V. High Frequency of JCV DNA Detection in Prostate Cancer Tissues. Cancer Genom. Proteom. 2015, 12, 189–200. [Google Scholar]
- Tognon, M.; Provenzano, M. New insights on the association between the prostate cancer and the small DNA tumour virus, BK polyomavirus. J. Transl. Med. 2015, 13, 387. [Google Scholar] [CrossRef]
- Delbue, S.; Comar, M.; Ferrante, P. Review on the role of the human Polyomavirus JC in the development of tumors. Infect. Agent Cancer 2017, 12, 10. [Google Scholar] [CrossRef]
- Levican, J.; Acevedo, M.; Leon, O.; Gaggero, A.; Aguayo, F. Role of BK human polyomavirus in cancer. Infect. Agent Cancer 2018, 13, 12. [Google Scholar] [CrossRef]
- Llewellyn, M.A.; Gordon, N.S.; Abbotts, B.; James, N.D.; Zeegers, M.P.; Cheng, K.K.; Macdonald, A.; Roberts, S.; Parish, J.L.; Ward, D.G.; et al. Defining the frequency of human papillomavirus and polyomavirus infection in urothelial bladder tumours. Sci. Rep. 2018, 8, 11290. [Google Scholar] [CrossRef]
- Prado, J.C.M.; Monezi, T.A.; Amorim, A.T.; Lino, V.; Paladino, A.; Boccardo, E. Human polyomaviruses and cancer: An overview. Clinics 2018, 73, e558s. [Google Scholar] [CrossRef]
- Ho, J.; Jedrych, J.J.; Feng, H.; Natalie, A.A.; Grandinetti, L.; Mirvish, E.; Crespo, M.M.; Yadav, D.; Fasanella, K.E.; Proksell, S.; et al. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J. Infect. Dis. 2015, 211, 1560–1565. [Google Scholar] [CrossRef]
- Nguyen, K.D.; Lee, E.E.; Yue, Y.; Stork, J.; Pock, L.; North, J.P.; Vandergriff, T.; Cockerell, C.; Hosler, G.A.; Pastrana, D.V.; et al. Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses. J. Am. Acad. Dermatol. 2017, 76, 932–940. [Google Scholar] [CrossRef]
- Vrancken, K.; Vervaeke, P.; Balzarini, J.; Liekens, S. Viruses as key regulators of angiogenesis. Rev. Med. Virol. 2011, 21, 181–200. [Google Scholar] [CrossRef] [Green Version]
- Danthi, P. Viruses and the Diversity of Cell Death. Annu. Rev. Virol. 2016, 3, 533–553. [Google Scholar] [CrossRef]
- Ming, X.; Jung, Y.S.; Babiuk, L.A.; Qian, Y. The host signaling pathways hijacked by oncogenic viruses. SM Vaccine Vaccin. 2017, 3, 1020. [Google Scholar]
- Fan, Y.; Sanyal, S.; Bruzzone, R. Breaking Bad: How Viruses Subvert the Cell Cycle. Front. Cell Infect. Microbiol. 2018, 8, 396. [Google Scholar] [CrossRef]
- Moreno-Altamirano, M.M.B.; Kolstoe, S.E.; Sanchez-Garcia, F.J. Virus Control of Cell Metabolism for Replication and Evasion of Host Immune Responses. Front. Cell Infect. Microbiol. 2019, 9, 95. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Hahn, W.C. Involvement of PP2A in viral and cellular transformation. Oncogene 2005, 24, 7746–7755. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10, eaag1796. [Google Scholar] [CrossRef]
- Kwun, H.J.; Shuda, M.; Camacho, C.J.; Gamper, A.M.; Thant, M.; Chang, Y.; Moore, P.S. Restricted protein phosphatase 2A targeting by Merkel cell polyomavirus small T antigen. J. Virol. 2015, 89, 4191–4200. [Google Scholar] [CrossRef]
- Kolupaeva, V.; Janssens, V. PP1 and PP2A phosphatases--cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013, 280, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Janssens, V.; Longin, S.; Goris, J. PP2A holoenzyme assembly: In cauda venenum (the sting is in the tail). Trends Biochem. Sci. 2008, 33, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Sontag, E. Protein phosphatase 2A: The Trojan Horse of cellular signaling. Cell Signal. 2001, 13, 7–16. [Google Scholar] [CrossRef]
- Cho, U.S.; Morrone, S.; Sablina, A.A.; Arroyo, J.D.; Hahn, W.C.; Xu, W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007, 5, e202. [Google Scholar] [CrossRef] [PubMed]
- Rundell, K.; Major, E.O.; Lampert, M. Association of cellular 56,000- and 32,000-molecular-weight protein with BK virus and polyoma virus t-antigens. J. Virol. 1981, 37, 1090–1093. [Google Scholar] [PubMed]
- Abdul-Sada, H.; Muller, M.; Mehta, R.; Toth, R.; Arthur, J.S.C.; Whitehouse, A.; Macdonald, A. The PP4R1 sub-unit of protein phosphatase PP4 is essential for inhibition of NF-kappaB by merkel polyomavirus small tumour antigen. Oncotarget 2017, 8, 25418–25432. [Google Scholar] [CrossRef] [PubMed]
- Sariyer, I.K.; Khalili, K.; Safak, M. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: Inhibition by small t antigen. Virology 2008, 375, 464–479. [Google Scholar] [CrossRef] [Green Version]
- Bollag, B.; Hofstetter, C.A.; Reviriego-Mendoza, M.M.; Frisque, R.J. JC virus small T antigen binds phosphatase PP2A and Rb family proteins and is required for efficient viral DNA replication activity. PLoS ONE 2010, 5, e10606. [Google Scholar] [CrossRef]
- Griffiths, D.A.; Abdul-Sada, H.; Knight, L.M.; Jackson, B.R.; Richards, K.; Prescott, E.L.; Peach, A.H.; Blair, G.E.; Macdonald, A.; Whitehouse, A. Merkel cell polyomavirus small T antigen targets the NEMO adaptor protein to disrupt inflammatory signaling. J. Virol. 2013, 87, 13853–13867. [Google Scholar] [CrossRef]
- Cheng, J.; Park, D.E.; Berrios, C.; White, E.A.; Arora, R.; Yoon, R.; Branigan, T.; Xiao, T.; Westerling, T.; Federation, A.; et al. Merkel cell polyomavirus recruits MYCL to the EP400 complex to promote oncogenesis. PLoS Pathog. 2017, 13, e1006668. [Google Scholar] [CrossRef]
- Shuda, M.; Kwun, H.J.; Feng, H.; Chang, Y.; Moore, P.S. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Investig. 2011, 121, 3623–3634. [Google Scholar] [CrossRef]
- Verhaegen, M.E.; Mangelberger, D.; Harms, P.W.; Vozheiko, T.D.; Weick, J.W.; Wilbert, D.M.; Saunders, T.L.; Ermilov, A.N.; Bichakjian, C.K.; Johnson, T.M.; et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Invest. Dermatol. 2015, 135, 1415–1424. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Patel, A.; Simonette, R.A.; Rady, P.; Tyring, S.K. Binding of the trichodysplasia spinulosa-associated polyomavirus small T antigen to protein phosphatase 2A: Elucidation of a potential pathogenic mechanism in a rare skin disease. JAMA Derm. 2014, 150, 1234–1236. [Google Scholar] [CrossRef]
- Wu, J.H.; Simonette, R.A.; Nguyen, H.P.; Rady, P.L.; Tyring, S.K. Molecular mechanisms supporting a pathogenic role for human polyomavirus 6 small T antigen: Protein phosphatase 2A targeting and MAPK cascade activation. J. Med. Virol. 2017, 89, 742–747. [Google Scholar] [CrossRef]
