Biomolecules of 2-Thiouracil, 4-Thiouracil and 2,4-Dithiouracil: A DFT Study of the Hydration, Molecular Docking and Effect in DNA:RNAMicrohelixes
Abstract
1. Introduction
- (i)
- How the distribution of water molecules around uracil, 2TU, 4TU and 2,4DTU nucleobases affect their molecular structures and charges.
- (ii)
- How the different electron distribution and charges of the thio-nucleobases under study, in special 2TU and 2,4DTU, interact with different pathogens and target proteins through a molecular docking study.
- (iii)
- How the electronegativity difference between sulphur and oxygen affects the intermolecular H-bonds between the two strands of the DNA:RNA helix, and thus in the helical parameters.
- (iv)
- How the positions of sulphur and the nucleosides in the strand affect the helical parameters in three different types of optimized microhelixes.
2. Computational Methodology
- (i)
- The AutoDock Tools graphical user interface [47] was used to remove the ligand and water molecules present in the targeted proteins and,
- (ii)
- Then polar hydrogen and Kollman charges were added in the targeted proteins.
- (iii)
- The AutoGrid 4.2 [47] was used to create affinity grid centered on the active site with grid size 126 × 126 × 126 with a spacing of 1.0 Å. The rigid targeted protein and flexible ligand docking were performed by using AutoDock 4.2 with the Lamarckian genetic algorithm applying the following protocol: Trials of 100 dockings, energy evaluations of 25,000,000, population size of 200, a mutation rate of 0.02, a crossover rate of 0.8 and an elitism value of 1.
3. Results and Discussion
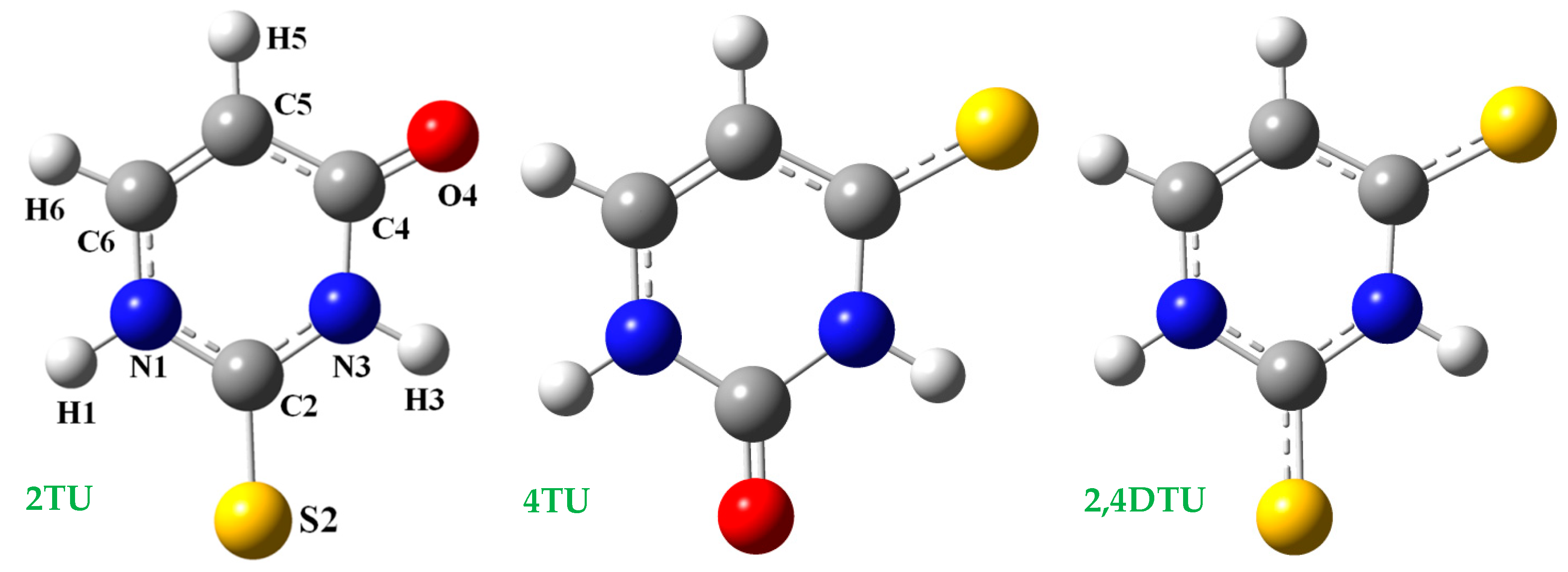
3.1. Geometry Optimization and Atomic Charges in the Isolated State
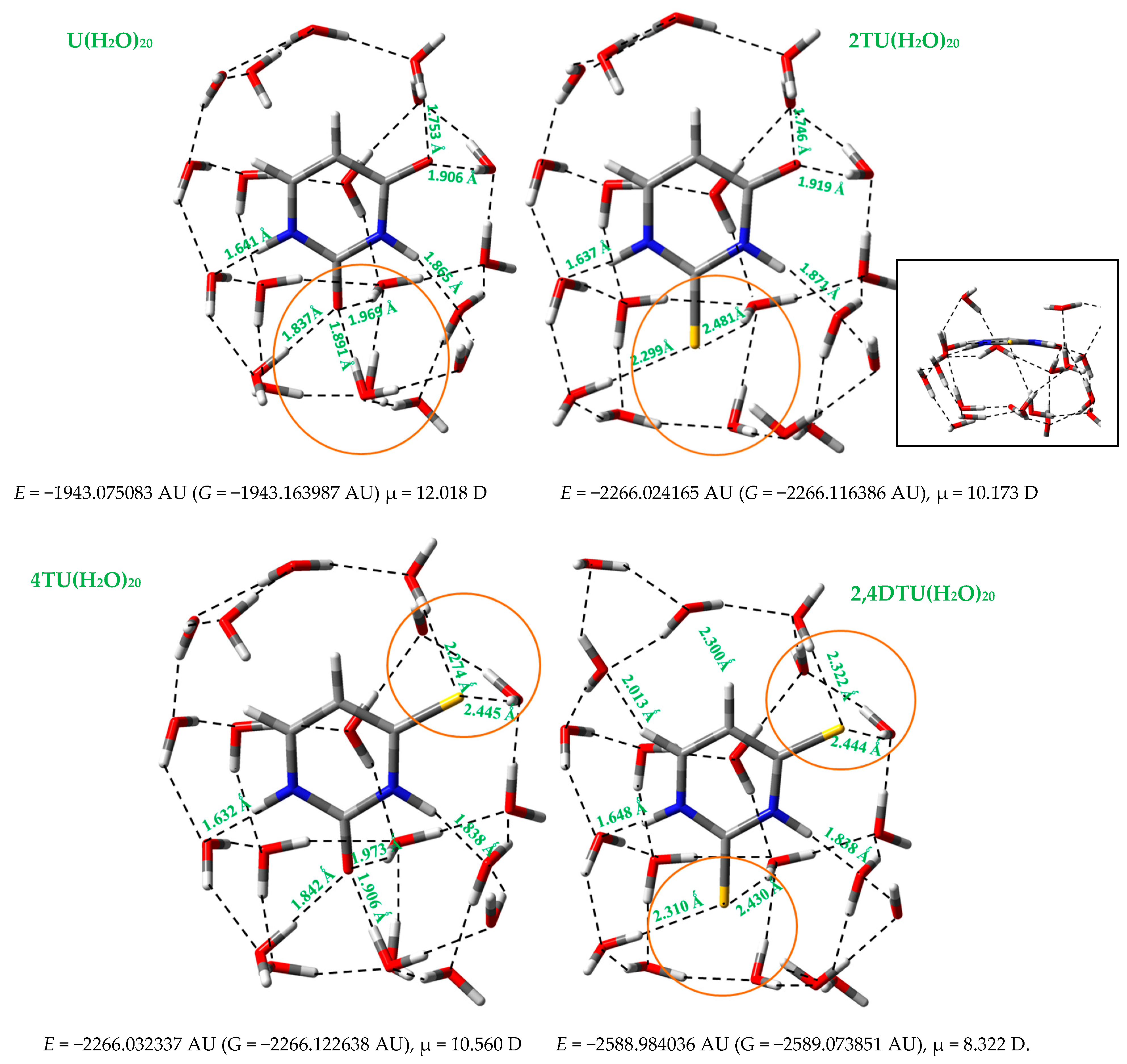
3.2. Hydration of the Nucleobases
- (i)
- A slight difference in the distribution of water molecules around the sulphur atoms as compared to that around the oxygen atoms, as indicated in a circle in orange color in Figure 2.
- (ii)
- Values of the deformation energies Edef and interaction energies are higher in the cluster with uracil U(H2O)20 than in the related clusters with 2TU [13], 4TU and 2,4DTU molecules.
- (i)
- In the isolated state 2TU was slightly less stable than 4TU. However, the hydration changed this stability and with several water molecules 2TU appeared more stable than 4TU.
