A Cerato-Platanin Family Protein FocCP1 Is Essential for the Penetration and Virulence of Fusarium oxysporum f. sp. cubense Tropical Race 4
Abstract
1. Introduction
2. Results
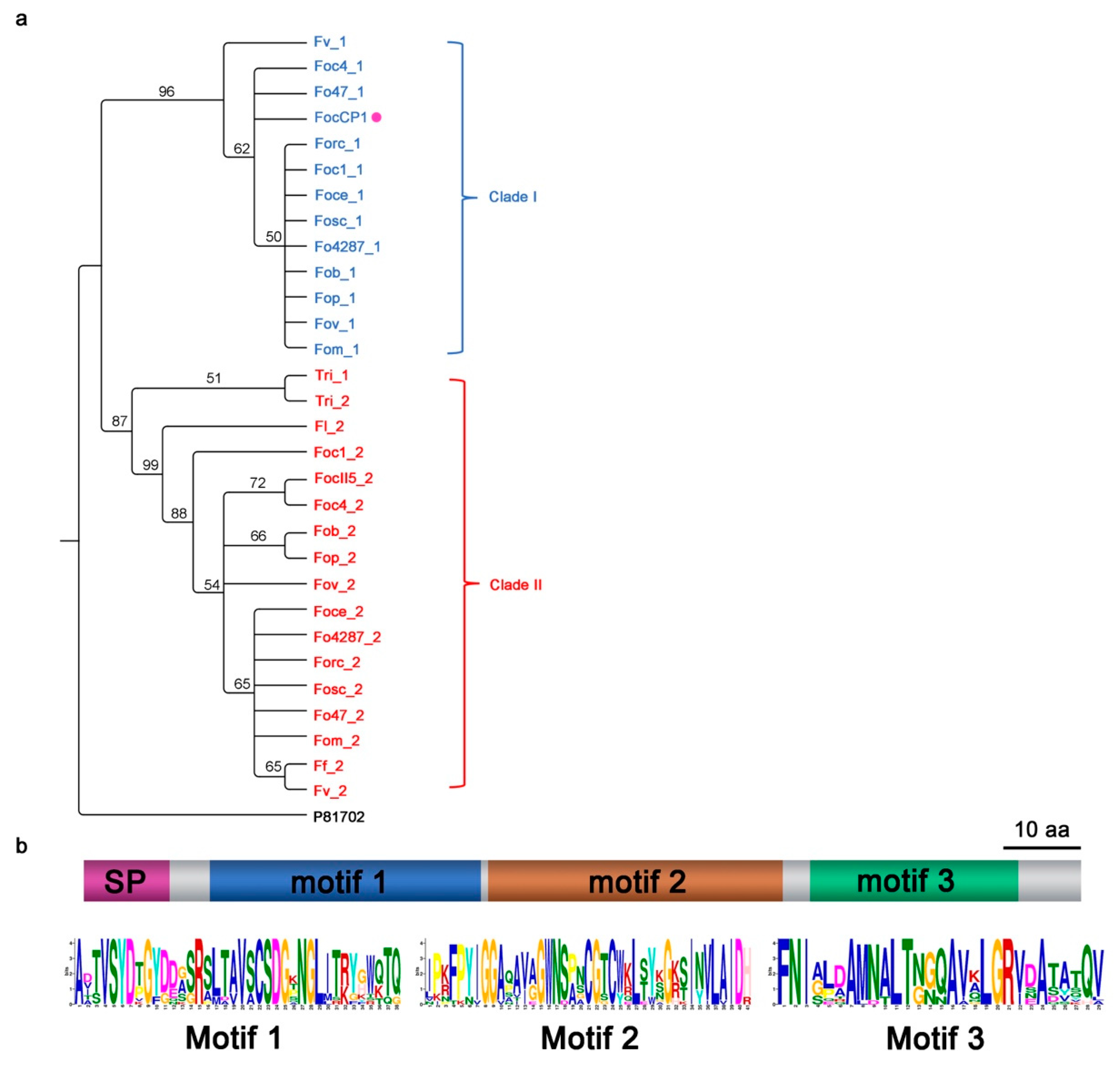
2.1. Identification and Characterization of a CP1 Homologue in Foc TR4
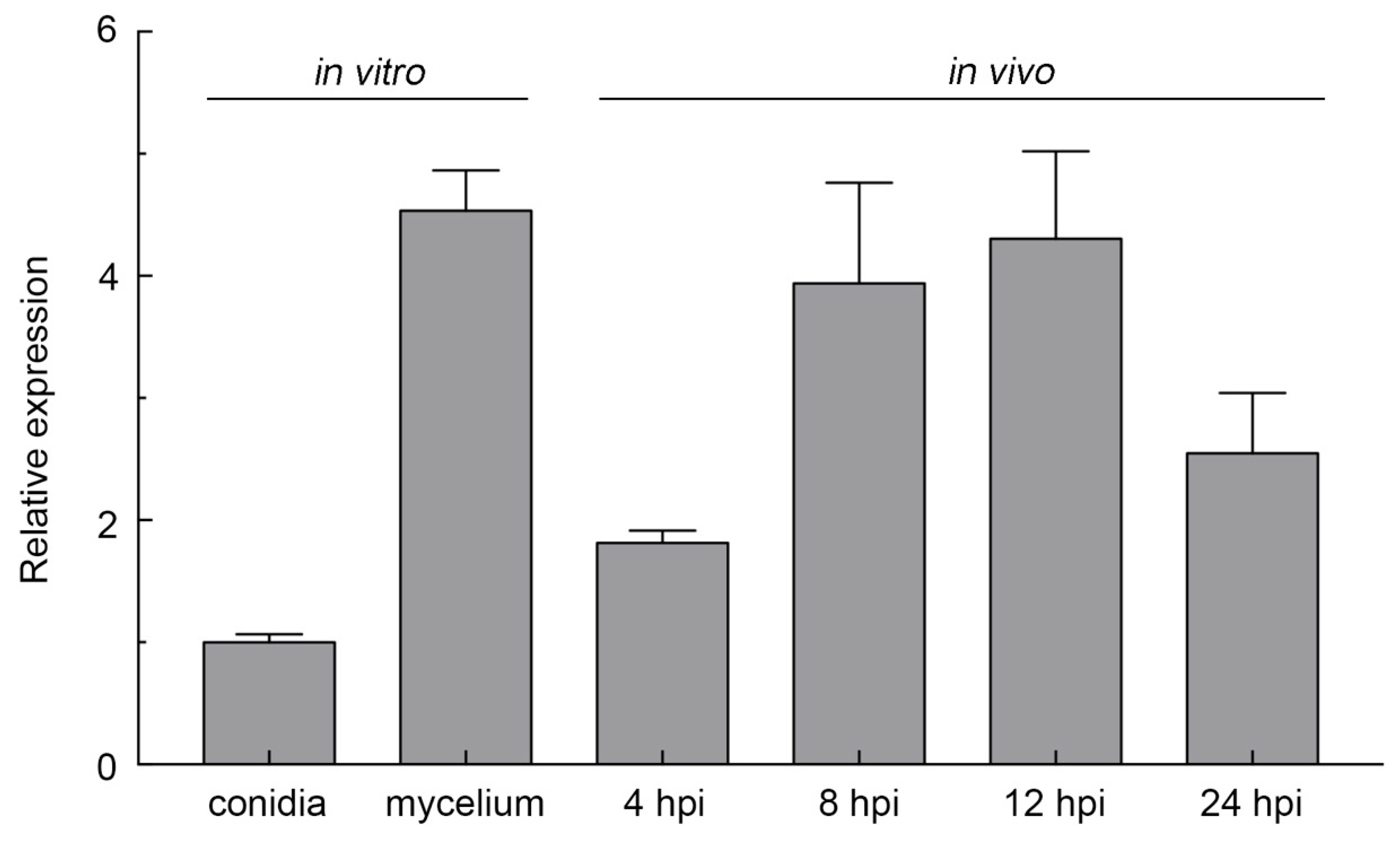
2.2. FocCP1 Is Highly Expressed during Spore Germination and Infection Progress
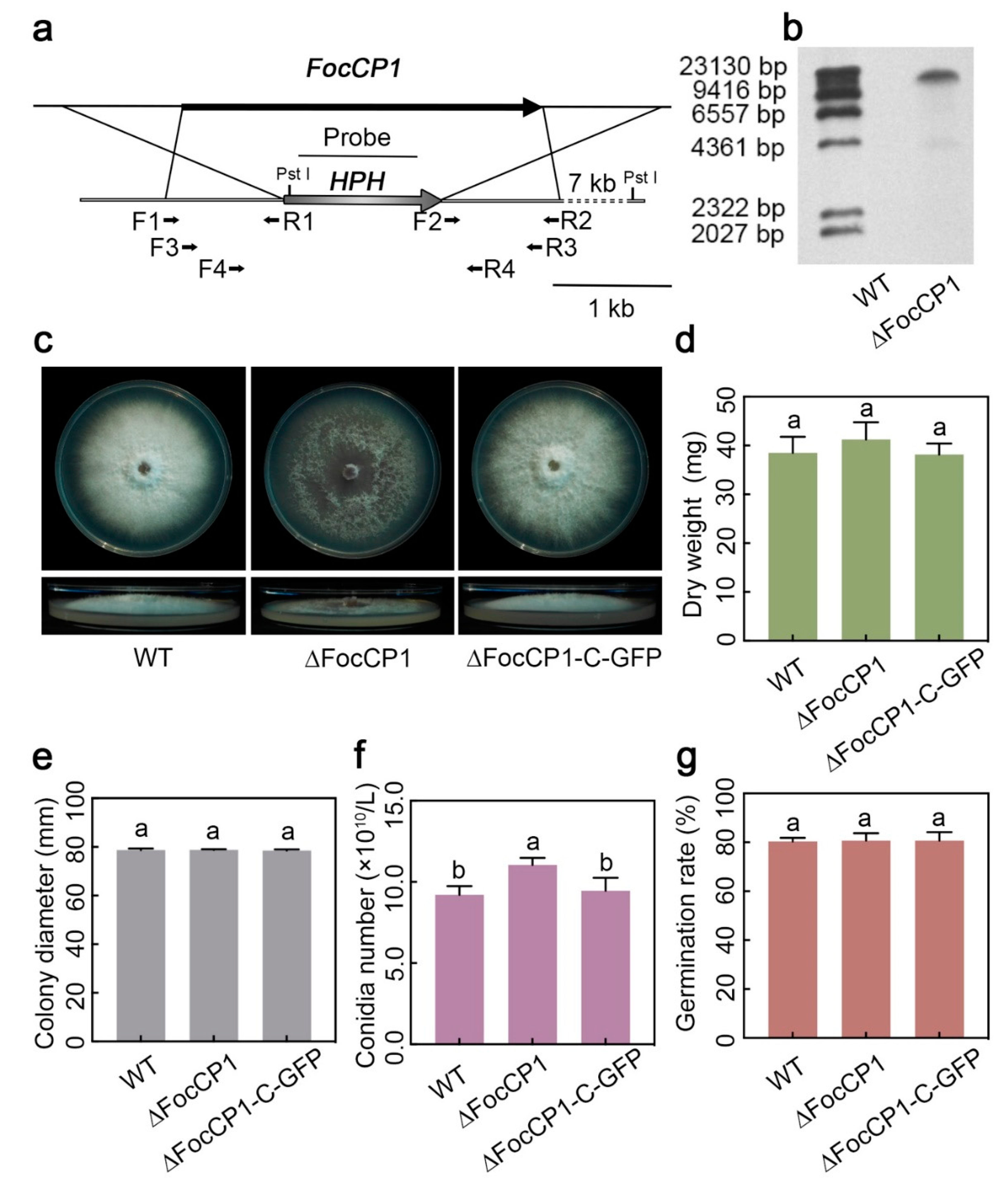
2.3. Generation of FocCP1-Mutated and FocCP1-Complemented Strains
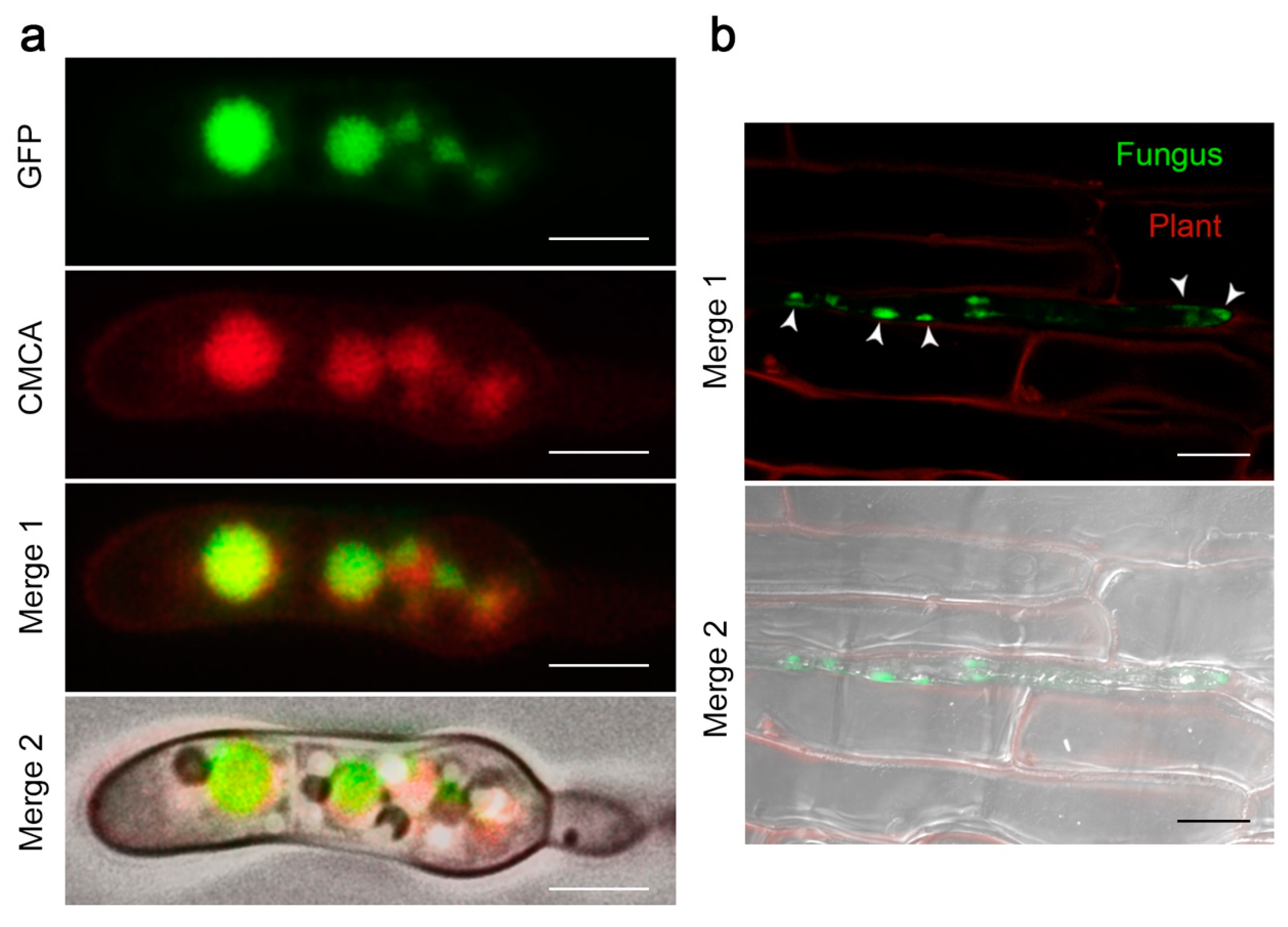
2.4. The FocCP1 Protein is Carried by Vacuoles and Predominantly Functions in the Apoplast
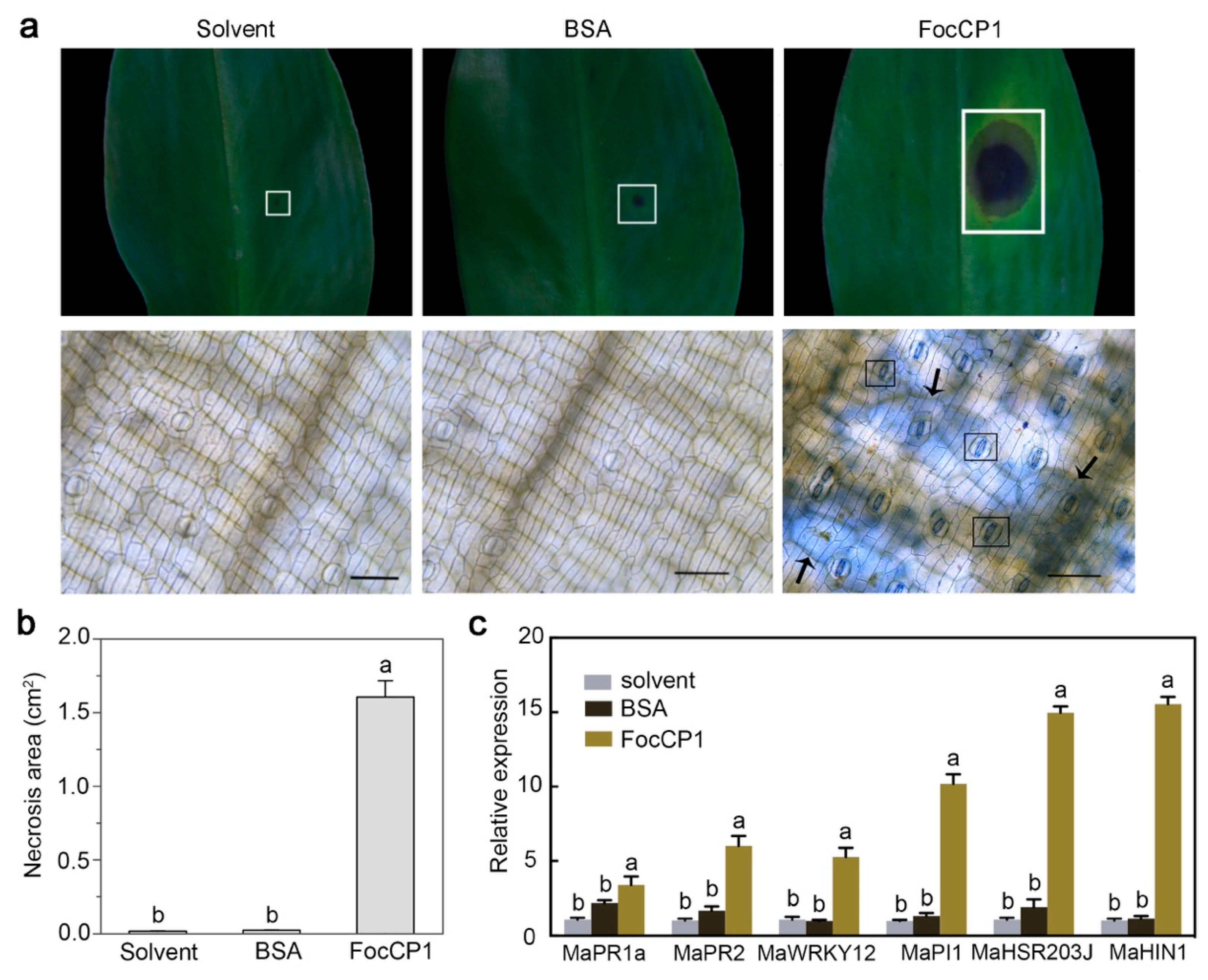
2.5. Recombinant FocCP1 Induces Necrosis in Banana Leaves
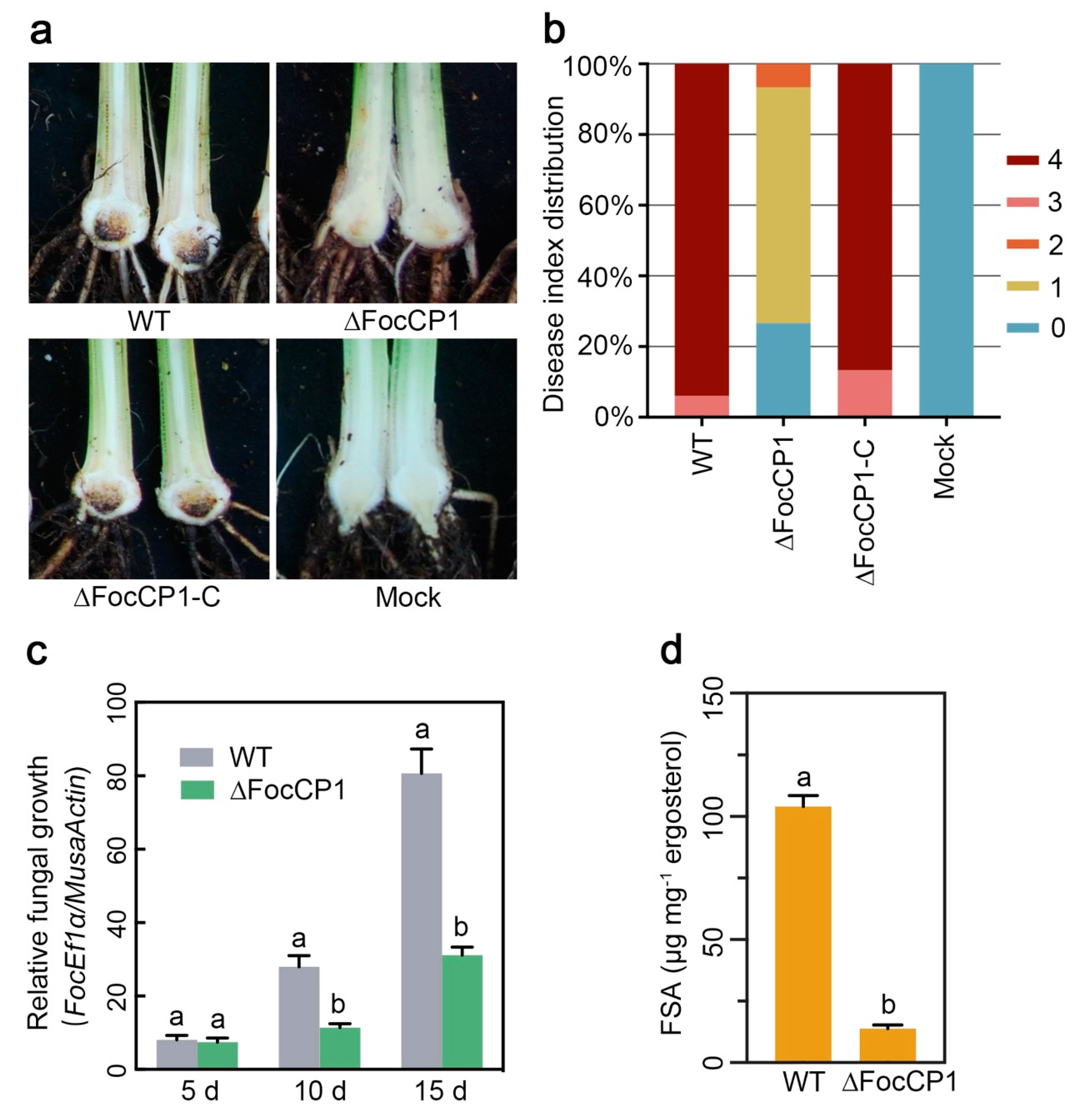
2.6. FocCP1 Contributes to the Pathogenicity of Foc TR4
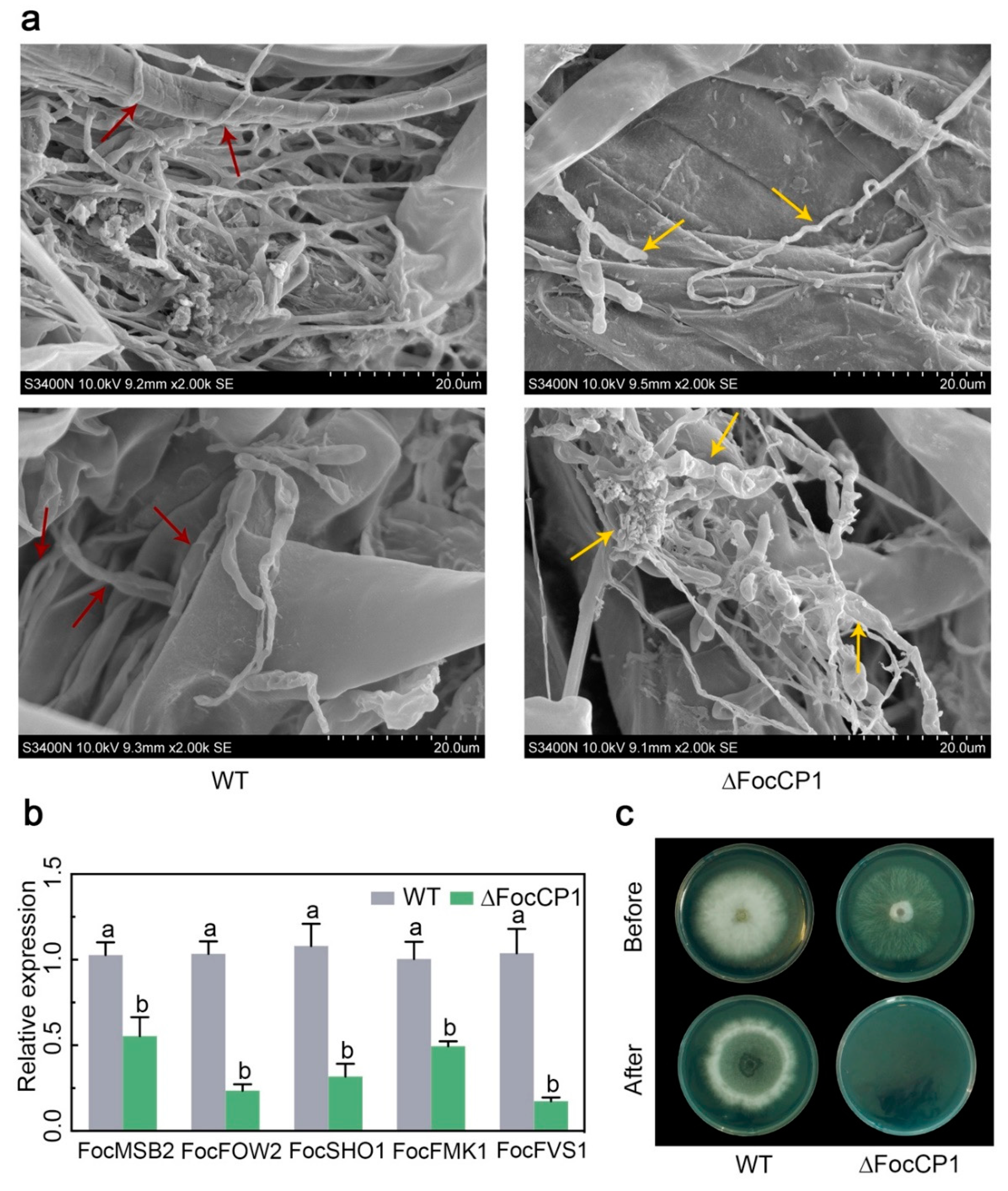
2.7. FocCP1 is Required for Successful Penetration of Banana Roots
3. Discussion
4. Materials and Methods
4.1. Fungal Strains, Plants, and Growth Conditions
4.2. Bioinformatics Analysis
4.3. Cloning, Expression, and Purification of the FocCP1 Protein
4.4. Histochemical Assays
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Construction of A Gene-Deletion Mutant and Complementary Strain
4.7. Pathogenicity Tests and Fungal Biomass Estimation
4.8. Analysis of FSA
4.9. Scanning Electron Microscopy Observation
4.10. Subcellular Localization Assays and Fluorescence Microscopy
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Foc TR4 | Fusarium oxysporum f. sp. cubense tropical race 4 |
| CP | cerato-platanin |
| FocCP1 | The first cerato-platanin protein from Foc TR4 |
| PAMPs | pathogen-associated molecular patterns |