- Wu, J.H.; Narayanan, D.; Simonette, R.A.; Rady, P.L.; Tyring, S.K. Dysregulation of the MEK/ERK/MNK1 signalling cascade by middle T antigen of the trichoydsplasia spinulosa polyomavirus. J. Eur. Acad. Derm. Venereol. 2017, 31, 1338–1341. [Google Scholar] [CrossRef]
- Joshi, S.; Platanias, L.C. Mnk kinase pathway: Cellular functions and biological outcomes. World J. Biol. Chem. 2014, 5, 321–333. [Google Scholar] [CrossRef]
- Roux, P.P.; Topisirovic, I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cell Biol. 2018, 38, e00070-18. [Google Scholar] [CrossRef]
- Knight, L.M.; Stakaityte, G.; Wood, J.J.; Abdul-Sada, H.; Griffiths, D.A.; Howell, G.J.; Wheat, R.; Blair, G.E.; Steven, N.M.; Macdonald, A.; et al. Merkel cell polyomavirus small T antigen mediates microtubule destabilization to promote cell motility and migration. J. Virol. 2015, 89, 35–47. [Google Scholar] [CrossRef]
- Stakaityte, G.; Nwogu, N.; Dobson, S.J.; Knight, L.M.; Wasson, C.W.; Salguero, F.J.; Blackbourn, D.J.; Blair, G.E.; Mankouri, J.; Macdonald, A.; et al. Merkel Cell Polyomavirus Small T Antigen Drives Cell Motility via Rho-GTPase-Induced Filopodium Formation. J. Virol. 2018, 92, e00940-17. [Google Scholar] [CrossRef]
- Brouhard, G.J.; Rice, L.M. Microtubule dynamics: An interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 2018, 19, 451–463. [Google Scholar] [CrossRef]
- Manna, T.; Thrower, D.A.; Honnappa, S.; Steinmetz, M.O.; Wilson, L. Regulation of microtubule dynamic instability in vitro by differentially phosphorylated stathmin. J. Biol. Chem. 2009, 284, 15640–15649. [Google Scholar] [CrossRef]
- DeMali, K.A.; Wennerberg, K.; Burridge, K. Integrin signaling to the actin cytoskeleton. Curr. Opin. Cell Biol. 2003, 15, 572–582. [Google Scholar] [CrossRef] [Green Version]
- Rathinam, R.; Berrier, A.; Alahari, S.K. Role of Rho GTPases and their regulators in cancer progression. Front. Biosci. 2011, 16, 2561–2571. [Google Scholar] [CrossRef]
- Spiering, D.; Hodgson, L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh. Migr. 2011, 5, 170–180. [Google Scholar] [CrossRef] [Green Version]
- Nwogu, N.; Boyne, J.R.; Dobson, S.J.; Poterlowicz, K.; Blair, G.E.; Macdonald, A.; Mankouri, J.; Whitehouse, A. Cellular sheddases are induced by Merkel cell polyomavirus small tumour antigen to mediate cell dissociation and invasiveness. PLoS Pathog. 2018, 14, e1007276. [Google Scholar] [CrossRef]
- Berrios, C.; Padi, M.; Keibler, M.A.; Park, D.E.; Molla, V.; Cheng, J.; Lee, S.M.; Stephanopoulos, G.; Quackenbush, J.; DeCaprio, J.A. Merkel Cell Polyomavirus Small T Antigen Promotes Pro-Glycolytic Metabolic Perturbations Required for Transformation. PLoS Pathog. 2016, 12, e1006020. [Google Scholar] [CrossRef]
- Engelman, J.A.; Luo, J.; Cantley, L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006, 7, 606–619. [Google Scholar] [CrossRef]
- Engelman, J.A. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat. Rev. Cancer 2009, 9, 5505–5562. [Google Scholar] [CrossRef]
- Buchkovich, N.J.; Yu, Y.; Zampieri, C.A.; Alwine, J.C. The TORrid affairs of viruses: Effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 2008, 6, 266–275. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef]
- Ersahin, T.; Tuncbag, N.; Cetin-Atalay, R. The PI3K/AKT/mTOR interactive pathway. Mol. BioSyst. 2015, 11, 1946–1954. [Google Scholar] [CrossRef]
- Yuan, H.; Veldman, T.; Rundell, K.; Schlegel, R. Simian virus 40 small tumor antigen activates AKT and telomerase and induces anchorage-independent growth of human epithelial cells. J. Virol. 2002, 76, 10685–10691. [Google Scholar] [CrossRef]
- Yu, Y.; Alwine, J.C. Interaction between simian virus 40 large T antigen and insulin receptor substrate 1 is disrupted by the K1 mutation, resulting in the loss of large T antigen-mediated phosphorylation of Akt. J. Virol. 2008, 82, 4521–4526. [Google Scholar] [CrossRef]
- Del Valle, L.; Wang, J.Y.; Lassak, A.; Peruzzi, F.; Croul, S.; Khalili, K.; Reiss, K. Insulin-like growth factor I receptor signaling system in JC virus T antigen-induced primitive neuroectodermal tumors--medulloblastomas. J. Neurovirol. 2002, 8, 138–147. [Google Scholar] [CrossRef]
- Link, A.; Shin, S.K.; Nagasaka, T.; Balaguer, F.; Koi, M.; Jung, B.; Boland, C.R.; Goel, A. JC virus mediates invasion and migration in colorectal metastasis. PLoS ONE 2009, 4, e8146. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Bader, A.G.; Kang, S.; Vogt, P.K. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 1475–1479. [Google Scholar] [CrossRef]
- Hafner, C.; Houben, R.; Baeurle, A.; Ritter, C.; Schrama, D.; Landthaler, M.; Becker, J.C. Activation of the PI3K/AKT pathway in Merkel cell carcinoma. PLoS ONE 2012, 7, e31255. [Google Scholar] [CrossRef]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Parikh, C.; Janakiraman, V.; Wu, W.I.; Foo, C.K.; Kljavin, N.M.; Chaudhuri, S.; Stawiski, E.; Lee, B.; Lin, J.; Li, H.; et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc. Natl. Acad. Sci. USA 2012, 109, 19368–19373. [Google Scholar] [CrossRef]
- Iwasaki, T.; Matsushita, M.; Nonaka, D.; Kuwamoto, S.; Kato, M.; Murakami, I.; Nagata, K.; Nakajima, H.; Sano, S.; Hayashi, K. Comparison of Akt/mTOR/4E-BP1 pathway signal activation and mutations of PIK3CA in Merkel cell polyomavirus-positive and Merkel cell polyomavirus-negative carcinomas. Hum. Pathol. 2015, 46, 210–216. [Google Scholar] [CrossRef]
- Nardi, V.; Song, Y.; Santamaria-Barria, J.A.; Cosper, A.K.; Lam, Q.; Faber, A.C.; Boland, G.M.; Yeap, B.Y.; Bergethon, K.; Scialabba, V.L.; et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin. Cancer Res. 2012, 18, 1227–1236. [Google Scholar] [CrossRef]
- Musa, J.; Orth, M.F.; Dallmayer, M.; Baldauf, M.; Pardo, C.; Rotblat, B.; Kirchner, T.; Leprivier, G.; Grunewald, T.G. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): A master regulator of mRNA translation involved in tumorigenesis. Oncogene 2016, 35, 4675–4688. [Google Scholar] [CrossRef]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hein, J.; Richardson, S.C.; Basse, P.H.; Toptan, T.; Moore, P.S.; Gjoerup, O.V.; Chang, Y. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J. Biol. Chem. 2011, 286, 17079–17090. [Google Scholar] [CrossRef]
