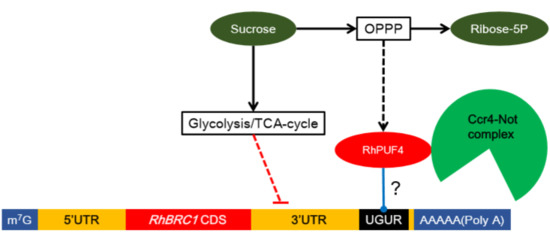
Posttranscriptional Regulation of RhBRC1 (Rosa hybrida BRANCHED1) in Response to Sugars is Mediated via its Own 3′ Untranslated Region, with a Potential Role of RhPUF4 (Pumilio RNA-Binding Protein Family)
Abstract
1. Introduction
2. Results
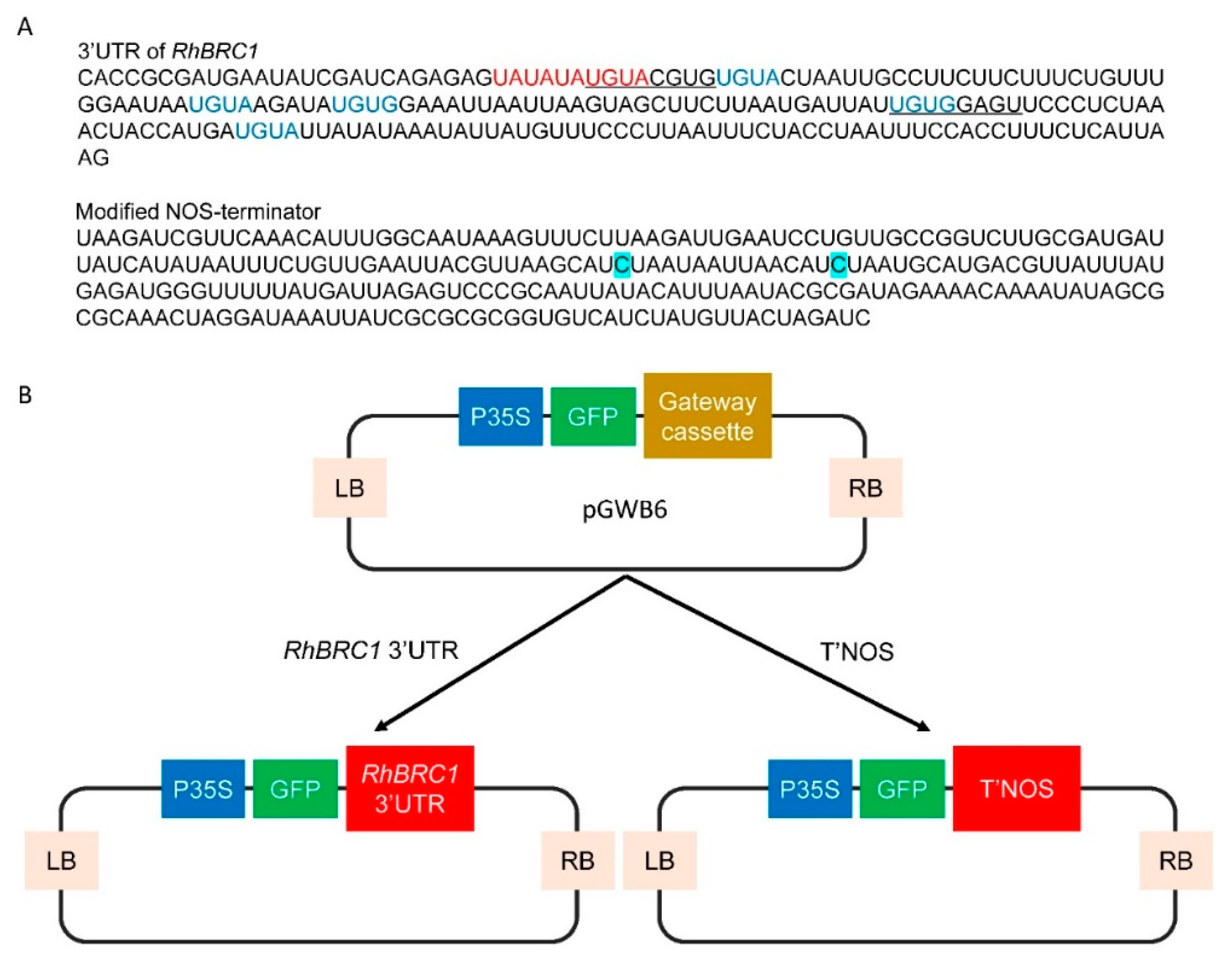
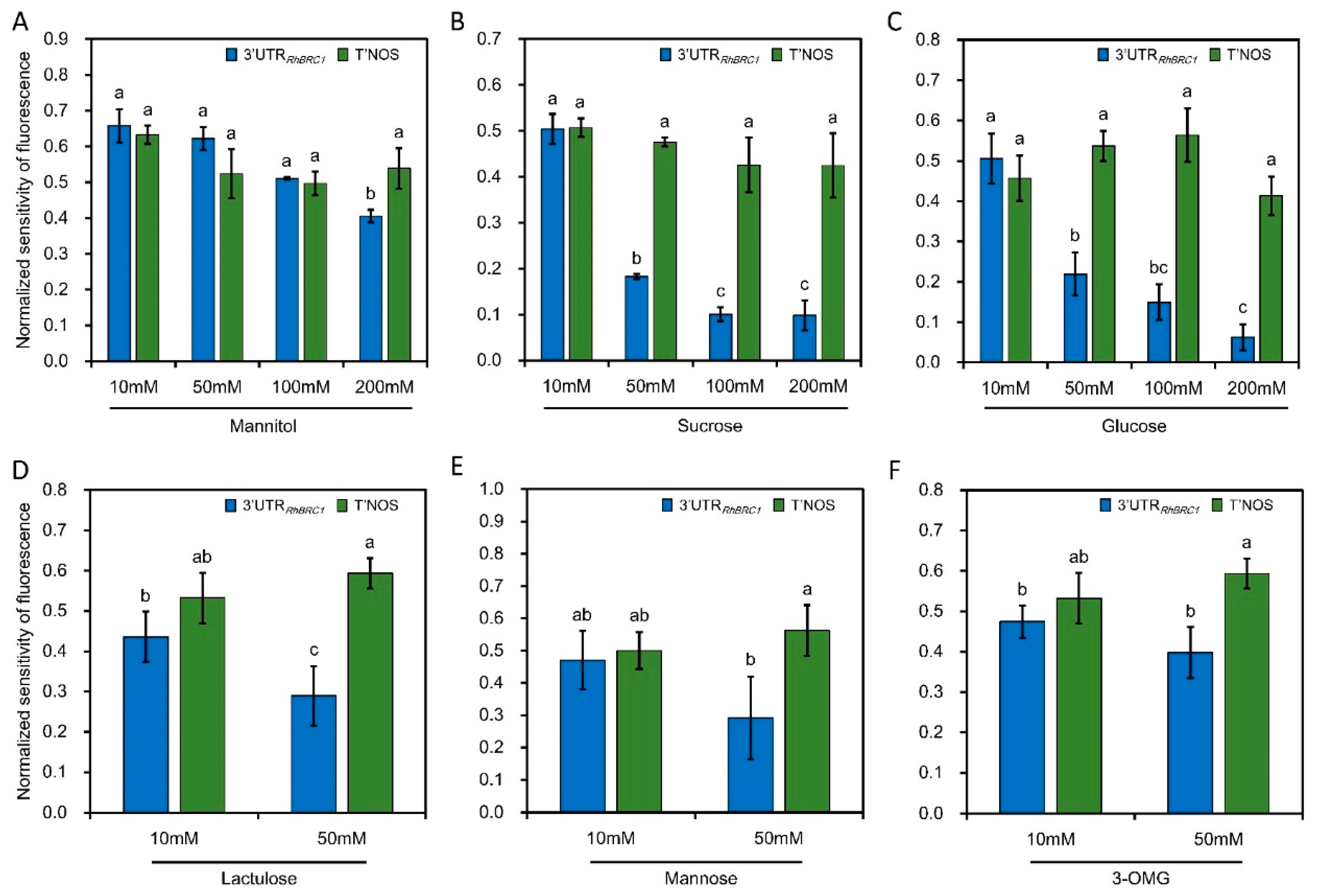
2.1. Sucrose and Glucose Influence the Expression of RhBRC1 through Its 3′UTR
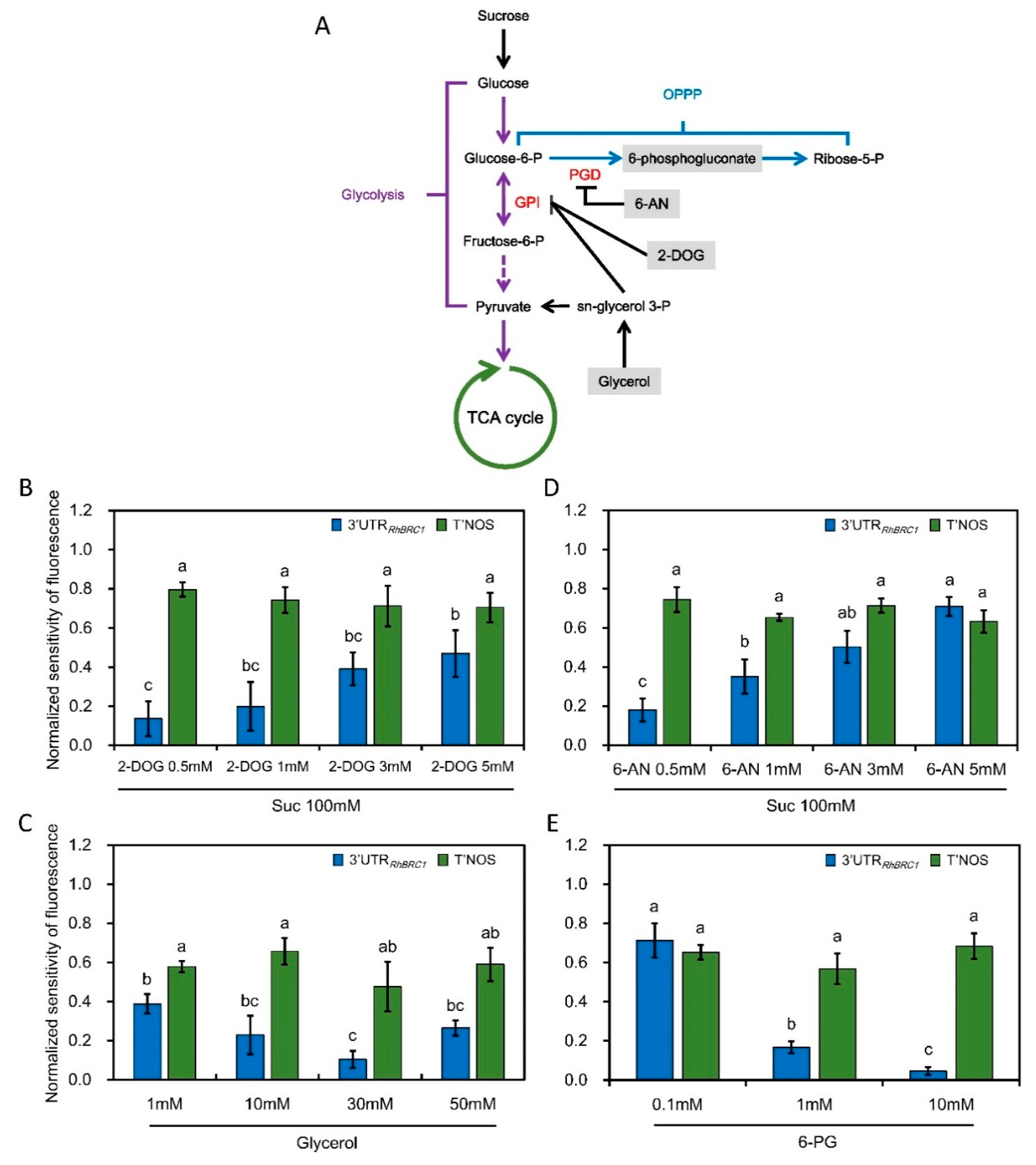
2.2. The 3′UTR of RhBRC1 Responds to Glycolysis/TCA-Cycle and OPPP Signaling
2.3. Identification of PUF Family Members in Rosa Chinensis
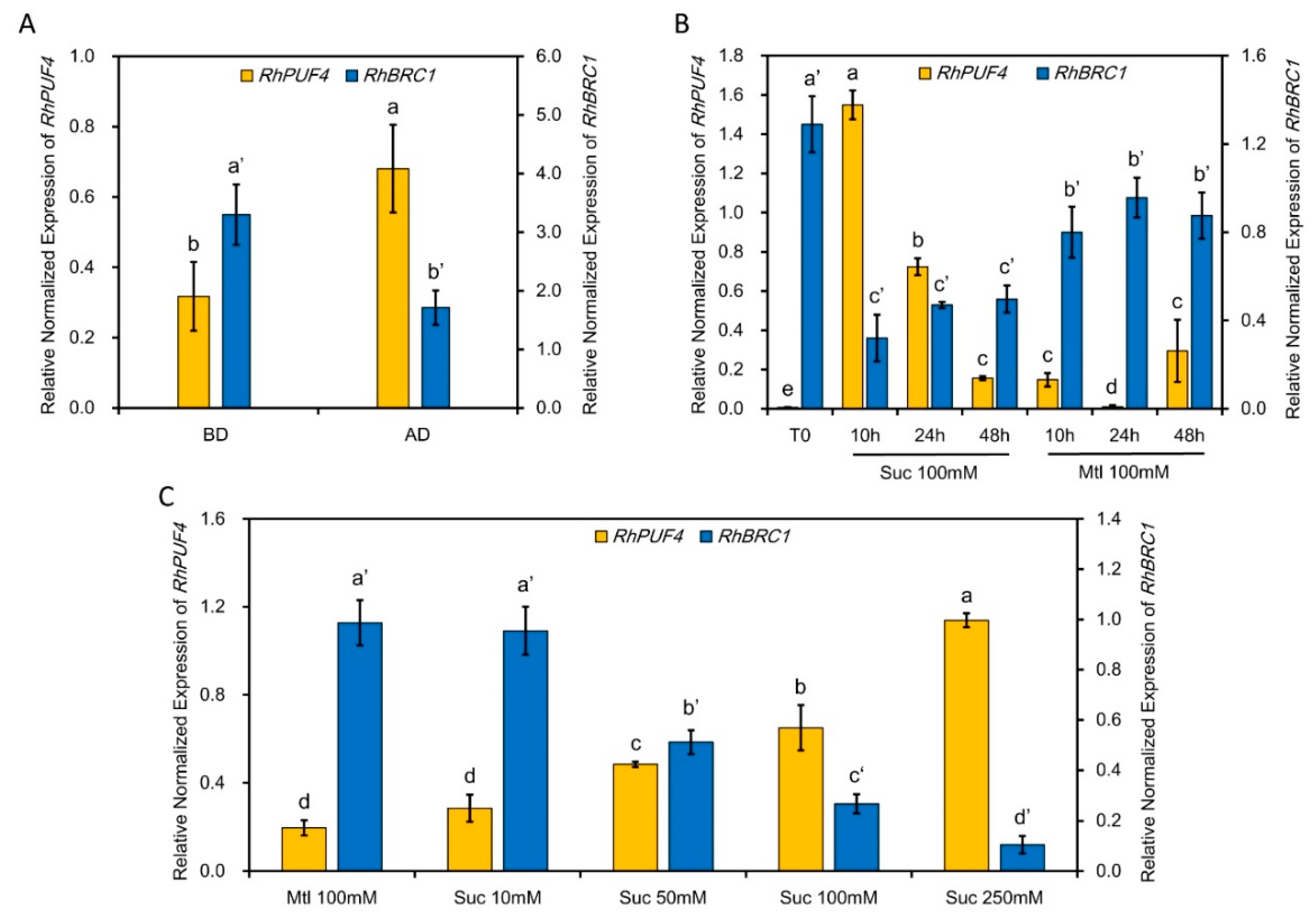
2.4. The Transcription Level of RhPUF4 Is Regulated by Sugar
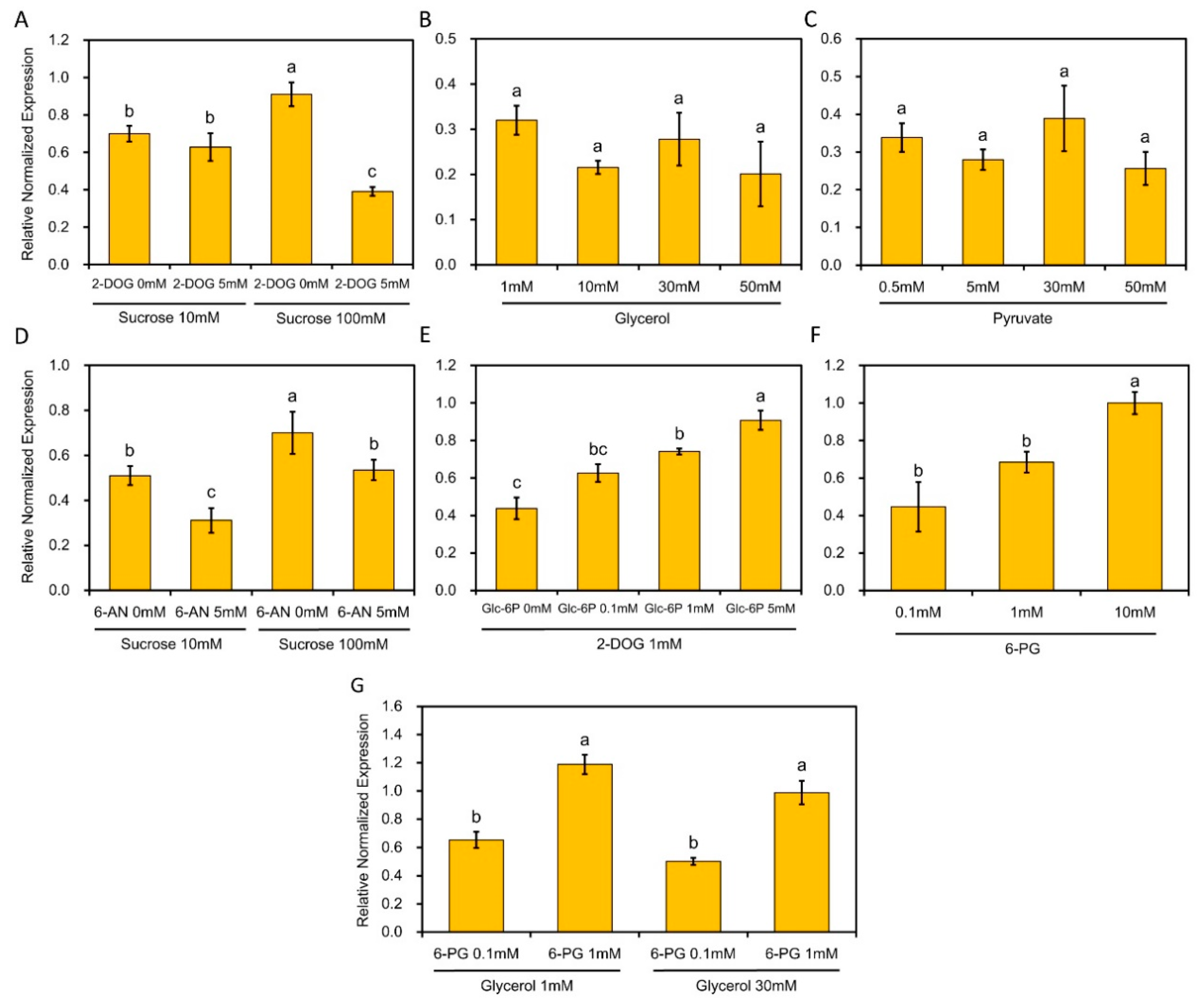
2.5. The Transcript Level of RhPUF4 Is More Likely to Be Sensitive to OPPP Signaling
2.6. RhPUF4 Could Bind to the 3′UTR of RhBRC1 and Promote Plant Growth
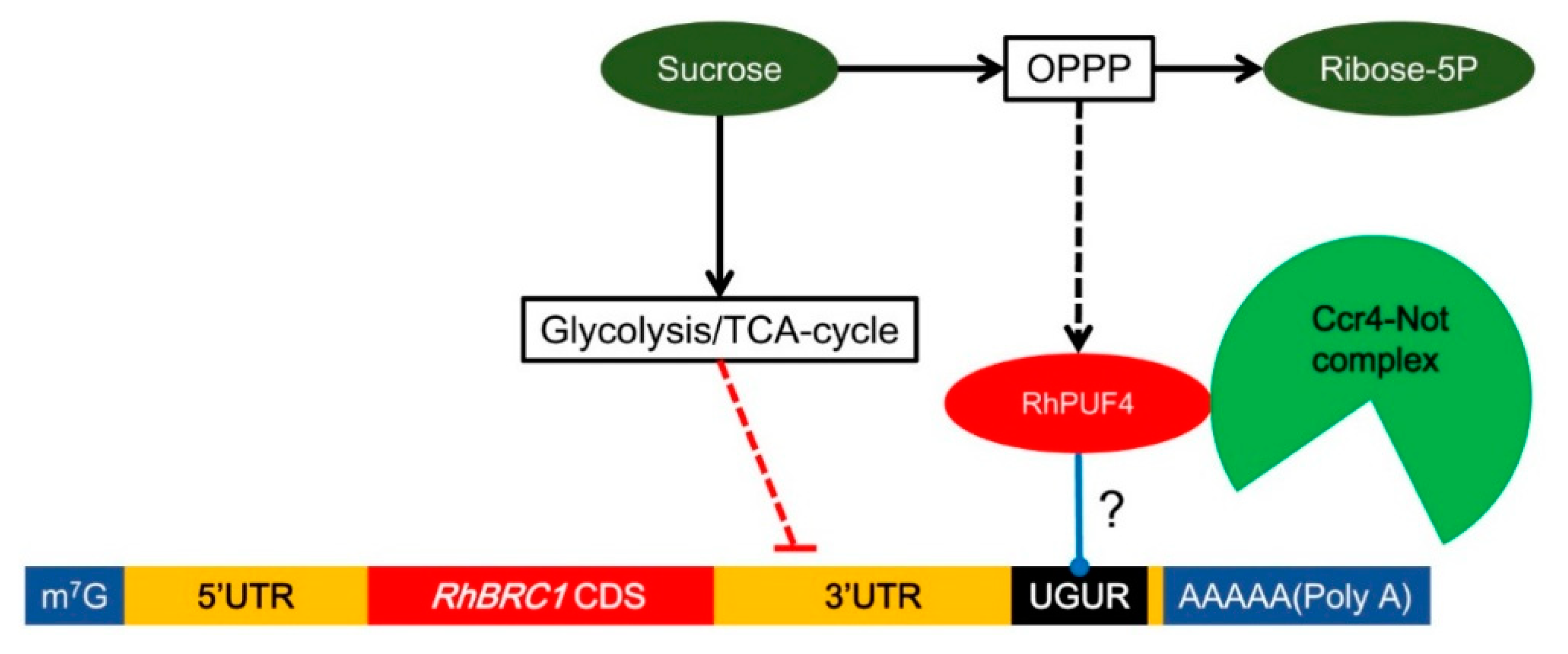
3. Discussion
3.1. Involvement of the 3′UTR Region in Sugar-Mediated Downregulation of RhBRC1
3.2. Posttranscriptional Regulation of RhBRC1 by Sucrose Is Mainly Mediated through the OPPP
3.3. Involvement of RhPUF4 in Posttranscription of RhBRC1 Mediated by the OPPP
4. Materials and Methods
4.1. Cloning and Transformation
4.2. Rosa Callus and Arabidopsis Transformation
4.3. Callus Treatment and GFP Quantification
4.4. Plant Culture and In-Vitro Cultivation of Axillary Buds
4.5. Rosa RNA Extraction
4.6. qRT-PCR
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiang, H.; Egli, D.B. Shade Induced Changes in Flower and Pod Number and Flower and Fruit Abscission in Soybean. Agrono. J. 1993, 85, 221–225. [Google Scholar] [CrossRef]
- Richards, R.A. Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 2000, 51, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant. Sci. 2015, 5, 741. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiao, Y. Axillary meristem initiation-a way to branch out. Curr. Opin. Plant. Biol. 2018, 41, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Le Moigne, M.A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.D.; Ogé, L.; Demotes-Mainard, S.; Hamama, L.; Davière, J.M.; Sakr, S. BRANCHED1:A key hub of shoot branching. Front. Plant Sci. 2019, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant. Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Seale, M.; Bennett, T.; Leyser, O. BRC1 expression regulates bud activation potential, but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 2017, 144, 1661–1673. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; UeguchisTanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant. J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant. Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant. Cell 1997, 9, 1607–1619. [Google Scholar]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. The Plant. J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant. J. 2002, 30, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genet. 1999, 162, 1927–1935. [Google Scholar]
- Wang, R.L.; Stec, A.; Hey, J.; Lukens, L.; Doebley, J. The limits of selection during maize domestication. Nature 1999, 398, 236–239. [Google Scholar] [CrossRef]
- Maurel, K.; Leite, G.B.; Bonhomme, M.; Guilliot, A.; Rageau, R.; Pétel, G.; Sakr, S. Trophic control of bud break in peach (Prunus persica) trees: A possible role of hexoses. Tree Physiol. 2004, 24, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, M.; Peuch, M.; Ameglio, T.; Rageau, R.; Guilliot, A.; Decourteix, M.; Alves, G.; Sakr, S.; Lacointe, A. Carbohydrate uptake from xylem vessels and its distribution among stem tissues and buds in walnut (Juglans regia L.). Tree Physiol. 2009, 30, 89–102. [Google Scholar] [CrossRef]
- Girault, T.; Abidi, F.; Sigogne, M.; Pelleschi-Travier, S.; Boumaza, R.; Sakr, S.; Leduc, N. Sugars are under light control during bud burst in Rosa sp. Plant Cell Environ. 2010, 33, 1339–1350. [Google Scholar] [CrossRef]
- Henry, C.; Rabot, A.; Laloi, M.; Mortreau, E.; Sigogne, M.; Leduc, N.; Lemoine, R.; Sakr, S.; Vian, A.; Pelleschi-Travier, S. Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant. Cell Environ. 2011, 34, 1776–1789. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Brutnell, T.P.; Finlayson, S.A. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant. Cell Environ. 2010, 33, 48–58. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Brutnell, T.P.; Hays, D.B.; Finlayson, S.A. Vegetative axillary bud dormancy induced by shade and defoliation signals in the grasses. Plant. Signal. Behav. 2010, 5, 317–319. [Google Scholar] [CrossRef][Green Version]