| HR | hypersensitive response |
| PTI | linear dichroism |
| ETI | effector-triggered immunity |
| SIX | secreted in xylem |
| SSCRP | small secreted cysteine-rich proteins |
References
- Garcia-Bastidas, F.; Ordonez, N.; Konkol, J.; Al-Qasim, M.; Naser, Z.; Abdelwali, M.; Salem, N.; Waalwijk, C.; Ploetz, R.C.; Kema, G.H.J. First Report of Fusarium oxysporum f. sp. cubense Tropical Race 4 Associated with Panama Disease of Banana outside. Southeast Asia Plant Dis. 2014, 98, 694. [Google Scholar] [CrossRef]
- Moore, N.; Pegg, K.; Allen, R.; Irwin, J. Vegetative compatibility and distribution of Fusarium oxysporum f. sp. cubense in Australia. Aust. J. Exp. Agric. 1993, 33, 797–802. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Ploetz, R.C. Fusarium Wilt of Banana Is Caused by Several Pathogens Referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Dita, M.A.; Waalwijk, C.; Buddenhagen, I.W.; Souza, M.T., Jr.; Kema, G.H.J. A molecular diagnostic for tropical race 4 of the banana fusarium wilt pathogen. Plant Pathol. 2010, 59, 348–357. [Google Scholar] [CrossRef]
- Butler, D. Fungus threatens top banana. Nature 2013, 504, 195–196. [Google Scholar] [CrossRef]
- Ploetz, R.; Freeman, S.; Konkol, J.; Al-Abed, A.; Naser, Z.; Shalan, K.; Barakat, R.; Israeli, Y. Tropical race 4 of Panama disease in the Middle East. Phytoparasitica 2015, 43, 283–293. [Google Scholar] [CrossRef]
- Ploetz, R.C. Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop. Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef]
- Schwessinger, B.; Zipfel, C. News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395. [Google Scholar] [CrossRef]