- Wu, J.H.; Simonette, R.A.; Hsiao, T.; Doan, H.Q.; Rady, P.L.; Tyring, S.K. Cutaneous Human Polyomavirus Small T Antigens and 4E-BP1 Targeting. Intervirology 2015, 58, 382–385. [Google Scholar] [CrossRef]
- Shuda, M.; Velasquez, C.; Cheng, E.; Cordek, D.G.; Kwun, H.J.; Chang, Y.; Moore, P.S. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci. USA 2015, 112, 5875–5882. [Google Scholar] [CrossRef] [Green Version]
- Velasquez, C.; Cheng, E.; Shuda, M.; Lee-Oesterreich, P.J.; Pogge von Strandmann, L.; Gritsenko, M.A.; Jacobs, J.M.; Moore, P.S.; Chang, Y. Mitotic protein kinase CDK1 phosphorylation of mRNA translation regulator 4E-BP1 Ser83 may contribute to cell transformation. Proc. Natl. Acad. Sci. USA 2016, 113, 8466–8471. [Google Scholar] [CrossRef] [Green Version]
- Bjornsti, M.A.; Houghton, P.J. Lost in translation: Dysregulation of cap-dependent translation and cancer. Cancer Cell 2004, 5, 519–523. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Enam, S.; Del Valle, L.; Lara, C.; Gan, D.D.; Ortiz-Hidalgo, C.; Palazzo, J.P.; Khalili, K. Association of human polyomavirus JCV with colon cancer: Evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2002, 62, 7093–7101. [Google Scholar]
- Gan, D.D.; Reiss, K.; Carrill, T.; Del Valle, L.; Croul, S.; Giordano, A.; Fishman, P.; Khalili, K. Involvement of Wnt signaling pathway in murine medulloblastoma induced by human neurotropic JC virus. Oncogene 2001, 20, 4864–4870. [Google Scholar] [CrossRef]
- Ripple, M.J.; Parker Struckhoff, A.; Trillo-Tinoco, J.; Li, L.; Margolin, D.A.; McGoey, R.; Del Valle, L. Activation of c-Myc and Cyclin D1 by JCV T-Antigen and beta-catenin in colon cancer. PLoS ONE 2014, 9, e106257. [Google Scholar] [CrossRef]
- Bhattacharyya, R.; Noch, E.K.; Khalili, K. A novel role of Rac1 GTPase in JCV T-antigen-mediated beta-catenin stabilization. Oncogene 2007, 26, 7628–7636. [Google Scholar] [CrossRef]
- Newton, A.C. Protein kinase C: Perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 208–230. [Google Scholar] [CrossRef]
- Garg, R.; Benedetti, L.G.; Abera, M.B.; Wang, H.; Abba, M.; Kazanietz, M.G. Protein kinase C and cancer: What we know and what we do not. Oncogene 2014, 33, 5225–5237. [Google Scholar] [CrossRef]
- Singh, R.M.; Cummings, E.; Pantos, C.; Singh, J. Protein kinase C and cardiac dysfunction: A review. Heart Fail. Rev. 2017, 22, 843–859. [Google Scholar] [CrossRef]
- Jain, K.; Basu, A. The Multifunctional Protein Kinase C-epsilon in Cancer Development and Progression. Cancers 2014, 6, 860–878. [Google Scholar] [CrossRef]
- Costa, A.; Mackelfresh, J.; Gilbert, L.; Bonner, M.Y.; Arbiser, J.L. Activation of Protein Kinase C epsilon in Merkel Cell Polyomavirus-Induced Merkel Cell Carcinoma. JAMA Derm. 2017, 153, 931–932. [Google Scholar] [CrossRef]
- Verma, S.; Ziegler, K.; Ananthula, P.; Co, J.K.; Frisque, R.J.; Yanagihara, R.; Nerurkar, V.R. JC virus induces altered patterns of cellular gene expression: Interferon-inducible genes as major transcriptional targets. Virology 2006, 345, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Sontag, E.; Sontag, J.M.; Garcia, A. Protein phosphatase 2A is a critical regulator of protein kinase C zeta signaling targeted by SV40 small t to promote cell growth and NF-kappaB activation. EMBO J. 1997, 16, 5662–5671. [Google Scholar] [CrossRef]
- Ugi, S.; Imamura, T.; Maegawa, H.; Egawa, K.; Yoshizaki, T.; Shi, K.; Obata, T.; Ebina, Y.; Kashiwagi, A.; Olefsky, J.M. Protein phosphatase 2A negatively regulates insulin’s metabolic signaling pathway by inhibiting Akt (protein kinase B) activity in 3T3-L1 adipocytes. Mol. Cell Biol. 2004, 24, 8778–8789. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Querbes, W.; Benmerah, A.; Tosoni, D.; Di Fiore, P.P.; Atwood, W.J. A JC virus-induced signal is required for infection of glial cells by a clathrin- and eps15-dependent pathway. J. Virol. 2004, 78, 250–256. [Google Scholar] [CrossRef]
- DuShane, J.K.; Wilczek, M.P.; Mayberry, C.L.; Maginnis, M.S. ERK Is a Critical Regulator of JC Polyomavirus Infection. J. Virol. 2018, 92, e01529-17. [Google Scholar] [CrossRef]
- DuShane, J.K.; Wilczek, M.P.; Crocker, M.A.; Maginnis, M.S. High-Throughput Characterization of Viral and Cellular Protein Expression Patterns During JC Polyomavirus Infection. Front. Microbiol. 2019, 10, 783. [Google Scholar] [CrossRef]
- Nelson, C.D.; Derdowski, A.; Maginnis, M.S.; O’Hara, B.A.; Atwood, W.J. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 2012, 428, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, S.; Moelling, K. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 1999, 286, 1741–1744. [Google Scholar] [CrossRef]
- Seamone, M.E.; Wang, W.; Acott, P.; Beck, P.L.; Tibbles, L.A.; Muruve, D.A. MAP kinase activation increases BK polyomavirus replication and facilitates viral propagation in vitro. J. Virol. Methods 2010, 170, 21–29. [Google Scholar] [CrossRef]
- Weston, C.R.; Davis, R.J. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007, 19, 142–149. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef]
- Kim, J.; Woolridge, S.; Biffi, R.; Borghi, E.; Lassak, A.; Ferrante, P.; Amini, S.; Khalili, K.; Safak, M. Members of the AP-1 family, c-Jun and c-Fos, functionally interact with JC virus early regulatory protein large T antigen. J. Virol. 2003, 77, 5241–5252. [Google Scholar] [CrossRef]
- Glenn, G.M.; Eckhart, W. Transcriptional regulation of early-response genes during polyomavirus infection. J. Virol. 1990, 64, 2193–2201. [Google Scholar] [Green Version]
- Sadowska, B.; Barrucco, R.; Khalili, K.; Safak, M. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J. Virol. 2003, 77, 665–672. [Google Scholar] [CrossRef]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef]
- Lasithiotaki, I.; Antoniou, K.M.; Derdas, S.P.; Sarchianaki, E.; Symvoulakis, E.K.; Psaraki, A.; Spandidos, D.A.; Stathopoulos, E.N.; Siafakas, N.M.; Sourvinos, G. The presence of Merkel cell polyomavirus is associated with deregulated expression of BRAF and Bcl-2 genes in non-small cell lung cancer. Int. J. Cancer 2013, 133, 604–611. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 2017, 41, 228–241. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Ulloa, E.; Lizano, M.; Sjoqvist, M.; Olmedo-Nieva, L.; Contreras-Paredes, A. Deregulation of the Notch pathway as a common road in viral carcinogenesis. Rev. Med. Virol. 2018, 28, e1988. [Google Scholar] [CrossRef]