- (ii)
- (iii)
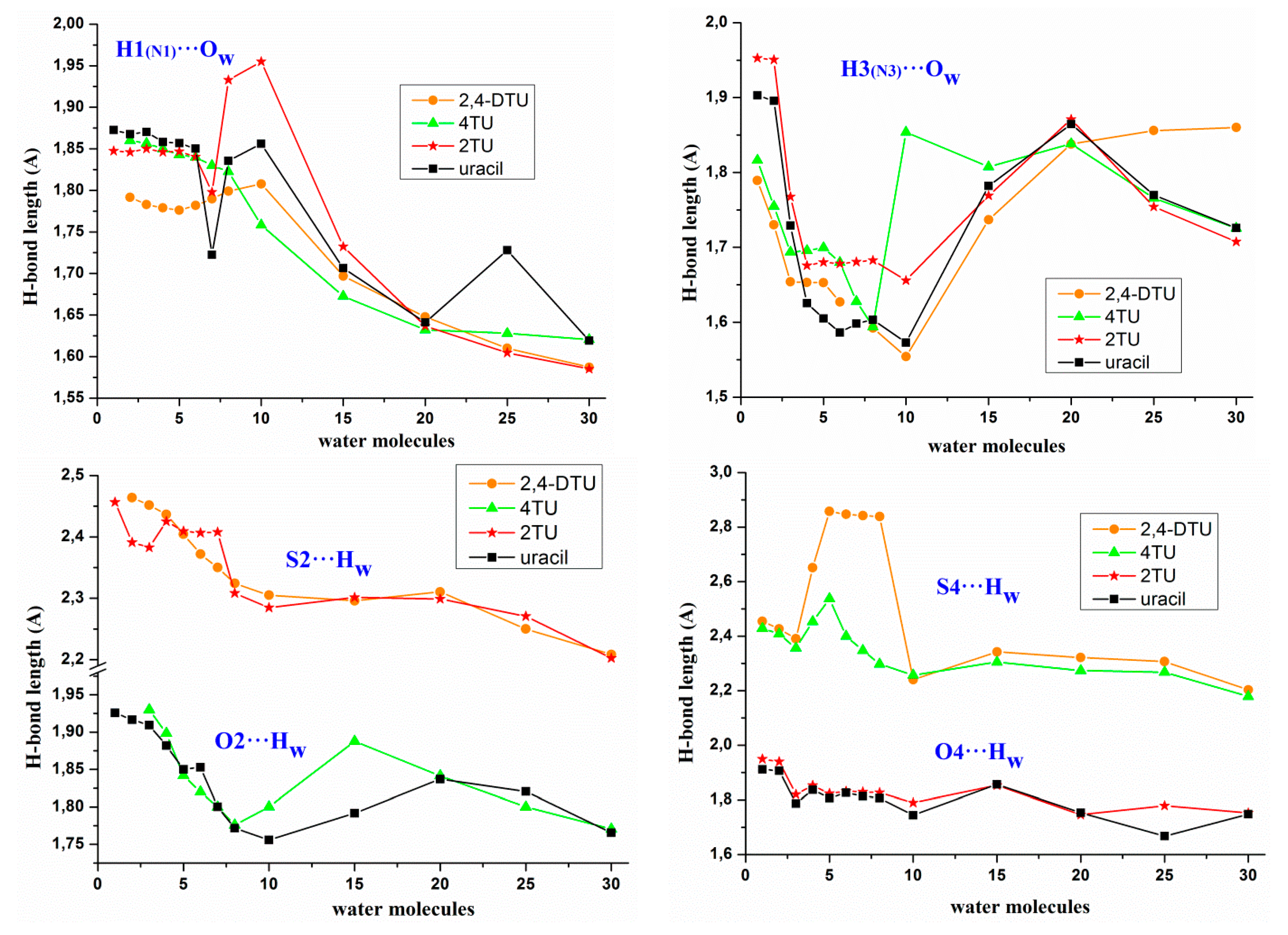
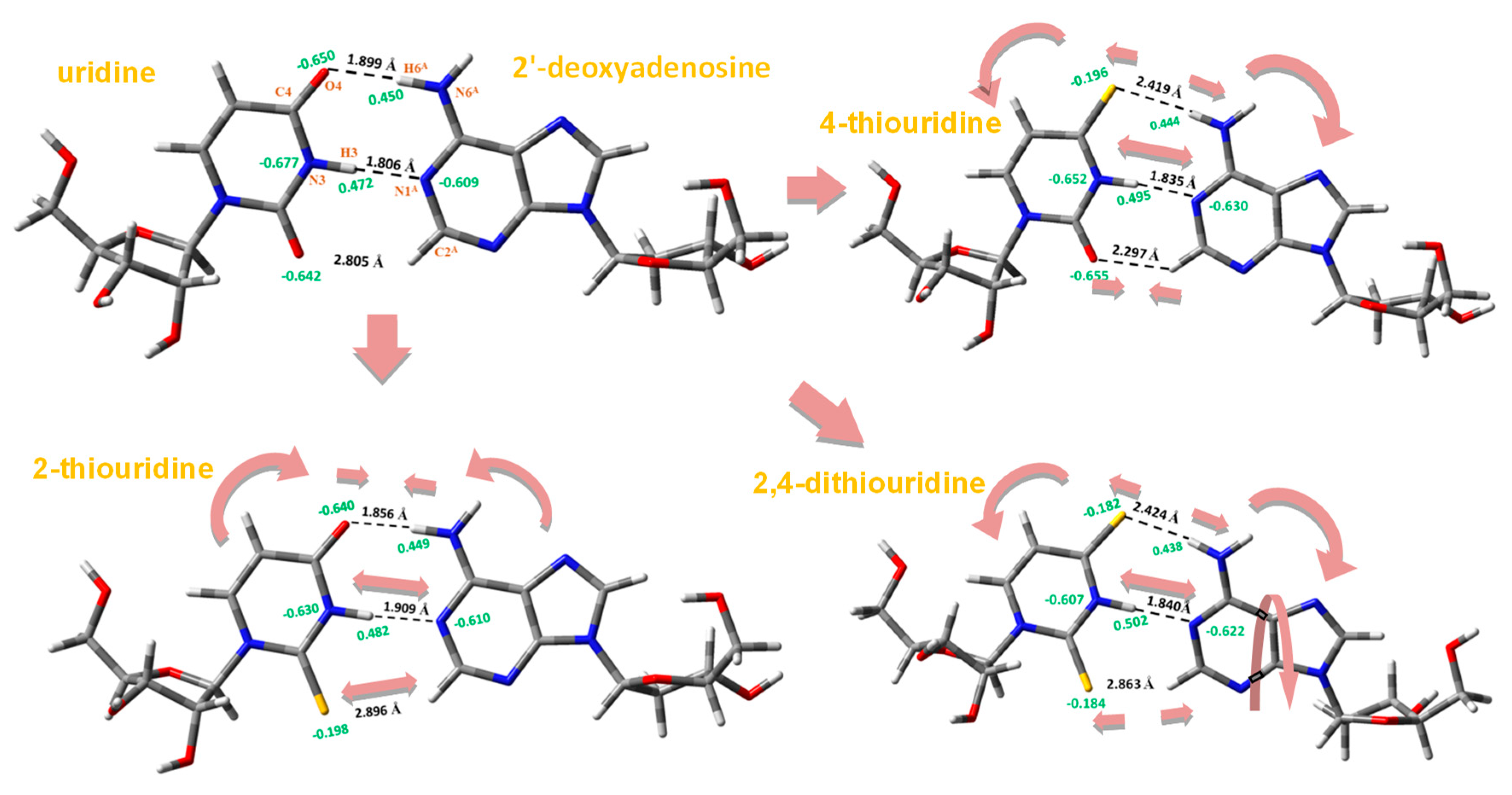
- A strengthening of the intermolecular H-bonds of the nucleobases appears with the hydration with a noticeable increment of this strengthening in the formation of the first hydration shell with about 10 water molecules, and a slight further increment in the second hydration shell with about 30 water molecules, Figure 3. However, as an exception the intermolecular H-bond H3(N3)···Ow was found stronger in the 1st hydration shell than in the 2nd hydration shell. It is also noted the large lengthening in the H3 (N3)···Ow H-bond and the small lengthening in O2···Hw, O4···Hw and S4···Hw H-bonds after the 1st hydration shell was raised. It seems that after the 1st hydration shell, the number of water···water interactions increase over the nucleobase···water, and the interactions of the new water molecules with the water molecules directly H-bonded to the nucleobase leads to a slight weakening of the nucleobase···water H-bonds, because the water···water interactions are remarkably stronger than nucleobase···water.