- Rafiqi, M.; Ellis, J.G.; Ludowici, V.A.; Hardham, A.R.; Dodds, P.N. Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr. Opin. Plant Biol. 2012, 15, 477–482. [Google Scholar] [CrossRef]
- Houterman, P.M.; Speijer, D.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C.; Rep, M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007, 8, 215–221. [Google Scholar] [CrossRef]
- Widinugraheni, S.; Nino-Sanchez, J.; van der Does, H.C.; van Dam, P.; Garcia-Bastidas, F.A.; Subandiyah, S.; Meijer, H.J.G.; Kistler, H.C.; Kema, G.H.J.; Rep, M. A SIX1 homolog in Fusarium oxysporum f. sp. cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS ONE 2018, 13, e0205896. [Google Scholar] [CrossRef]
- Li, C.; Zuo, C.; Deng, G.; Kuang, R.; Yang, Q.; Hu, C.; Sheng, O.; Zhang, S.; Ma, L.; Wei, Y.; et al. Contamination of bananas with beauvericin and fusaric acid produced by Fusarium oxysporum f. sp. cubense. PLoS ONE 2013, 8, e70226. [Google Scholar] [CrossRef]
- Ding, Z.; Yang, L.; Wang, G.; Guo, L.; Liu, L.; Wang, J.; Huang, J. Fusaric acid is a virulence factor of Fusarium oxysporum f. sp. cubense on banana plantlets. Trop. Plant Pathol. 2018, 43, 297–305. [Google Scholar] [CrossRef]
- Pazzagli, L.; Cappugi, G.; Manao, G.; Camici, G.; Santini, A.; Scala, A. Purification, Characterization, and Amino Acid Sequence of Cerato-platanin, a New Phytotoxic Protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 1999, 274, 24959–24964. [Google Scholar] [CrossRef]
- Boddi, S.; Comparini, C.; Calamassi, R.; Pazzagli, L.; Cappugi, G.; Scala, A. Cerato-platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani. FEMS Microbiol. Lett. 2004, 233, 341–346. [Google Scholar] [CrossRef]