- Kebrom, T.; Chandler, P.; Swain, S.; King, R.; Richards, R.; Spielmeyer, W. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant. Physiol. 2012, 160, 308–318. [Google Scholar] [CrossRef]
- Rabot, A.; Henry, C.; Ben Baaziz, K.; Mortreau, E.; Azri, W.; Lothier, J.; Hamama, L.; Boummaza, R.; Leduc, N.; Pelleschi-Travier, S.; et al. Insight into the role of sugars in bud burst under light in the rose. Plant. Cell Physiol. 2012, 53, 1068–1082. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.G.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant. J. 2017, 92, 611–623. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Evers, J.B. Sugar as a key component of the shoot branching regulation network. Plant. Cell Environ 2015, 38, 1455–1456. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B.; et al. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Mullet, J.E. Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant. Cell Environ. 2015, 38, 1471–1478. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Tanaka, M.; Mori, H. Auxin–cytokinin interactions in the control of shoot branching. Plant. Mol. Biol. 2009, 69, 429. [Google Scholar] [CrossRef]
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant. Physiol. 2009, 150, 482–493. [Google Scholar] [CrossRef]
- Dun, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012, 158, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Lejay, L.; Wirth, J.; Pervent, M.; Cross, J.M.F.; Tillard, P.; Gojon, A. Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant. Physiol. 2008, 146, 2036–2053. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dédaldéchamp, F.; Perez-Garcia, M.D.; Ogé, L.; Hamama, L.; Atanassova, R. The Sugar-Signaling Hub: Overview of Regulators and Interaction with the Hormonal and Metabolic Network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef] [PubMed]
- Sheu, J.J.; Jan, S.P.; Lee, H.T.; Yu, S.M. Control of transcription and mRNA turnover as mechanisms of metabolic repression of α-amylase gene expression. Plant. J. 1994, 5, 655–664. [Google Scholar] [CrossRef]