- Carbone, M.; Burck, C.; Rdzanek, M.; Rudzinski, J.; Cutrone, R.; Bocchetta, M. Different susceptibility of human mesothelial cells to polyomavirus infection and malignant transformation. Cancer Res. 2003, 63, 6125–6129. [Google Scholar]
- Wardhani, L.O.; Matsushita, M.; Kuwamoto, S.; Nonaka, D.; Nagata, K.; Kato, M.; Kitamura, Y.; Hayashi, K. Expression of Notch 3 and Jagged 1 Is Associated With Merkel Cell Polyomavirus Status and Prognosis in Merkel Cell Carcinoma. Anticancer Res. 2019, 39, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012, 487, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef] [PubMed]
- McMillan, R.; Matsui, W. Molecular pathways: The hedgehog signaling pathway in cancer. Clin. Cancer Res. 2012, 18, 4883–4888. [Google Scholar] [CrossRef] [PubMed]
- Kuromi, T.; Matsushita, M.; Iwasaki, T.; Nonaka, D.; Kuwamoto, S.; Nagata, K.; Kato, M.; Akizuki, G.; Kitamura, Y.; Hayashi, K. Association of expression of the hedgehog signal with Merkel cell polyomavirus infection and prognosis of Merkel cell carcinoma. Hum. Pathol. 2017, 69, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Brunner, M.; Thurnher, D.; Pammer, J.; Heiduschka, G.; Petzelbauer, P.; Schmid, C.; Schneider, S.; Erovic, B.M. Expression of hedgehog signaling molecules in Merkel cell carcinoma. Head Neck 2010, 32, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Singh, R.R.; Vega, F. Aberrant activation of the hedgehog signaling pathway in malignant hematological neoplasms. Am. J. Pathol. 2012, 180, 2–11. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, P.; Foiani, M.; Kumar, A. ATM and ATR signaling at a glance. J. Cell Sci. 2016, 129, 1285. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Darbinyan, A.; White, M.K.; Akan, S.; Radhakrishnan, S.; Del Valle, L.; Amini, S.; Khalili, K. Alterations of DNA damage repair pathways resulting from JCV infection. Virology 2007, 364, 73–86. [Google Scholar] [CrossRef] [Green Version]
- Orba, Y.; Suzuki, T.; Makino, Y.; Kubota, K.; Tanaka, S.; Kimura, T.; Sawa, H. Large T antigen promotes JC virus replication in G2-arrested cells by inducing ATM- and ATR-mediated G2 checkpoint signaling. J. Biol. Chem. 2010, 285, 1544–1554. [Google Scholar] [CrossRef]
- Huang, J.L.; Lin, C.S.; Chang, C.C.; Lu, Y.N.; Hsu, Y.L.; Wong, T.Y.; Wang, Y.F. Human JC virus small tumour antigen inhibits nucleotide excision repair and sensitises cells to DNA-damaging agents. Mutagenesis 2015, 30, 475–485. [Google Scholar] [CrossRef]
- White, M.K.; Bellizzi, A.; Ibba, G.; Pietropaolo, V.; Palamara, A.T.; Wollebo, H.S. The DNA damage response promotes polyomavirus JC infection by nucleus to cytoplasm NF- kappaB activation. Virol. J. 2017, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Abend, J.R.; Low, J.A.; Imperiale, M.J. Global effects of BKV infection on gene expression in human primary kidney epithelial cells. Virology 2010, 397, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Zhao, L.; Gamez, M.; Imperiale, M.J. Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog. 2012, 8, e1002898. [Google Scholar] [CrossRef]
- Justice, J.L.; Needham, J.M.; Thompson, S.R. BK polyomavirus activates the DNA damage response to prolong S phase. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Verhalen, B.; Justice, J.L.; Imperiale, M.J.; Jiang, M. Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection. J. Virol. 2015, 89, 5032–5039. [Google Scholar] [CrossRef]
- Trojanek, J.; Croul, S.; Ho, T.; Wang, J.Y.; Darbinyan, A.; Nowicki, M.; Del Valle, L.; Skorski, T.; Khalili, K.; Reiss, K. T-antigen of the human polyomavirus JC attenuates faithful DNA repair by forcing nuclear interaction between IRS-1 and Rad51. J. Cell Physiol. 2006, 206, 35–46. [Google Scholar] [CrossRef]
- Hein, J.; Boichuk, S.; Wu, J.; Cheng, Y.; Freire, R.; Jat, P.S.; Roberts, T.M.; Gjoerup, O.V. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 2009, 83, 117–127. [Google Scholar] [CrossRef]
- White, M.K.; Skowronska, A.; Gordon, J.; Del Valle, L.; Deshmane, S.L.; Giordano, A.; Khalili, K. Analysis of a mutant p53 protein arising in a medulloblastoma from a mouse transgenic for the JC virus early region. Anticancer Res. 2006, 26, 4079–4092. [Google Scholar]
- Aragon, L. The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative. Annu. Rev. Genet. 2018, 52, 89–107. [Google Scholar] [CrossRef]
- Swingle, M.; Ni, L.; Honkanen, R.E. Small-molecule inhibitors of ser/thr protein phosphatases: Specificity, use and common forms of abuse. Methods Mol. Biol. 2007, 365, 23–38. [Google Scholar]
- Chowdhury, D.; Keogh, M.C.; Ishii, H.; Peterson, C.L.; Buratowski, S.; Lieberman, J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 2005, 20, 801–809. [Google Scholar] [CrossRef]
- Adams, T.E.; Epa, V.C.; Garrett, T.P.; Ward, C.W. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol. Life Sci 2000, 57, 1050–1093. [Google Scholar] [CrossRef]
- Mauro, L.; Salerno, M.; Morelli, C.; Boterberg, T.; Bracke, M.E.; Surmacz, E. Role of the IGF-I receptor in the regulation of cell-cell adhesion: Implications in cancer development and progression. J. Cell Physiol. 2003, 194, 108–116. [Google Scholar] [CrossRef]
- Belfiore, A.; Frasca, F.; Pandini, G.; Sciacca, L.; Vigneri, R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009, 30, 586–623. [Google Scholar] [CrossRef]
- Trojanek, J.; Ho, T.; Del Valle, L.; Nowicki, M.; Wang, J.Y.; Lassak, A.; Peruzzi, F.; Khalili, K.; Skorski, T.; Reiss, K. Role of the insulin-like growth factor I/insulin receptor substrate 1 axis in Rad51 trafficking and DNA repair by homologous recombination. Mol. Cell Biol. 2003, 23, 7510–7524. [Google Scholar] [CrossRef]
- Lassak, A.; Del Valle, L.; Peruzzi, F.; Wang, J.Y.; Enam, S.; Croul, S.; Khalili, K.; Reiss, K. Insulin receptor substrate 1 translocation to the nucleus by the human JC virus T-antigen. J. Biol. Chem. 2002, 277, 17231–17238. [Google Scholar] [CrossRef]