- Gomes, E.V.; Costa, M.D.N.; De Paula, R.G.; De Azevedo, R.R.; Da Silva, F.L.; Noronha, E.F.; Ulhoa, C.J.; Monteiro, V.N.; Cardoza, R.E.; Gutierrez, S.; et al. The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci. Rep. 2015, 5, 17998. [Google Scholar] [CrossRef]
- Frías, M.; Gonzalez, C.; Brito, N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, H.; Li, G.; Li, W.; Wang, X.; Song, F. Ectopic expression of MgSM1, a Cerato-platanin family protein fromMagnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 2009, 7, 763–777. [Google Scholar] [CrossRef]
- Crutcher, F.K.; Moran-Diez, M.E.; Ding, S.-L.; Liu, J.; Horwitz, B.A.; Mukherjee, P.K.; Kenerley, C.M. A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 2015, 119, 476–486. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Isordia-Jasso, M.I.; Islas-Osuna, M.A.; Delgado-Sanchez, P.; Jimenez-Bremont, J.F.; Rodriguez-Kessler, M.; Rosales-Saavedra, M.T.; Herrera-Estrella, A.; Casas-Flores, S. The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 2015, 6, 77. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae-Secreted Protein MSP1 Induces Cell Death and Elicits Defense Responses in Rice. Mol. Plant Microbe Interact. 2016, 29, 299–312. [Google Scholar] [CrossRef]
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef]
- Chen, H.; Kovalchuk, A.; Asiegbu, F.O.; Keriö, S. Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 2013, 105, 1479–1488. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Singh, K.B.; Taylor, J.M. ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2018, 217, 1764–1778. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci.USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef]
- Haq, S.K.; Atif, S.M.; Khan, R.H. Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch. Biochem. Biophys. 2004, 431, 145–159. [Google Scholar] [CrossRef]