- (iv)
- The H1(N1)···Ow H-bond appeared slightly stronger than H3(N3)···Ow. With the progress of the hydration, the shortening in the H1(N1)···Ow H-bond was also slightly larger than in H3(N3)···Ow.
- (v)
- The shortening of the S···Ow H-bonds appeared higher than O···Ow H-bonds. Thus for example, this shortening in 2TU and 4TUmolecules as compared to uracil was 0.160 Å with O2 and 0.163 Å with O4, while in 2,4DTU was 0.255 Å with S2 and 0.251 Å with S4.
- (vi)
- The C5-H···Ow and C6-H···Ow H-bonds of hydrophobic sites were very weak in the beginning of the hydration. However, with the progress of the hydration and the formation of a water net that compressed the structure, these H-bonds appeared with values around 2.0–2.3 Å, and thus became prominent. The C6-H···Ow H-bond appeared slightly stronger than C5-H···Ow in all the clusters, e.g., in the cluster U(H2O)20 C5-H···Ow = 2.307 Å and C6-H···Ow = 2.292 Å, while in U(H2O)30 the values were 2.365 and 2.184 Å, respectively. In 2TU, 4TU and 2,4DTU these H-bonds appeared slightly shorter, e.g., in 2,4DTU(H2O)20 the value of C5-H···Ow = 2.300 Å and C6-H···Ow = 2.013 Å, while in 2,4DTU(H2O)30 the values were 2.236 and 2.203 Å, respectively.
- (vii)
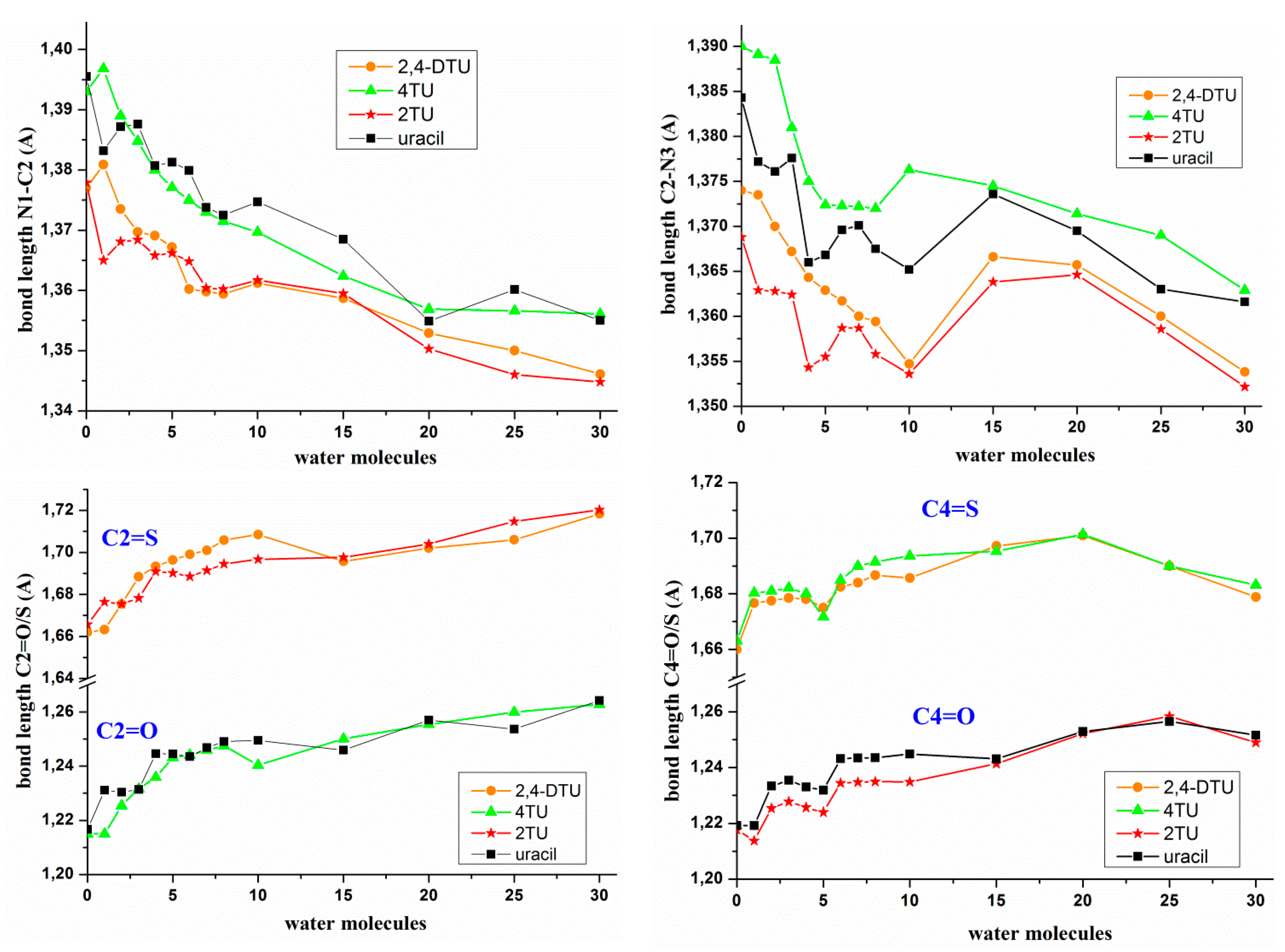
- On hydration, the molecular structure of the nucleobase was changed noticeably with longer C=O/C=S bond lengths and shorter adjacent N1–C2 and C2–N3 bond lengths, Figure 4, in addition to other transformations in the structure of the nucleobase. The largest change in the bond lengths appeared in the formation of the 1st hydration shell with a further slight and prolonged lengthening/shortening of the bond length with the progress of the hydration. Similar features have been observed by us in the hydration of other related nucleobases [41,42]. It was noted a similar trend with the hydration of the C=O and C=S bonds, Figure 4.
- (viii)
- The C2–N3 bond length changed in an irregular way with the hydration, with a shortening in the 1st hydration shell with 8–10 water molecules and then a lengthening of C2–N3 bond up to 20 water molecules. In the formation of the 2nd hydration shell with up to 30 water molecules a new and noticeable shortening of C2–N3 bond appeared again.
- (ix)
- An opening of the ipso angles on C2 and C4 (corresponding to the N1–C2–N3 and N3–C4–C5 angles), and related closing of the neighboring angles C6-N1–C2 and N3–C4–C5 was observed with the hydration. For example, in the uracil molecule, an opening of 4° in angle N1–C2–N3 and 3° in angle N3–C4–C5 was calculated from the isolated state to the cluster with 30 water molecules, and in contrast a closing of 2° in angle C6-N1–C2 and 4° in angle C2–N3–C4. However, with the hydration slightly lower values were found in 2TU, 4TU and 2,4DTU molecules, e.g., in 2,4DTU was found an opening of 3.4° in N1–C2–N3 and 2.3° in N3–C4–C5, and a closing of 1.4° in C6-N1–C2 and 3.4° in C2–N3–C4.