- Reiss, K.; Del Valle, L.; Lassak, A.; Trojanek, J. Nuclear IRS-1 and cancer. J. Cell Physiol. 2012, 227, 2992–3000. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Diaz, J.; Tsang, S.H.; Buck, C.B.; You, J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013, 87, 9173–9188. [Google Scholar] [CrossRef]
- Tsang, S.H.; Wang, X.; Li, J.; Buck, C.B.; You, J. Host DNA damage response factors localize to merkel cell polyomavirus DNA replication sites to support efficient viral DNA replication. J. Virol. 2014, 88, 3285–3297. [Google Scholar] [CrossRef]
- Li, J.; Diaz, J.; Wang, X.; Tsang, S.H.; You, J. Phosphorylation of Merkel cell polyomavirus large tumor antigen at serine 816 by ATM kinase induces apoptosis in host cells. J. Biol. Chem. 2015, 290, 1874–1884. [Google Scholar] [CrossRef]
- Genovese, C.; Trani, D.; Caputi, M.; Claudio, P.P. Cell cycle control and beyond: Emerging roles for the retinoblastoma gene family. Oncogene 2006, 25, 5201–5209. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [Green Version]
- Dick, F.A.; Rubin, S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013, 14, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.G.; Schwarz, J.K.; Cress, W.D.; Nevins, J.R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 1993, 365, 349–352. [Google Scholar] [CrossRef]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Logan, N.; Graham, A.; Zhao, X.; Fisher, R.; Maiti, B.; Leone, G.; La Thangue, N.B. E2F-8: An E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 2005, 24, 5000–5004. [Google Scholar] [CrossRef]
- Brehm, A.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998, 391, 597–601. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef]
- Dyson, N.; Buchkovich, K.; Whyte, P.; Harlow, E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell 1989, 58, 249–255. [Google Scholar] [CrossRef]
- Dyson, N.; Bernards, R.; Friend, S.H.; Gooding, L.R.; Hassell, J.A.; Major, E.O.; Pipas, J.M.; Vandyke, T.; Harlow, E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J. Virol. 1990, 64, 1353–1356. [Google Scholar] [Green Version]
- Harris, K.F.; Christensen, J.B.; Imperiale, M.J. BK virus large T antigen: Interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J. Virol. 1996, 70, 2378–2386. [Google Scholar]
- Bollag, B.; Prins, C.; Snyder, E.L.; Frisque, R.J. Purified JC virus T and T’ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology 2000, 274, 165–178. [Google Scholar] [CrossRef]
- Shuda, M.; Feng, H.; Kwun, H.J.; Rosen, S.T.; Gjoerup, O.; Moore, P.S.; Chang, Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 2008, 105, 16272–16277. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Rozenblatt-Rosen, O.; Paulson, K.G.; Nghiem, P.; DeCaprio, J.A. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J. Virol. 2013, 87, 6118–6126. [Google Scholar] [CrossRef]
- Borchert, S.; Czech-Sioli, M.; Neumann, F.; Schmidt, C.; Wimmer, P.; Dobner, T.; Grundhoff, A.; Fischer, N. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J. Virol. 2014, 88, 3144–3160. [Google Scholar] [CrossRef]
- Akiyama, T.; Ohuchi, T.; Sumida, S.; Matsumoto, K.; Toyoshima, K. Phosphorylation of the retinoblastoma protein by cdk2. Proc. Natl. Acad. Sci. USA 1992, 89, 7900–7904. [Google Scholar] [CrossRef] [PubMed]
- Hesbacher, S.; Pfitzer, L.; Wiedorfer, K.; Angermeyer, S.; Borst, A.; Haferkamp, S.; Scholz, C.J.; Wobser, M.; Schrama, D.; Houben, R. RB1 is the crucial target of the Merkel cell polyomavirus Large T antigen in Merkel cell carcinoma cells. Oncotarget 2016, 7, 32956–32968. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Simonette, R.A.; Nguyen, H.P.; Doan, H.Q.; Rady, P.L.; Tyring, S.K. Emerging differential roles of the pRb tumor suppressor in trichodysplasia spinulosa-associated polyomavirus and Merkel cell polyomavirus pathogeneses. J. Clin. Virol. 2016, 76, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Nguyen, H.P.; Rady, P.L.; Tyring, S.K. Molecular insight into the viral biology and clinical features of trichodysplasia spinulosa. Br. J. Derm. 2016, 174, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Kurimchak, A.; Grana, X. PP2A holoenzymes negatively and positively regulate cell cycle progression by dephosphorylating pocket proteins and multiple CDK substrates. Gene 2012, 499, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jayadeva, G.; Kurimchak, A.; Garriga, J.; Sotillo, E.; Davis, A.J.; Haines, D.S.; Mumby, M.; Grana, X. B55alpha PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J. Biol. Chem. 2010, 285, 29863–29873. [Google Scholar] [CrossRef] [PubMed]
- Kazem, S.; van der Meijden, E.; Wang, R.C.; Rosenberg, A.S.; Pope, E.; Benoit, T.; Fleckman, P.; Feltkamp, M.C. Polyomavirus-associated Trichodysplasia spinulosa involves hyperproliferation, pRB phosphorylation and upregulation of p16 and p21. PLoS ONE 2014, 9, e108947. [Google Scholar] [CrossRef] [PubMed]
- Bollag, B.; Kilpatrick, L.H.; Tyagarajan, S.K.; Tevethia, M.J.; Frisque, R.J. JC virus T’135, T’136 and T’165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. J. Neurovirol. 2006, 12, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.F.; Chang, E.; Christensen, J.B.; Imperiale, M.J. BK virus as a potential co-factor in human cancer. Dev. Biol. Stand. 1998, 94, 81–91. [Google Scholar]
- McCabe, M.T.; Low, J.A.; Imperiale, M.J.; Day, M.L. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene 2006, 25, 2727–2735. [Google Scholar] [CrossRef] [Green Version]
- McCabe, M.T.; Low, J.A.; Daignault, S.; Imperiale, M.J.; Wojno, K.J.; Day, M.L. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006, 66, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Li, M.S.; Nagasaka, T.; Shin, S.K.; Fuerst, F.; Ricciardiello, L.; Wasserman, L.; Boland, C.R. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology 2006, 130, 1950–1961. [Google Scholar] [CrossRef] [PubMed]