- Pontier, D.; Deslandes, L.; Bezombes-Marion, I.; Balagué, C.; Tronchet, M.; Roby, D. Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 2001, 26, 495–507. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Di Pietro, A. The Membrane Mucin Msb2 Regulates Invasive Growth and Plant Infection in Fusarium oxysporum. Plant Cell 2011, 23, 1171–1185. [Google Scholar] [CrossRef]
- Imazaki, I.; Kurahashi, M.; Iida, Y.; Tsuge, T. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol. Microbiol. 2007, 63, 737–753. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Di Pietro, A. The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum. Mol. Plant Pathol. 2015, 16, 593–603. [Google Scholar] [CrossRef]
- Rispail, N.; Di Pietro, A. Fusarium oxysporum Ste12 Controls Invasive Growth and Virulence Downstream of the Fmk1 MAPK Cascade. Mol. Plant-Microbe Interact. 2009, 22, 830–839. [Google Scholar] [CrossRef]
- Iida, Y.; Fujiwara, K.; Yoshioka, Y.; Tsuge, T. Mutation of FVS1, encoding a protein with a sterile alpha motif domain, affects asexual reproduction in the fungal plant pathogen Fusarium oxysporum. FEMS Microbiol. Lett. 2014, 351, 104–112. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Yuan, J.; Qiu, D. The Verticillium dahliae SnodProt1-Like Protein VdCP1 Contributes to Virulence and Triggers the Plant Immune System. Front. Plant Sci. 2017, 8, 1880. [Google Scholar] [CrossRef]
- Pazzagli, L.; Seidl-Seiboth, V.; Barsottini, M.; Vargas, W.A.; Scala, A.; Mukherjee, P.K. Cerato-platanins: Elicitors and effectors. Plant Sci. 2014, 228, 79–87. [Google Scholar] [CrossRef]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 2017, 7, 41037. [Google Scholar] [CrossRef]