- (x)
- The uracil ring was full planar in the isolated state of all the molecules under study. At the beginning of the hydration this planarity remained maintained. However, in the first hydration shell this planarity of ring disappeared, and becomes out-of-plane around 1–2°, and this deformation remarkably increased with further hydration. The maximum deformation appeared in the clusters with about 15–20 water molecules, with changes around 7–10°. Thus, in U (H2O)20 the N1–C2–N3–C4 torsional angle had the large value of −8.1°, the torsional angle C2–N3–C4–C5 had 11.4° and N3–C4–C5-C6 had −9.9°. Since the attraction between the water molecules and the sulphur atoms was lower than the attraction between the water molecules and the oxygen atoms, the ring deformation was noticeably less in the thiouracil molecules as compared to uracil. Thus, for example in 2,4DTU (H2O)20 the torsional angles N1–C2–N3–C4, C2–N3–C4–C5 and N3–C4–C5-C6 had the values 0.2°, 5.7° and −8.6°, respectively.
- (xi)
- The uracil molecule had the highest negative charges on N1, N3, O2 and O4 atoms, as compared to in 2TU, 4TU and 2,4DTU molecules, Table 2 and Figure S5. This feature shows that in uracil molecule the intermolecular H-bonds through these atoms (N1H, N3H, O2, O4) with the water molecules were stronger than in 2TU, 4TU and 2,4DTU molecules. Moreover, the lower attraction of the thiouracil molecules with water could facilitate the removing of these water molecules when in the primer of the helix begin the establishment of the WC pairs. The lower attraction of the thiouracil molecules with other molecules, such as adenine could also deform the growing of the RNA helix, as explained in Section 3.4.
- (xii)
- The sulphur atom in S2 position remarkably reduced the negative charge on N1, ca. 0.06e, but this reduction on N3 atom was small when the sulphur atom was in position S4, as compared to the uracil molecule. Thus, the NBO negative charge on N1 atom had the lowest value in the 2,4DTU molecule. This negative charge on N1 was also reduced with the hydration.
- (xiii)
- The negative charge on the N3 atom was reduced in a similar amount, ca. 0.06e, by the sulphur atoms in S2 and S4 positions. Thus, in 2,4DTU molecule this charge was reduced to two times and therefore N3 atom appeared with the lowest charge and close to that on N1.
- (xiv)
- The charge on O2/S2 atom was little affected by the substitution of O4 by S4 atom or vice versa. Similarly, the charge on O4/S4 was also little affected by the substitution of O2 by S2 atom or vice versa. Since the negative charge on the nitrogen atoms was reduced with the hydration, the negative charge on the oxygen atoms was increased by a similar amount with the hydration. The trend of this increase in negative charge was similar in uracil, 2TU, 4TU and 2,4DTU molecules.
