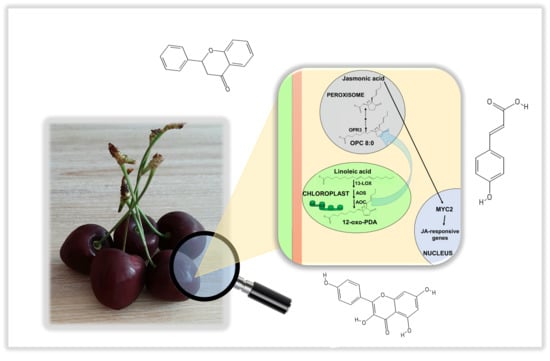
Identification of Jasmonic Acid Biosynthetic Genes in Sweet Cherry and Expression Analysis in Four Ancient Varieties from Tuscany
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of JA Biosynthetic Genes in Sweet Cherries
2.2. Total and Targeted Quantification of Phenolics in the Four Ancient Varieties
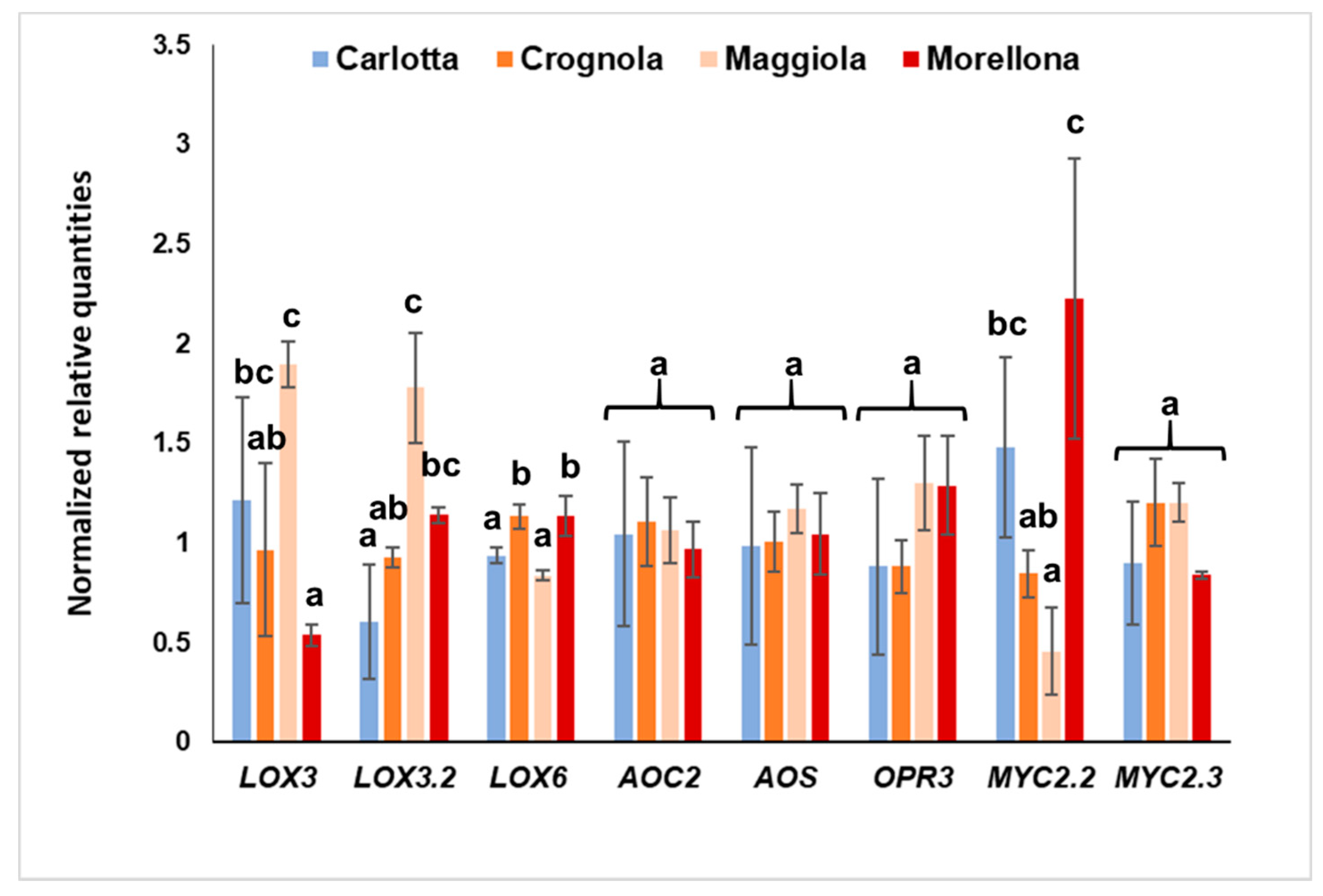
2.3. Gene Expression Analysis of JA Biosynthetic Genes in the Four Ancient Varieties
3. Materials and Methods
3.1. Fruit Harvesting
3.2. Chemical Assays, HPLC Analyses and Statistics
3.3. RNA Extraction, Primer Design and Real-Time PCR Data Analysis
3.4. Bioinformatics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berni, R.; Romi, M.; Cantini, C.; Hausman, J.-F.; Guerriero, G.; Cai, G. Functional Molecules in Locally-Adapted Crops: The Case Study of Tomatoes, Onions, and Sweet Cherry Fruits From Tuscany in Italy. Front. Plant Sci. 2019, 9, 1983. [Google Scholar] [CrossRef]
- Berni, R.; Hoque, M.Z.; Legay, S.; Cai, G.; Siddiqui, K.S.; Hausman, J.-F.; Andre, C.M.; Guerriero, G. Tuscan Varieties of Sweet Cherry Are Rich Sources of Ursolic and Oleanolic Acid: Protein Modeling Coupled to Targeted Gene Expression and Metabolite Analyses. Molecules 2019, 24, 1590. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Tomiyama, A.; Seto, H. Changes of Endogenous Jasmonic Acid and Methyl Jasmonate in Apples and Sweet Cherries during Fruit Development. J. Am. Soc. Hortic. Sci. 2000, 125, 282–287. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J. Metabolism of the plant hormone jasmonate: A sentinel for tissue damage and master regulator of stress response. Phytochem. Rev. 2018, 17, 51–80. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Forner, S.; Strnad, M.; Hause, B. Jasmonates in flower and seed development. Biochimie 2013, 95, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Saracoglu, O.; Ozturk, B.; Yildiz, K.; Kucuker, E. Pre-harvest methyl jasmonate treatments delayed ripening and improved quality of sweet cherry fruits. Sci. Hortic. 2017, 226, 19–23. [Google Scholar] [CrossRef]
- Asghari, M.; Hasanlooe, A.R. Methyl jasmonate effectively enhanced some defense enzymes activity and Total Antioxidant content in harvested “Sabrosa” strawberry fruit. Food Sci. Nutr. 2015, 4, 377–383. [Google Scholar] [CrossRef]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 20. [Google Scholar] [CrossRef]
- Mendoza, D.; Cuaspud, O.; Arias, J.P.; Ruiz, O.; Arias, M. Effect of salicylic acid and methyl jasmonate in the production of phenolic compounds in plant cell suspension cultures of Thevetia peruviana. Biotechnol. Rep. 2018, 19, e00273. [Google Scholar] [CrossRef] [PubMed]
- Onofrio, C.D.; Cox, A.; Davies, C.; Boss, P.K. Induction of secondary metabolism in grape cell cultures by jasmonates. Funct. Plant Biol. 2009, 36, 323–338. [Google Scholar] [CrossRef]
- Zaheer, M.; Reddy, V.D.; Giri, C.C. Enhanced daidzin production from jasmonic and acetyl salicylic acid elicited hairy root cultures of Psoralea corylifolia L. (Fabaceae). Nat. Prod. Res. 2016, 30, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Díaz, M.; Lobos, T.; Cardemil, L.; Nunes-Nesi, A.; Retamales, J.; Jaakola, L.; Alberdi, M.; Ribera-Fonseca, A. Methyl Jasmonate: An Alternative for Improving the Quality and Health Properties of Fresh Fruits. Molecules 2016, 21, 567. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Mattheis, J.P.; Fellman, J.K. A role for jasmonates in climacteric fruit ripening. Planta 1998, 204, 444–449. [Google Scholar] [CrossRef]