- Frias, M.; Brito, N.; Gonzalez, C. The Botrytis cinerea cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 2013, 14, 191–196. [Google Scholar] [CrossRef]
- Djonović, S.; Pozo, M.J.; Dangott, L.J.; Howell, C.R.; Kenerley, C.M. Sm1, a Proteinaceous Elicitor Secreted by the Biocontrol FungusTrichoderma virensInduces Plant Defense Responses and Systemic Resistance. Mol. Plant Microbe Interact. 2006, 19, 838–853. [Google Scholar] [CrossRef]
- Quarantin, A.; Glasenapp, A.; Schäfer, W.; Favaron, F.; Sella, L. Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 2016, 109, 220–229. [Google Scholar] [CrossRef]
- Li, S.; Dong, Y.; Li, L.; Zhang, Y.; Yang, X.; Zeng, H.; Shi, M.; Pei, X.; Qiu, D.; Yuan, Q. The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants. Int. J. Mol. Sci. 2019, 20, 2849. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Hydrophobins: Multipurpose Proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef]
- Pazzagli, L.; Zoppi, C.; Carresi, L.; Tiribilli, B.; Sbrana, F.; Schiff, S.; Pertinhez, T.A.; Scala, A.; Cappugi, G. Characterization of ordered aggregates of cerato-platanin and their involvement in fungus–host interactions. Biochim. Biophys. Acta (BBA) Gen. Subj. 2009, 1790, 1334–1344. [Google Scholar] [CrossRef]
- Frischmann, A.; Neudl, S.; Gaderer, R.; Bonazza, K.; Zach, S.; Gruber, S.; Spadiut, O.; Friedbacher, G.; Grothe, H.; Seidl-Seiboth, V. Self-assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungusTrichoderma atroviride. J. Biol. Chem. 2013, 288, 4278–4287. [Google Scholar] [CrossRef]
- Doehlemann, G.; Requena, N.; Schaefer, P.; Brunner, F.; O’Connell, R.; Parker, J.E. Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytol. 2014, 204, 803–814. [Google Scholar] [CrossRef]