- Chan, M.T.; Yu, S.M. The 3′ untranslated region of a rice α-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl. Acad. Sci. USA 1998, 95, 6543–6547. [Google Scholar] [CrossRef]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc. Natl. Acad. Sci. USA 1999, 96, 10512–10517. [Google Scholar] [CrossRef]
- Nicolai, M.; Roncato, M.A.; Canoy, A.S.; Rouquie, D.; Sarda, X.; Freyssinet, G.; Robaglia, C. Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant. Physiol. 2006, 141, 663–673. [Google Scholar] [CrossRef]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533. [Google Scholar] [CrossRef]
- Wang, M.; Ogé, L.; Perez-Garcia, M.D.; Hamama, L.; Sakr, S. The PUF Protein Family: Overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 2018, 19, 410. [Google Scholar] [CrossRef]
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Tam, P.P.; Barrette-Ng, I.H.; Simon, D.M.; Tam, M.W.; Ang, A.L.; Muench, D.G. The PUF family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010, 10, 44. [Google Scholar] [CrossRef]
- Friend, K.; Campbell, Z.T.; Cooke, A.; Kroll-Conner, P.; Wickens, M.P.; Kimble, J. A conserved PUF–Ago–eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2014, 19, 176–183. [Google Scholar] [CrossRef]
- Van Etten, J.; Schagat, T.L.; Hrit, J.; Weidmann, C.A.; Brumbaugh, J.; Coon, J.J.; Goldstrohm, A.C. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012, 287, 36370–36383. [Google Scholar] [CrossRef]
- Miles, W.O.; Tschöp, K.; Herr, A.; Ji, J.Y.; Dyson, N.J. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012, 26, 356–368. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Mendell, J.T.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Francischini, C.W.; Quaggio, R.B. Molecular characterization of Arabidopsis thaliana PUF proteins–binding specificity and target candidates. FEBS J. 2009, 276, 5456–5470. [Google Scholar] [CrossRef]
- Zhang, C.; Muench, D.G. A nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 2015, 290, 30108–30118. [Google Scholar] [CrossRef]
- Chan, M.T.; Yu, S.M. The 3′ untranslated region of a rice α-amylase gene mediates sugar-dependent abundance of mRNA. Plant. J. 1998, 15, 685–695. [Google Scholar] [CrossRef]
- Wick, A.N.; Drury, D.R.; Nakada, H.I.; Wolfe, J.B. Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 1957, 224, 963–969. [Google Scholar]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181. [Google Scholar] [CrossRef]