- Concha, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.A.; Schwab, W.; Figueroa, C.R. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Balbontín, C.; Gutiérrez, C.; Wolff, M.; Figueroa, C.R.; Balbontín, C.; Gutiérrez, C.; Wolff, M.; Figueroa, C.R. Effect of abscisic acid and methyl jasmonate preharvest applications on fruit quality and cracking tolerance of sweet cherry. Chil. J. Agric. Res. 2018, 78, 438–446. [Google Scholar] [CrossRef]
- Yao, H.; Tian, S. Effects of pre- and post-harvest application of salicylic acid or methyl jasmonate on inducing disease resistance of sweet cherry fruit in storage. Postharvest Biol. Technol. 2005, 35, 253–262. [Google Scholar] [CrossRef]
- Berni, R.; Cantini, C.; Romi, M.; Hausman, J.-F.; Guerriero, G.; Cai, G. Agrobiotechnology Goes Wild: Ancient Local Varieties as Sources of Bioactives. Int. J. Mol. Sci. 2018, 19, 2248. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011, 6, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, D.; Chu, W.; Zhu, D.; Yan, H.; Xiang, Y. Retention and Molecular Evolution of Lipoxygenase Genes in Modern Rosid Plants. Front. Genet. 2016, 7, 176. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, M.G.; Dinh, S.T.; Oh, Y.; Gaquerel, E.; Baldwin, I.T.; Galis, I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC Plant Biol. 2013, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, A.; Lenglet, A.; Wolfender, J.-L.; Farmer, E.E. Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis. Plants 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, M.A.; Veldink, G.A.; Vliegenthart, J.F. Fatty acid hydroperoxide lyase: A plant cytochrome p450 enzyme involved in wound healing and pest resistance. Chembiochem 2001, 2, 494–504. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, Y.; Peng, F.; He, N.; Li, Y.; Zhao, D. Changes of Aroma Components in Hongdeng Sweet Cherry During Fruit Development. Agric. Sci. China 2007, 6, 1376–1382. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Berni, R.; Piasecki, E.; Legay, S.; Hausman, J.-F.; Siddiqui, K.S.; Cai, G.; Guerriero, G. Identification of the laccase-like multicopper oxidase gene family of sweet cherry (Prunus avium L.) and expression analysis in six ancient Tuscan varieties. Sci. Rep. 2019, 9, 3557. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed]
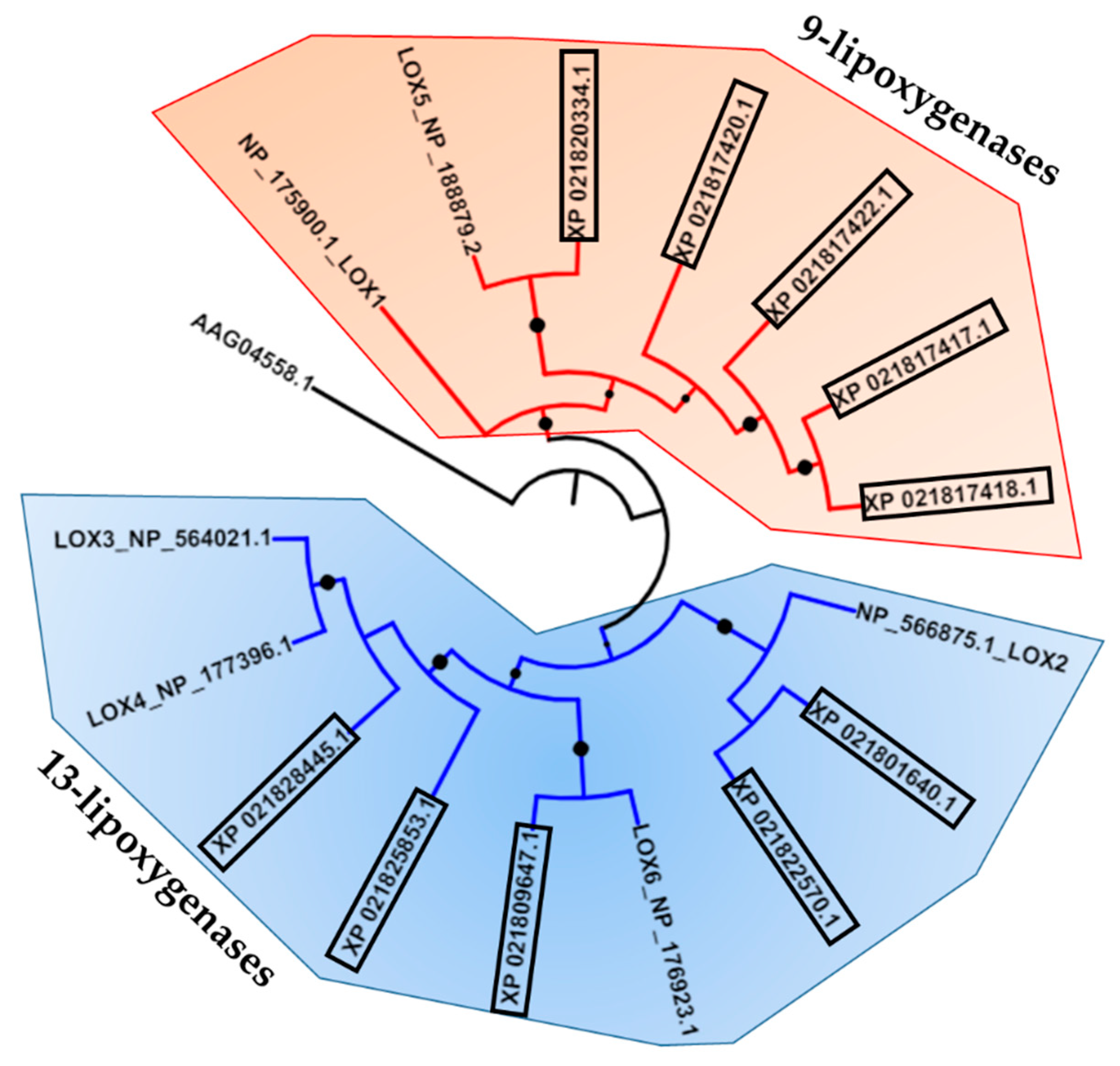
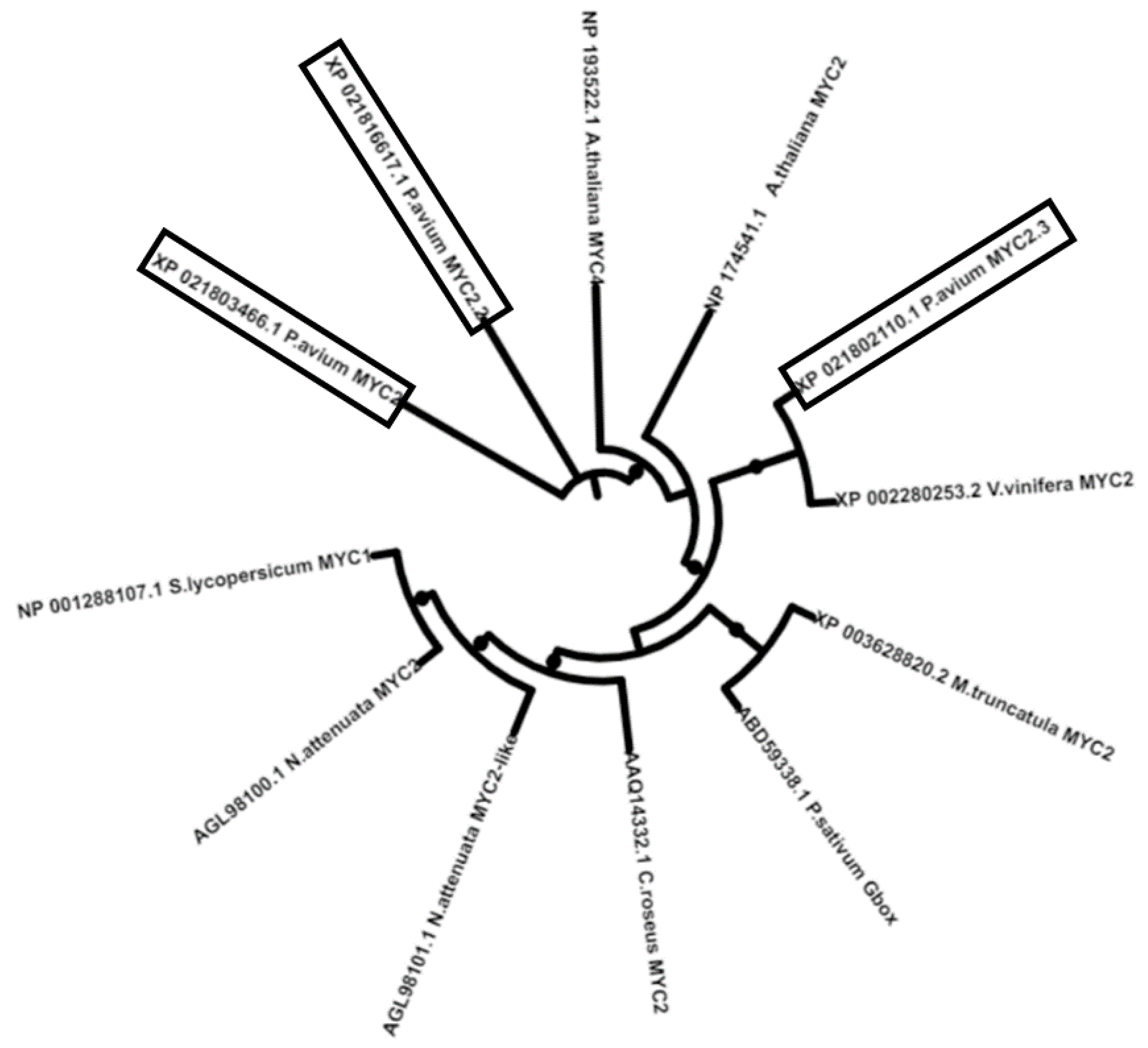
| Proposed Nomenclature | GDR Code | NCBI Accession (Best Hit) |
|---|---|---|
| PavLOX2 | Pav_sc0001040.1_g410.1.mk | XP_021822553.1 |
| PavLOX2.2 | Pav_sc0001040.1_g480.1.mk | XP_021822570.1 |
| PavLOX2.3 | Pav_sc0005842.1_g020.1.mk/Pav_sc0001040.1_g230.1.br | XP_021801640.1/XP_021822570.1 |
| PavLOX3 | Pav_sc0001305.1_g640.1.mk | XP_021825853.1 |
| PavLOX3.2 | Pav_sc0001580.1_g270.1.mk | XP_021828445.1 |
| PavLOX6 | Pav_sc0000351.1_g320.1.mk | XP_021809647.1 |
| PavAOS | Pav_sc0000890.1_g1300.1.mk/Pav_sc0004356.1_g050.1.mk | XP_021820947.1/XP_021800682.1 |
| PavAOC | Pav_sc0000618.1_g380.1.mk | XP_021804158.1 |
| PavAOC 2 | Pav_sc0000567.1_g750.1.mk | XP_021814377.1 |
| PavOPR3 | Pav_sc0000129.1_g860.1.mk | XP_021804971.1 |
| PavMYC2 | Pav_sc0000107.1_g180.1.mk | XP_021803466.1 |
| PavMYC2.2 | Pav_sc0000652.1_g730.1.mk | XP_021816617.1 |