- (xv)
- The dipole moment (µ) was remarkably changed depending on the spatial distribution of the water molecules. Due to the large polarity of the water molecules, small variations in their distribution can create large differences in µ. With the progress of the hydration an irregular trend of change in the values of μ was observed in all the molecules under study, Figure S5. The highest value appeared in the cluster with 20 water molecules.
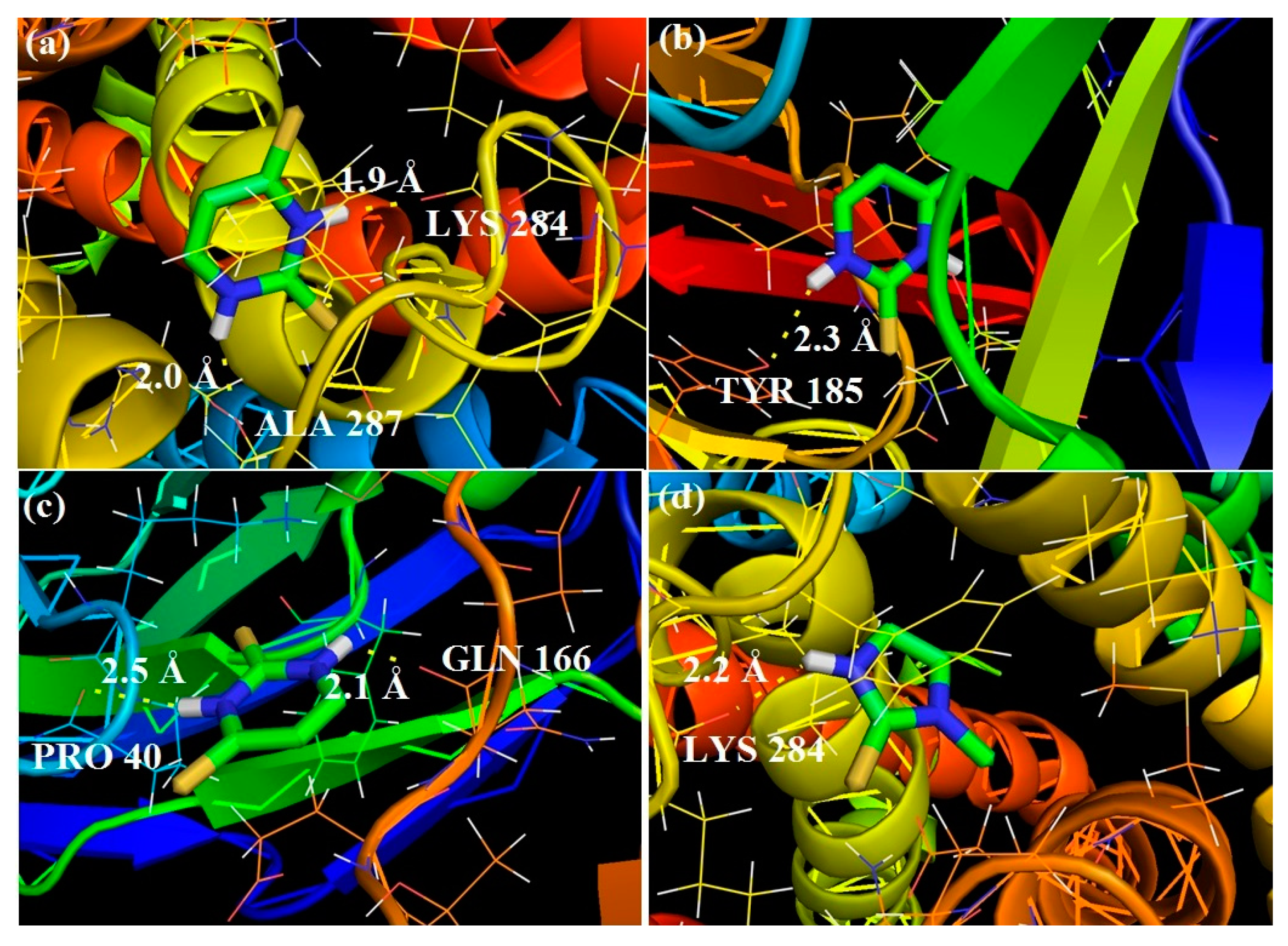
3.3. Molecular Docking Analysis
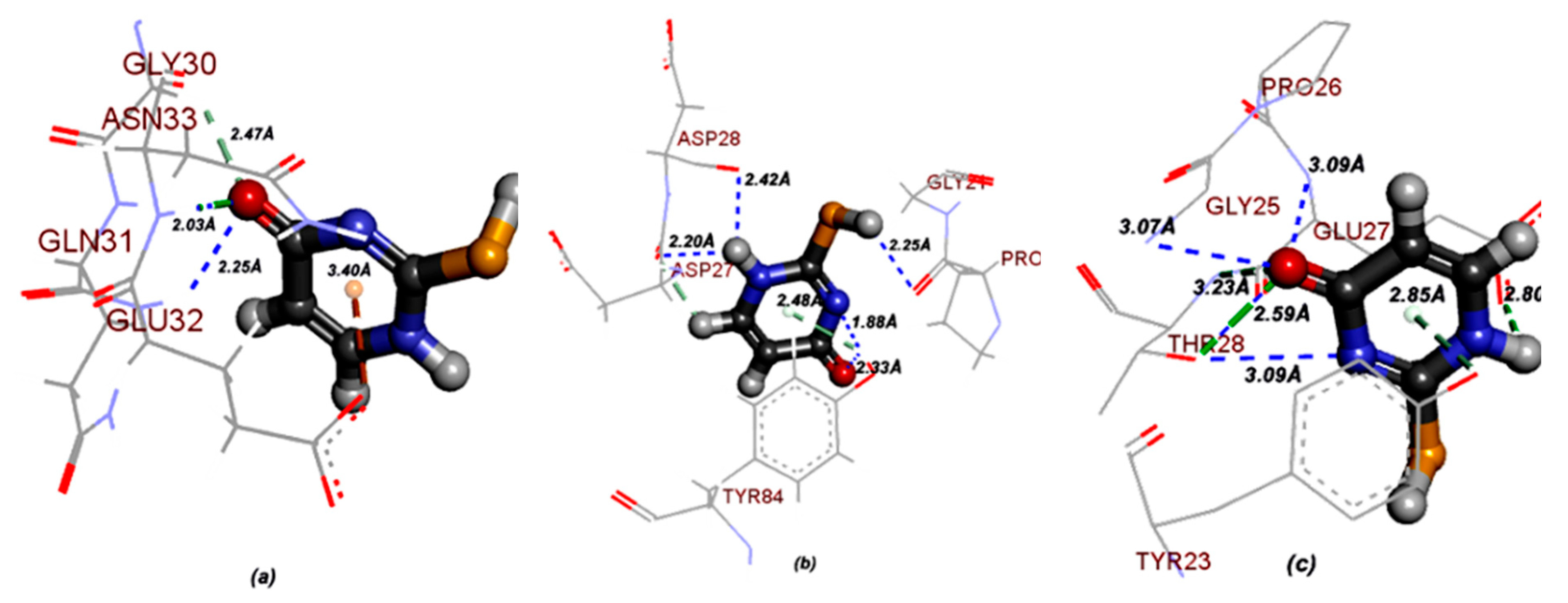
3.3.1. Molecular Docking of 2TU
3.3.2. Molecular Docking of 2,4DTU
Target Proteins Studied