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium Wilt of Banana: Current Knowledge on Epidemiology and Research Needs toward Sustainable Disease Management. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef]
- Ma, L.J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Dai, Y.; Cao, Z.; Huang, L.; Liu, S.; Shen, Z.; Wang, Y.; Wang, H.; Zhang, H.; Li, D.; Song, F. CCR4-Not Complex Subunit Not2 Plays Critical Roles in Vegetative Growth, Conidiation and Virulence in Watermelon Fusarium Wilt Pathogen Fusarium oxysporum f. sp. niveum. Front. Microbiol. 2016, 7, 1449. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O.; Dufayard, J.-F. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- LaVallie, E.R.; Diblasio, E.A.; Kovacic, S.; Grant, K.L.; Schendel, P.F.; McCoy, J.M. A Thioredoxin Gene Fusion Expression System That Circumvents Inclusion Body Formation in the E. coli Cytoplasm. Nat. Biotechnol. 1993, 11, 187–193. [Google Scholar] [CrossRef]
- Rate, D.N.; Cuenca, J.V.; Bowman, G.R.; Guttman, D.S.; Greenberg, J.T. The Gain-of-Function Arabidopsis acd6 Mutant Reveals Novel Regulation and Function of the Salicylic Acid Signaling Pathway in Controlling Cell Death, Defenses, and Cell Growth. Plant Cell 1999, 11, 1695–1708. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, C.; Liu, X.; Ma, Z. A transcription factor FgSte12 is required for pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a Magnaporthe grisea Serine/Threonine Kinase Gene, MgATG1, in Appressorium Turgor and Pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef]