| PavMYC2.3 | Pav_sc0006499.1_g050.1.mk | XP_021802110.1 |
| Variety | Total Antioxidants (mmol Fe2+ mmol/100g FW) | Polyphenols (mg GAE/100g FW) | Flavonoids (mg QeE/100g FW) | Anthocyanins (mg CyE/100g FW) |
|---|---|---|---|---|
| ‘Carlotta’ | 1.73 ± 0.01 b | 159.18 ± 0.41 b | 49.33 ± 0.71 b | 34.46 ± 0.81 b |
| ‘Morellona’ | 2.22 ± 0.02 c | 313.11 ± 3.55 c | 97.86 ± 1.56 c | 65.27 ± 1.03 d |
| ‘Maggiola’ | 1.43 ± 0.02 a | 137.70 ± 2.01 a | 40.06 ± 1.23 a | 32.41 ± 0.91 a |
| ‘Crognola’ | 3.07 ± 0.01 d | 387.11 ± 1.29 d | 102.05 ± 2.42 d | 58.46 ± 1.11 c |
| Variety | Chlorogenic Acid (μg/g FW) | p-Coumaric Acid (μg/g FW) | (+)-Catechin (μg/g FW) | Rutin (μg/g FW) | Cyanidin-3-Glucoside (μg/g FW) |
|---|---|---|---|---|---|
| ‘Carlotta’ | 179.42 ± 0.83 b | 29.02 ± 0.57 a | 122.83 ± 4.96 b | 32.54 ± 0.63 b | 59.50 ± 1.02 b |
| ‘Morellona’ | 276.38 ± 0.98 c | 51.17 ± 1.11 b | 172.55 ± 1.10 c | 39.05 ± 0.43 b | 80.62 ± 0.77 c |
| ‘Maggiola’ | 99.23 ± 0.57 a | 76.50 ± 0.88 c | 46.74 ± 0.99 a | 29.11 ± 0.72 a | 31.26 ± 1.47 a |
| ‘Crognola’ | 312.67 ± 1.11 d | 123.49 ± 0.64 d | 219.44 ± 2.49 d | 99.78 ± 0.55 c | 149.77 ± 1.29 d |
| Name | Sequence (5’→3’) | R2 | Tm (°C) | Amplicon Size (bp) | Efficiency (%) |
|---|---|---|---|---|---|
| Pav_LOX3 Fwd | TCTTGACCTCATTGGGAACC | 0.995 | 79.65 | 85 | 104.03 |
| Pav_LOX3 Rev | ACCTGCTTGGATGGTGAATC | ||||
| Pav_LOX3.2 Fwd | GCCATCAGTGAAGATTTGGTG | 0.995 | 81.39 | 106 | 96.7 |
| Pav_LOX3.2 Rev | CCTCCTTGATCTTGTTCCTCAC | ||||
| Pav_LOX6 Fwd | CAGCATGGTGAAAGAGGTTC | 0.992 | 79.51 | 86 | 91.51 |
| Pav_LOX6 Rev | AAGCAAATCTATCCCCTTT | ||||
| Pav_AOS Fwd | GGAGATGTTGTTCGGGTTTC | 0.995 | 78.98 | 74 | 104.42 |
| Pav_AOS Rev | CTACAAACTCCTCCGCT | ||||