- Ksiaa, F.; Ziadi, S.; Mokni, M.; Korbi, S.; Trimeche, M. The presence of JC virus in gastric carcinomas correlates with patient’s age, intestinal histological type and aberrant methylation of tumor suppressor genes. Mod. Pathol. 2010, 23, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Ksiaa, F.; Ziadi, S.; Gacem, R.B.; Dhiab, M.B.; Trimeche, M. Correlation between DNA methyltransferases expression and Epstein-Barr virus, JC polyomavirus and Helicobacter pylori infections in gastric carcinomas. Neoplasma 2014, 61, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, P.; Myszka, A.; Ramsey, D.; Kielan, W.; Sasiadek, M.M. Detection of viral DNA sequences in sporadic colorectal cancers in relation to CpG island methylation and methylator phenotype. Tumour Biol. 2011, 32, 653–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef] [Green Version]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The p53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem. 2016, 85, 375–404. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Bollag, B.; Chuke, W.F.; Frisque, R.J. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: Identification of sequences important for efficient transformation. J. Virol. 1989, 63, 863–872. [Google Scholar] [PubMed]
- Staib, C.; Pesch, J.; Gerwig, R.; Gerber, J.K.; Brehm, U.; Stangl, A.; Grummt, F. p53 inhibits JC virus DNA replication in vivo and interacts with JC virus large T-antigen. Virology 1996, 219, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Reiss, K.; Khalili, K. Viruses and cancer: Lessons from the human polyomavirus, JCV. Oncogene 2003, 22, 6517–6523. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, C.V.; Das, G.C. Interaction of human polyomavirus BK with the tumor-suppressor protein p53. Oncogene 1996, 13, 323–332. [Google Scholar]
- Caller, L.G.; Davies, C.T.R.; Antrobus, R.; Lehner, P.J.; Weekes, M.P.; Crump, C.M. Temporal proteomic analysis of BK polyomavirus infection reveals virus-induced G2 arrest and highly effective evasion of innate immune sensing. J. Virol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.L.; Verhalen, B.; Kumar, R.; Lefkowitz, E.J.; Imperiale, M.J.; Jiang, M. Quantitative Proteomic Analysis of Enriched Nuclear Fractions from BK Polyomavirus-Infected Primary Renal Proximal Tubule Epithelial Cells. J. Proteome Res. 2015, 14, 4413–4424. [Google Scholar] [CrossRef] [Green Version]
- Florenes, V.A.; Maelandsmo, G.M.; Forus, A.; Andreassen, A.; Myklebost, O.; Fodstad, O. MDM2 gene amplification and transcript levels in human sarcomas: Relationship to TP53 gene status. J. Natl. Cancer Inst. 1994, 86, 1297–1302. [Google Scholar] [CrossRef]
- Sasaki, Y.; Negishi, H.; Idogawa, M.; Yokota, I.; Koyama, R.; Kusano, M.; Suzuki, H.; Fujita, M.; Maruyama, R.; Toyota, M.; et al. p53 negatively regulates the hepatoma growth factor HDGF. Cancer Res. 2011, 71, 7038–7047. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [Green Version]
- Park, D.E.; Cheng, J.; Berrios, C.; Montero, J.; Cortes-Cros, M.; Ferretti, S.; Arora, R.; Tillgren, M.L.; Gokhale, P.C.; DeCaprio, J.A. Dual inhibition of MDM2 and MDM4 in virus-positive Merkel cell carcinoma enhances the p53 response. Proc. Natl. Acad. Sci. USA 2019, 116, 1027–1032. [Google Scholar] [CrossRef]
- Pei, D.; Zhang, Y.; Zheng, J. Regulation of p53: A collaboration between Mdm2 and Mdmx. Oncotarget 2012, 3, 228–235. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef]
- Benedict, C.A.; Norris, P.S.; Ware, C.F. To kill or be killed: Viral evasion of apoptosis. Nat. Immunol 2002, 3, 1013–1018. [Google Scholar] [CrossRef]
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040. [Google Scholar] [CrossRef] [Green Version]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef]
- Basile, A.; Darbinian, N.; Kaminski, R.; White, M.K.; Gentilella, A.; Turco, M.C.; Khalili, K. Evidence for modulation of BAG3 by polyomavirus JC early protein. J. Gen. Virol. 2009, 90, 1629–1640. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Wang, C.; Tjian, R. Positive and negative regulation of transcription in vitro: Enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell 1987, 50, 847–861. [Google Scholar] [CrossRef]
- Sariyer, I.K.; Merabova, N.; Patel, P.K.; Knezevic, T.; Rosati, A.; Turco, M.C.; Khalili, K. Bag3-induced autophagy is associated with degradation of JCV oncoprotein, T-Ag. PLoS ONE 2012, 7, e45000. [Google Scholar] [CrossRef]
- Kroemer, G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat. Med. 1997, 3, 614–620. [Google Scholar] [CrossRef]
- Sahi, H.; Koljonen, V.; Kavola, H.; Haglund, C.; Tukiainen, E.; Sihto, H.; Bohling, T. Bcl-2 expression indicates better prognosis of Merkel cell carcinoma regardless of the presence of Merkel cell polyomavirus. Virchows Arch. 2012, 461, 553–559. [Google Scholar] [CrossRef]
- Arora, R.; Shuda, M.; Guastafierro, A.; Feng, H.; Toptan, T.; Tolstov, Y.; Normolle, D.; Vollmer, L.L.; Vogt, A.; Domling, A.; et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci Transl Med. 2012, 4, 133ra56. [Google Scholar] [CrossRef]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell Sci 2019, 132, jcs223826. [Google Scholar] [CrossRef]
- Schrama, D.; Hesbacher, S.; Becker, J.C.; Houben, R. Survivin downregulation is not required for T antigen knockdown mediated cell growth inhibition in MCV infected merkel cell carcinoma cells. Int. J. Cancer 2013, 132, 2980–2982. [Google Scholar] [CrossRef]
- Jiang, Y.; Saavedra, H.I.; Holloway, M.P.; Leone, G.; Altura, R.A. Aberrant regulation of survivin by the RB/E2F family of proteins. J. Biol. Chem. 2004, 279, 40511–40520. [Google Scholar] [CrossRef]
- Batinica, M.; Akgul, B.; Silling, S.; Mauch, C.; Zigrino, P. Correlation of Merkel cell polyomavirus positivity with PDGFRalpha mutations and survivin expression in Merkel cell carcinoma. J. Derm. Sci 2015, 79, 43–49. [Google Scholar] [CrossRef]
- Pina-Oviedo, S.; Urbanska, K.; Radhakrishnan, S.; Sweet, T.; Reiss, K.; Khalili, K.; Del Valle, L. Effects of JC virus infection on anti-apoptotic protein survivin in progressive multifocal leukoencephalopathy. Am. J. Pathol. 2007, 170, 1291–1304. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Mani, A.; Gelmann, E.P. The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 2005, 23, 4776–4789. [Google Scholar] [CrossRef]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef]
- Luo, H. Interplay between the virus and the ubiquitin-proteasome system: Molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Reviriego-Mendoza, M.M.; Frisque, R.J. Interaction and co-localization of JC virus large T antigen and the F-box protein beta-transducin-repeat containing protein. Virology 2011, 410, 119–128. [Google Scholar] [CrossRef]