- Guo, L.; Wenner, N.; Kuldau, G.A. FvSO regulates vegetative hyphal fusion, asexual growth, fumonisin B1 production, and virulence in Fusarium verticillioides. Fungal Biol. 2015, 119, 1158–1169. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatilepPZP family ofAgrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Wu, B.; Yang, J.; Bi, F.; Dong, T.; Yang, Q.; Hu, C.; Xiang, D.; Chen, H.; Huang, H.; et al. A Cerato-Platanin Family Protein FocCP1 Is Essential for the Penetration and Virulence of Fusarium oxysporum f. sp. cubense Tropical Race 4. Int. J. Mol. Sci. 2019, 20, 3785. https://doi.org/10.3390/ijms20153785
Liu S, Wu B, Yang J, Bi F, Dong T, Yang Q, Hu C, Xiang D, Chen H, Huang H, et al. A Cerato-Platanin Family Protein FocCP1 Is Essential for the Penetration and Virulence of Fusarium oxysporum f. sp. cubense Tropical Race 4. International Journal of Molecular Sciences. 2019; 20(15):3785. https://doi.org/10.3390/ijms20153785
Chicago/Turabian StyleLiu, Siwen, Bo Wu, Jing Yang, Fangcheng Bi, Tao Dong, Qiaosong Yang, Chunhua Hu, Dandan Xiang, Hongrui Chen, Huoqing Huang, and et al. 2019. "A Cerato-Platanin Family Protein FocCP1 Is Essential for the Penetration and Virulence of Fusarium oxysporum f. sp. cubense Tropical Race 4" International Journal of Molecular Sciences 20, no. 15: 3785. https://doi.org/10.3390/ijms20153785
APA StyleLiu, S., Wu, B., Yang, J., Bi, F., Dong, T., Yang, Q., Hu, C., Xiang, D., Chen, H., Huang, H., Shao, C., Chen, Y., Yi, G., Li, C., & Guo, X. (2019). A Cerato-Platanin Family Protein FocCP1 Is Essential for the Penetration and Virulence of Fusarium oxysporum f. sp. cubense Tropical Race 4. International Journal of Molecular Sciences, 20(15), 3785. https://doi.org/10.3390/ijms20153785