- Lange, K.; Proft, E.R. Inhibition of the 6-phosphogluconate dehydrogenase in the rat kidney by 6-aminonicotinamide. Naunyn Schmiedeberg Arch. Pharmacol. 1970, 267, 177–180. [Google Scholar] [CrossRef]
- Hothersall, J.S.; Gordge, M.; Noronha-Dutra, A.A. Inhibition of NADPH supply by 6-aminonicotinamide: Effect on glutathione, nitric oxide and superoxide in J774 cells. FEBS Lett. 1998, 434, 97–100. [Google Scholar] [CrossRef]
- Aubert, S.; Gout, E.; Bligny, R.; Douce, R. Multiple effects of glycerol on plant cell metabolism. Phosphorus-31 nuclear magnetic resonance studies. J. Biol. Chem. 1994, 269, 21420–21427. [Google Scholar]
- Jung, S.; Staton, M.; Lee, T.; Blenda, A.; Svancara, R.; Abbott, A.; Main, D. GDR (Genome Database for Rosaceae): Integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res. 2007, 36, D1034–D1040. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic acids Res. 2018, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Hibrand Saint-Oyant, L.; Ruttink, T.; Hamama, L.; Kirov, I.; Lakhwani, D.; Zhou, N.N.; Bourke, P.M.; Daccord, N.; Leus, L.; Schulz, D.; et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 2018, 4, 473–484. [Google Scholar] [CrossRef]
- Abbasi, N.; Park, Y.I.; Choi, S.B. Pumilio Puf domain RNA-binding proteins in Arabidopsis. Plant. Signal. Behav. 2011, 6, 364–368. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42(W1), W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xia, K.; Liang, Z.; Chen, K.; Gao, C.; Zhang, M. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 2016, 6, 32158. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Daimon, Y.; Kurotani, K.I.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant. Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G.; et al. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, X.; Liu, P.; Li, D.; Chen, T.; Gu, X.; Sun, J. MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PLoS Genet. 2017, 13, e1006833. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, Y.Q. Insights into the mechanism of plant development: Interactions of miRNAs pathway with phytohormone response. Biochem. Biophys. Res. Commun. 2009, 384, 1–5. [Google Scholar] [CrossRef]