- Lau, A.W.; Fukushima, H.; Wei, W. The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front. Biosci. 2012, 17, 2197–2212. [Google Scholar] [CrossRef]
- Kwun, H.J.; Wendzicki, J.A.; Shuda, Y.; Moore, P.S.; Chang, Y. Merkel cell polyomavirus small T antigen induces genome instability by E3 ubiquitin ligase targeting. Oncogene 2017, 36, 6784–6792. [Google Scholar] [CrossRef]
- Kwun, H.J.; Shuda, M.; Feng, H.; Camacho, C.J.; Moore, P.S.; Chang, Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe 2013, 14, 125–135. [Google Scholar] [CrossRef]
- Kwun, H.J.; Chang, Y.; Moore, P.S. Protein-mediated viral latency is a novel mechanism for Merkel cell polyomavirus persistence. Proc. Natl. Acad. Sci. USA 2017, 114, E4040–E4047. [Google Scholar] [CrossRef] [Green Version]
- Dye, K.N.; Welcker, M.; Clurman, B.E.; Roman, A.; Galloway, D.A. Merkel cell polyomavirus Tumor antigens expressed in Merkel cell carcinoma function independently of the ubiquitin ligases Fbw7 and beta-TrCP. PLoS Pathog. 2019, 15, e1007543. [Google Scholar] [CrossRef]
- McDonough, H.; Patterson, C. CHIP: A link between the chaperone and proteasome systems. Cell Stress Chaperones 2003, 8, 303–308. [Google Scholar] [CrossRef]
- Zhan, S.; Wang, T.; Ge, W. Multiple functions of the E3 ubiquitin ligase CHIP in immunity. Int. Rev. Immunol 2017, 36, 300–312. [Google Scholar] [CrossRef]
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef]
- Christian, F.; Smith, E.L.; Carmody, R.J. The Regulation of NF-kappaB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Henkel, T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef]
- Matsushita, M.; Iwasaki, T.; Nonaka, D.; Kuwamoto, S.; Nagata, K.; Kato, M.; Kitamura, Y.; Hayashi, K. Higher Expression of Activation-induced Cytidine Deaminase Is Significantly Associated with Merkel Cell Polyomavirus-negative Merkel Cell Carcinomas. Yonago Acta Med. 2017, 60, 145–153. [Google Scholar] [CrossRef] [Green Version]
- McCool, K.W.; Miyamoto, S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012, 246, 311–326. [Google Scholar] [CrossRef]
- Wollebo, H.S.; Melis, S.; Khalili, K.; Safak, M.; White, M.K. Cooperative roles of NF-kappaB and NFAT4 in polyomavirus JC regulation at the KB control element. Virology 2012, 432, 146–154. [Google Scholar] [CrossRef]
- Wu, S.Y.; Chiang, C.M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007, 282, 13141–13145. [Google Scholar] [CrossRef]
- Taniguchi, Y. The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. Int. J. Mol. Sci. 2016, 17, 1849. [Google Scholar] [CrossRef]
- Belkina, A.C.; Denis, G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 2012, 12, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Hajmirza, A.; Emadali, A.; Gauthier, A.; Casasnovas, O.; Gressin, R.; Callanan, M.B. BET Family Protein BRD4: An Emerging Actor in NFkappaB Signaling in Inflammation and Cancer. Biomedicines 2018, 6, 16. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Schowalter, R.M.; Jiao, J.; Buck, C.B.; You, J. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 2012, 8, e1003021. [Google Scholar] [CrossRef]
- Wollebo, H.S.; Bellizzi, A.; Cossari, D.H.; Salkind, J.; Safak, M.; White, M.K. The Brd4 acetyllysine-binding protein is involved in activation of polyomavirus JC. J. Neurovirol 2016, 22, 615–625. [Google Scholar] [CrossRef] [Green Version]
- Suresh, R.; Mosser, D.M. Pattern recognition receptors in innate immunity, host defense, and immunopathology. Adv. Physiol Educ 2013, 37, 284–291. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Shahzad, N.; Shuda, M.; Gheit, T.; Kwun, H.J.; Cornet, I.; Saidj, D.; Zannetti, C.; Hasan, U.; Chang, Y.; Moore, P.S.; et al. The T antigen locus of Merkel cell polyomavirus downregulates human Toll-like receptor 9 expression. J. Virol. 2013, 87, 13009–13019. [Google Scholar] [CrossRef]
- Jouhi, L.; Koljonen, V.; Bohling, T.; Haglund, C.; Hagstrom, J. The expression of Toll-like receptors 2, 4, 5, 7 and 9 in Merkel cell carcinoma. Anticancer Res. 2015, 35, 1843–1849. [Google Scholar]
- Giacobbi, N.S.; Gupta, T.; Coxon, A.T.; Pipas, J.M. Polyomavirus T antigens activate an antiviral state. Virology 2015, 476, 377–385. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, S.; Otte, J.; Enam, S.; Del Valle, L.; Khalili, K.; Gordon, J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J. Virol. 2003, 77, 10638–10644. [Google Scholar] [CrossRef]
- Assetta, B.; De Cecco, M.; O’Hara, B.; Atwood, W.J. JC Polyomavirus Infection of Primary Human Renal Epithelial Cells Is Controlled by a Type I IFN-Induced Response. mBio 2016, 7, e00903-16. [Google Scholar] [CrossRef] [Green Version]
- An, P.; Saenz Robles, M.T.; Duray, A.M.; Cantalupo, P.G.; Pipas, J.M. Human polyomavirus BKV infection of endothelial cells results in interferon pathway induction and persistence. PLoS Pathog. 2019, 15, e1007505. [Google Scholar] [CrossRef]
- Grinde, B.; Gayorfar, M.; Rinaldo, C.H. Impact of a polyomavirus (BKV) infection on mRNA expression in human endothelial cells. Virus Res. 2007, 123, 86–94. [Google Scholar] [CrossRef]
- Malmgaard, L. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 2004, 24, 439–454. [Google Scholar] [CrossRef]
- Hotter, D.; Kirchhoff, F. Interferons and beyond: Induction of antiretroviral restriction factors. J. Leukoc Biol. 2018, 103, 465–477. [Google Scholar] [CrossRef]
- Chen, I.Y.; Ichinohe, T. Response of host inflammasomes to viral infection. Trends Microbiol 2015, 23, 55–63. [Google Scholar] [CrossRef]
- Lupfer, C.; Malik, A.; Kanneganti, T.D. Inflammasome control of viral infection. Curr. Opin. Virol. 2015, 12, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, A.; Wornle, M.; Motamedi, N.; Anders, H.J.; Grone, E.F.; Nitschko, H.; Kurktschiev, P.; Debiec, H.; Kretzler, M.; Cohen, C.D.; et al. Activation of innate immune defense mechanisms contributes to polyomavirus BK-associated nephropathy. Kidney Int. 2012, 81, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, A.; Merkle, M.; Motamedi, N.; Nitschko, H.; Koppel, S.; Wornle, M. BK virus infection activates the TNFalpha/TNF receptor system in Polyomavirus-associated nephropathy. Mol. Cell Biochem 2016, 411, 191–199. [Google Scholar] [CrossRef]