| Pav_AOC2 Fwd | CAATCTCTCGCATTCCTTCC | 0.996 | 80.32 | 71 | 92.44 |
| Pav_AOC2 Rev | AGTTCTTGGAGTTTGGGAA | ||||
| Pav_OPR3 Fwd | CAAGTGGTGGAGCATTATCG | 0.987 | 85.01 | 89 | 104.42 |
| Pav_OPR3 Rev | AGTTTTGAGCCCCAGTCTTG | ||||
| Pav_MYC2.2 Fwd | CCGCTCTGTTGTTCCAAATG | 0.982 | 80.05 | 106 | 95.12 |
| Pav_MYC2.2 Rev | TAGCCTCCAATTCCTCAACC | ||||
| Pav_MYC2.3 Fwd | GGGTGAAGGGTTTTACAAGG | 0.997 | 83.31 | 96 | 91.58 |
| Pav_MYC2.3 Rev | GGACTTTTTTCCTGTACTCC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berni, R.; Cai, G.; Xu, X.; Hausman, J.-F.; Guerriero, G. Identification of Jasmonic Acid Biosynthetic Genes in Sweet Cherry and Expression Analysis in Four Ancient Varieties from Tuscany. Int. J. Mol. Sci. 2019, 20, 3569. https://doi.org/10.3390/ijms20143569
Berni R, Cai G, Xu X, Hausman J-F, Guerriero G. Identification of Jasmonic Acid Biosynthetic Genes in Sweet Cherry and Expression Analysis in Four Ancient Varieties from Tuscany. International Journal of Molecular Sciences. 2019; 20(14):3569. https://doi.org/10.3390/ijms20143569
Chicago/Turabian StyleBerni, Roberto, Giampiero Cai, Xuan Xu, Jean-Francois Hausman, and Gea Guerriero. 2019. "Identification of Jasmonic Acid Biosynthetic Genes in Sweet Cherry and Expression Analysis in Four Ancient Varieties from Tuscany" International Journal of Molecular Sciences 20, no. 14: 3569. https://doi.org/10.3390/ijms20143569
APA StyleBerni, R., Cai, G., Xu, X., Hausman, J.-F., & Guerriero, G. (2019). Identification of Jasmonic Acid Biosynthetic Genes in Sweet Cherry and Expression Analysis in Four Ancient Varieties from Tuscany. International Journal of Molecular Sciences, 20(14), 3569. https://doi.org/10.3390/ijms20143569