- Zhou, M.; Luo, H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant. Signal. Behav. 2014, 9, 1375–1391. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant. J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant. Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Liang, Y.; Seale, M.; Ward, S.; Müller, D.; Leyser, O. Strigolactone regulates shoot development through a core signalling pathway. Biol. Open 2016, 5, 1806–1820. [Google Scholar] [CrossRef] [PubMed]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.I. Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 2005, 8, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009, 21, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102. [Google Scholar] [CrossRef]
- Corradetti, M.N.; Guan, K.L. Upstream of the mammalian target of rapamycin: Do all roads pass through mTOR? Oncogene 2006, 25, 6347. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef]
- Van Leene, J.; Han, C.; Gadeyne, A.; Eeckhout, D.; Matthijs, C.; Cannoot, B.; De Winne, N.; Persiau, G.; Van De Slijke, E.; Van de Cotte, B.; et al. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 2019, 5, 316. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Guo, W.; Wang, Q.; Chen, Z.; Huang, S.; Zhao, F.; Yao, M.; Zhao, Y.; He, X. MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology 2015, 149, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, C.; Grieco, D.; Costanzo, V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011, 30, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012, 69, 3613–3634. [Google Scholar] [CrossRef] [PubMed]
- Valley, C.T.; Porter, D.F.; Qiu, C.; Campbell, Z.T.; Hall, T.M.T.; Wickens, M. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc. Natl. Acad. Sci. USA 2012, 109, 6054–6059. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, L.J.; Gay, A.C.; Pon, L.A. PUF3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 2007, 176, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Maj, T.; Kryczek, I.; Li, W.; Wu, K.; Zhao, L.; Wei, S.; Crespo, J.; Wan, S.; Vatan, L.; et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2106, 17, 95. [Google Scholar] [CrossRef]