- Darbinyan, A.; Kaminski, R.; White, M.K.; Darbinian-Sarkissian, N.; Khalili, K. Polyomavirus JC infection inhibits differentiation of oligodendrocyte progenitor cells. J. Neurosci Res. 2013, 91, 116–127. [Google Scholar] [CrossRef]
- Richards, K.F.; Guastafierro, A.; Shuda, M.; Toptan, T.; Moore, P.S.; Chang, Y. Merkel cell polyomavirus T antigens promote cell proliferation and inflammatory cytokine gene expression. J. Gen. Virol. 2015, 96, 3532–3544. [Google Scholar] [CrossRef]
- Rasheed, K.; Abdulsalam, I.; Fismen, S.; Grimstad, O.; Sveinbjornsson, B.; Moens, U. CCL17/TARC and CCR4 expression in Merkel cell carcinoma. Oncotarget 2018, 9, 31432–31447. [Google Scholar] [CrossRef]
- Monnier, J.; Samson, M. Prokineticins in angiogenesis and cancer. Cancer Lett 2010, 296, 144–149. [Google Scholar] [CrossRef]
- Lauttia, S.; Sihto, H.; Kavola, H.; Koljonen, V.; Bohling, T.; Joensuu, H. Prokineticins and Merkel cell polyomavirus infection in Merkel cell carcinoma. Br. J. Cancer 2014, 110, 1446–1455. [Google Scholar] [CrossRef]
- Sever, R.; Glass, C.K. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol. 2013, 5, a016709. [Google Scholar] [CrossRef]
- Mazaira, G.I.; Zgajnar, N.R.; Lotufo, C.M.; Daneri-Becerra, C.; Sivils, J.C.; Soto, O.B.; Cox, M.B.; Galigniana, M.D. The Nuclear Receptor Field: A Historical Overview and Future Challenges. Nucl. Recept. Res. 2018, 5, 101320. [Google Scholar] [CrossRef]
- Sherman, M.H.; Downes, M.; Evans, R.M. Nuclear receptors as modulators of the tumor microenvironment. Cancer Prev Res. 2012, 5, 3–10. [Google Scholar] [CrossRef]
- Long, M.D.; Campbell, M.J. Pan-cancer analyses of the nuclear receptor superfamily. Nucl. Recept. Res. 2015, 2. [Google Scholar] [CrossRef]
- Dhiman, V.K.; Bolt, M.J.; White, K.P. Nuclear receptors in cancer––uncovering new and evolving roles through genomic analysis. Nat. Rev. Genet. 2018, 19, 160–174. [Google Scholar] [CrossRef]
- Moens, U.; Van Ghelue, M.; Johansen, B.; Seternes, O.M. Concerted expression of BK virus large T- and small t-antigens strongly enhances oestrogen receptor-mediated transcription. J. Gen. Virol. 1999, 80, 585–594. [Google Scholar] [CrossRef]
- Moens, U.; Subramaniam, N.; Johansen, B.; Johansen, T.; Traavik, T. A steroid hormone response unit in the late leader of the noncoding control region of the human polyomavirus BK confers enhanced host cell permissivity. J. Virol. 1994, 68, 2398–2408. [Google Scholar]
- Pitson, S.M. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem Sci 2011, 36, 97–107. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Bhat, V.K.; Bernhart, E.; Plastira, I.; Fan, K.; Ghaffari-Tabrizi-Wizsy, N.; Wadsack, C.; Rechberger, G.; Eichmann, T.; Asslaber, M.; Spassova, I.; et al. Pharmacological Inhibition of Serine Palmitoyl Transferase and Sphingosine Kinase-1/-2 Inhibits Merkel Cell Carcinoma Cell Proliferation. J. Invest. Derm. 2019, 139, 807–817. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479, 609–618. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Noch, E.; Sariyer, I.K.; Gordon, J.; Khalili, K. JC virus T-antigen regulates glucose metabolic pathways in brain tumor cells. PLoS ONE 2012, 7, e35054. [Google Scholar] [CrossRef]
- Kumar, S.H.; Rangarajan, A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J. Virol. 2009, 83, 8565–8574. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.A.; Grosse, Y.; Lauby-Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Straif, K. Carcinogenicity of malaria and of some polyomaviruses. Lancet. Oncol 2012, 13, 339–340. [Google Scholar] [CrossRef]
- Arora, R.; Chang, Y.; Moore, P.S. MCV and Merkel cell carcinoma: A molecular success story. Curr. Opin. Virol. 2012, 2, 489–498. [Google Scholar] [CrossRef]
- Baez, C.F.; Brandao Varella, R.; Villani, S.; Delbue, S. Human Polyomaviruses: The Battle of Large and Small Tumor Antigens. Virology 2017, 8, 1178122x17744785. [Google Scholar] [CrossRef]
| Virus | LT | sT | T’135 | T’136 | T’165 |
|---|---|---|---|---|---|
| BKPyV | p53, pRb | ABCA13, ANKRD30B, ATP2A2, BAG2, BAG3, BAG5, cathepsin BCCND3, CD44, CDK2, CDKN1A, CNP, CSRP1, DnaJC7, DP1, E2F3, E2F4, E2F5, GLIPR2, HSDL2, Hsp70, HSPA4L, HSPBP1, NAGK, Nse2/Mms21, PCNA, PP2CA, PP2R1α, PPM1B, RB1, RBL1, RBL2, SCCPHD, SEC61B, SMC5, SQRDL, SRP9, SRRM2, STUB1, TGFBI | |||
| JCPyV | AP1, BAG3, BRN1, β-catenin, CEBP, Hsp70, IRS-1, LEF1, NF2, Oct6, pRb, p53, Purα, SKP1, YB-1 | PP2Cα | Hsp70, pRb | pRb | pRb |
| WUPyV | p53, RB1 | ||||
| MCPyV | ABCA13, ABCD3, AP2A1, ATM, BAG2, BAG3, BAG5, Brd4, CREBBP, CK2β, DDX24, DnaJC7, DP1, E2F3, E2F4, EMD, FAM71E2, GTF3C1, HDLBP, Hsp70, IκBIP, KPNA2, KPNA3, KPNA4, MAP4, MED14, P4HA3, PGAM5, PIP4K2 β, PP2AR1α, PTRF, RB1, RTN4, SALL2, SDPR, SGPL1, SRP14, SRPRB, STUB1, TCEB1, TRIM38, TSPYL1, Vam6p, VAPA, VAPB, VPS11, USMG5 | ABHD12, ACBD3, ADAM9, AIP, ANKRD13A, ATP2A2, BAG2, BAG3, BAG5, cadherin 1, CCHC, CD44, CDC20, CDH, CNP, COPG2, DnaJA1, DnaJB4, DnaJC7, EFEMP2, eIF4EBP1, emerin, Fbxw7, Hsp70, IGF2R, IκBIP, LOX, MBOAT7, MMP14, MPZL1, MTCH2, myoferlin, NEMO, Notch2, NSD1, P4HB, PDGFRβ, PGRMC2, PRAF2, PPP2CA, PPP2CB, PPP2R1A, PP2R1B, PP4R1, PPM1A, PPM1B, PPM1G, PSMC2, PSMC3, PSMC4, PTTPG1IP, Rab18, RNH1, RPL21, RPs27L, SPARC, SQRDL, SRPRB, STUB1, SURF4, TIMM8A, TMEM165, TMX3, TOLLIP, USMG5, VKORC1, YAP1 | |||
| HPyV6 | p53, RB1 | PP2Cα, PP2R1α | |||
| HPyV7 | p53, RB1 | ||||
| TSPyV | p53, RB1 | PP2Cα | |||
| MWPyV | pRb | PP2R1α |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moens, U.; Macdonald, A. Effect of the Large and Small T-Antigens of Human Polyomaviruses on Signaling Pathways. Int. J. Mol. Sci. 2019, 20, 3914. https://doi.org/10.3390/ijms20163914
Moens U, Macdonald A. Effect of the Large and Small T-Antigens of Human Polyomaviruses on Signaling Pathways. International Journal of Molecular Sciences. 2019; 20(16):3914. https://doi.org/10.3390/ijms20163914
Chicago/Turabian StyleMoens, Ugo, and Andrew Macdonald. 2019. "Effect of the Large and Small T-Antigens of Human Polyomaviruses on Signaling Pathways" International Journal of Molecular Sciences 20, no. 16: 3914. https://doi.org/10.3390/ijms20163914