- Tang, H.; Lee, M.; Sharpe, O.; Salamone, L.; Noonan, E.J.; Hoang, C.D.; Levine, S.; Robinson, W.H.; Shrager, J.B. Oxidative stress-responsive microRNA-320 regulates glycolysis in diverse biological systems. FASEB J. 2012, 26, 4710–4721. [Google Scholar] [CrossRef]
- Miller, J.E.; Reese, J.C. Ccr4-Not complex: The control freak of eukaryotic cells. Critical reviews Biochem. Mol. Biol. 2012, 47, 315–333. [Google Scholar] [CrossRef]
- Collart, M.A.; Panasenko, O.O. The Ccr4–not complex. Gene 2012, 492, 42–53. [Google Scholar] [CrossRef]
- Arae, T.; Morita, K.; Imahori, R.; Suzuki, Y.; Yasuda, S.; Sato, T.; Yamaguchi, J.; Chiba, Y. Identification of Arabidopsis CCR4-NOT Complexes with Pumilio RNA-Binding Proteins, APUM5 and APUM2. Plant. Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Toyooka, K.; Matsuoka, K.; Jinbo, T.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Hamama, L.; Voisine, L.; Pierre, S.; Cesbron, D.; Ogé, L.; Lecerf, M.; Cailleux, S.; Bosselut, J.; Foucrier, S.; Foucher, F.; et al. Improvement of in vitro donor plant competence to increase de novo shoot organogenesis in rose genotypes. Sci. Hortic. 2019, 252, 85–95. [Google Scholar] [CrossRef]
- Mohan, M.M.; Ibrahim, S.M. Callus induction from leaf bit explants of rose. Res. Crops 2000, 1, 71–73. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef]
- Chua, S.L.; Too, W.C.S.; Khoo, B.Y.; Few, L.L. UBC and YWHAZ as suitable reference genes for accurate normalisation of gene expression using MCF7, HCT116 and HepG2 cell lines. Cytotechnology 2011, 63, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]


| Gene Name | Sequence | |
|---|---|---|
| PrRhPUF1. | Forward | 5′ GAGGAACATGAGTGGAGGTCT 3′ |
| Reverse | 5′ CATTTGAAGGCTAAGGGTCAG 3′ | |
| PrRhPUF2 | Forward | 5′ TGCCCTACCAGAACGGTTTA 3′ |
| Reverse | 5′ CAGCAAGAGCCTGACAACACT 3′ | |
| PrRhPUF3 | Forward | 5′ ATGGCTTAGGTGGGTTTGGT 3′ |
| Reverse | 5′ ACTGACAATGCCGTCTGGAA 3′ | |
| PrRhPUF4 | Forward | 5′ CTTGAAACAGCCACTACGGA 3′ |
| Reverse | 5′ GGTCATCACAAGTCTCCAACAC 3′ | |
| PrRhPUF5 | Forward | 5′ TCAGGTCCTCTTCTTGTCCG 3′ |
| Reverse | 5′ TCCCTTTCAGTGCCTTATTCC 3′ | |
| PrRhPUF6 | Forward | 5′ ATGCAGCACATGCTCTGG 3′ |
| Reverse | 5′ TAAGTTGGTTCGTCAATTCGT 3′ | |
| PrRhPUF7 | Forward | 5′ AGCGTCCAATCATGCCACTAG 3′ |
| Reverse | 5′ ATACTGGTCCTGAGCAAGAGCA 3′ | |
| PrRhPUF8 | Forward | 5′ TAGTGGCAGTTCAGGCAATC 3′ |
| Reverse | 5′ TCCATCCGTCCCTGTTAGTC 3′ | |
| PrRhPUF9 | Forward | 5′ TCTTGCACTAAGATGCCAATG 3′ |
| Reverse | 5′ CAGCTTATCTCGATGTCTCCC 3′ | |
| PrRhPUF10 | Forward | 5′ ATACAAAGCCATTGCCTCAG 3′ |
| Reverse | 5′ CTTGCAGATCAATCGGTCTC 3′ | |
| PrRhPUF11 | Forward | 5′ TGCACAATATGGTGCGAGTG 3′ |
| Reverse | 5′ CCTCTTTGAAAACAGACGCCT 3′ | |
| PrRhPUF12 | Forward | 5′ GCAGCGATAACCAGTTAGGC 3′ |
| Reverse | 5′ TCTCTCAGCTCCAAACATATGC 3′ | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Ogé, L.; Voisine, L.; Perez-Garcia, M.-D.; Jeauffre, J.; Hibrand Saint-Oyant, L.; Grappin, P.; Hamama, L.; Sakr, S. Posttranscriptional Regulation of RhBRC1 (Rosa hybrida BRANCHED1) in Response to Sugars is Mediated via its Own 3′ Untranslated Region, with a Potential Role of RhPUF4 (Pumilio RNA-Binding Protein Family). Int. J. Mol. Sci. 2019, 20, 3808. https://doi.org/10.3390/ijms20153808
Wang M, Ogé L, Voisine L, Perez-Garcia M-D, Jeauffre J, Hibrand Saint-Oyant L, Grappin P, Hamama L, Sakr S. Posttranscriptional Regulation of RhBRC1 (Rosa hybrida BRANCHED1) in Response to Sugars is Mediated via its Own 3′ Untranslated Region, with a Potential Role of RhPUF4 (Pumilio RNA-Binding Protein Family). International Journal of Molecular Sciences. 2019; 20(15):3808. https://doi.org/10.3390/ijms20153808
Chicago/Turabian StyleWang, Ming, Laurent Ogé, Linda Voisine, Maria-Dolores Perez-Garcia, Julien Jeauffre, Laurence Hibrand Saint-Oyant, Philippe Grappin, Latifa Hamama, and Soulaiman Sakr. 2019. "Posttranscriptional Regulation of RhBRC1 (Rosa hybrida BRANCHED1) in Response to Sugars is Mediated via its Own 3′ Untranslated Region, with a Potential Role of RhPUF4 (Pumilio RNA-Binding Protein Family)" International Journal of Molecular Sciences 20, no. 15: 3808. https://doi.org/10.3390/ijms20153808
APA StyleWang, M., Ogé, L., Voisine, L., Perez-Garcia, M.-D., Jeauffre, J., Hibrand Saint-Oyant, L., Grappin, P., Hamama, L., & Sakr, S. (2019). Posttranscriptional Regulation of RhBRC1 (Rosa hybrida BRANCHED1) in Response to Sugars is Mediated via its Own 3′ Untranslated Region, with a Potential Role of RhPUF4 (Pumilio RNA-Binding Protein Family). International Journal of Molecular Sciences, 20(15), 3808. https://doi.org/10.3390/ijms20153808