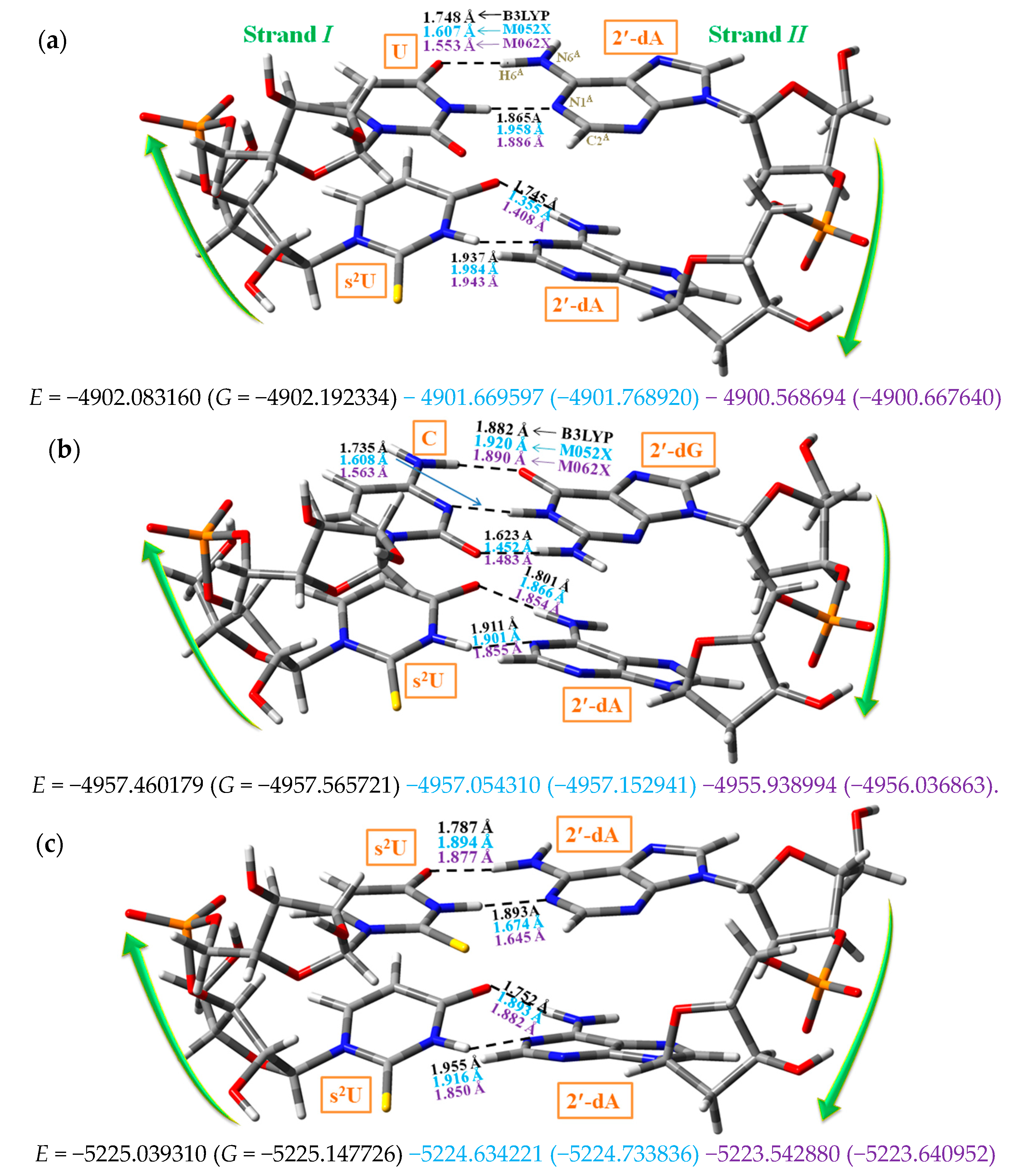
3.4. WC Base Pairs with the Nucleosides 2-Thiouridine, 4-Thiouridine and 2,4-Dithiouridine
Effects of the Sulphur Atom on the Geometrical Parameters of the WC Pair
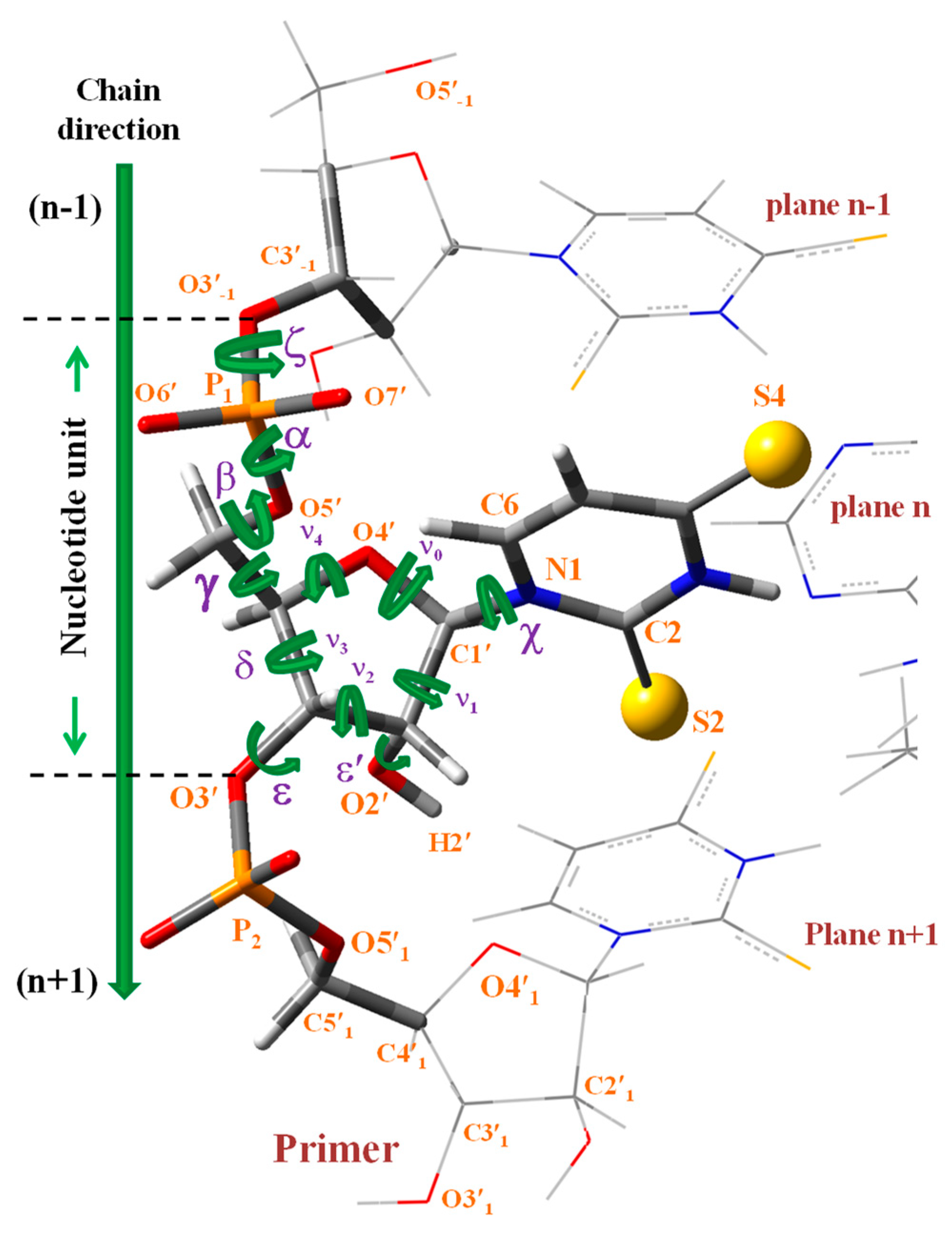
3.5. MicrohelixesDNA:RNA with the Sulphur Atom
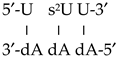 with the two strands was represented by only one of the strands (the RNA), such as this notation: 5′-U s2U U-3′. In this way the helix:
with the two strands was represented by only one of the strands (the RNA), such as this notation: 5′-U s2U U-3′. In this way the helix: 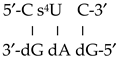 was represented by this notation: 5′-C s4U C-3′, and the helix:
was represented by this notation: 5′-C s4U C-3′, and the helix: 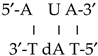 (where T = thymidine) by 5′-A U A-3′, and so on with the remaining structures included in these Table 6 and Tables S1–S4, i.e., in the notation used for the microhelixes the RNA strand was only represented.
(where T = thymidine) by 5′-A U A-3′, and so on with the remaining structures included in these Table 6 and Tables S1–S4, i.e., in the notation used for the microhelixes the RNA strand was only represented.3.5.1. Different Inter- and Intra-Molecular H-Bonds Found in the Simulated Microhelixes
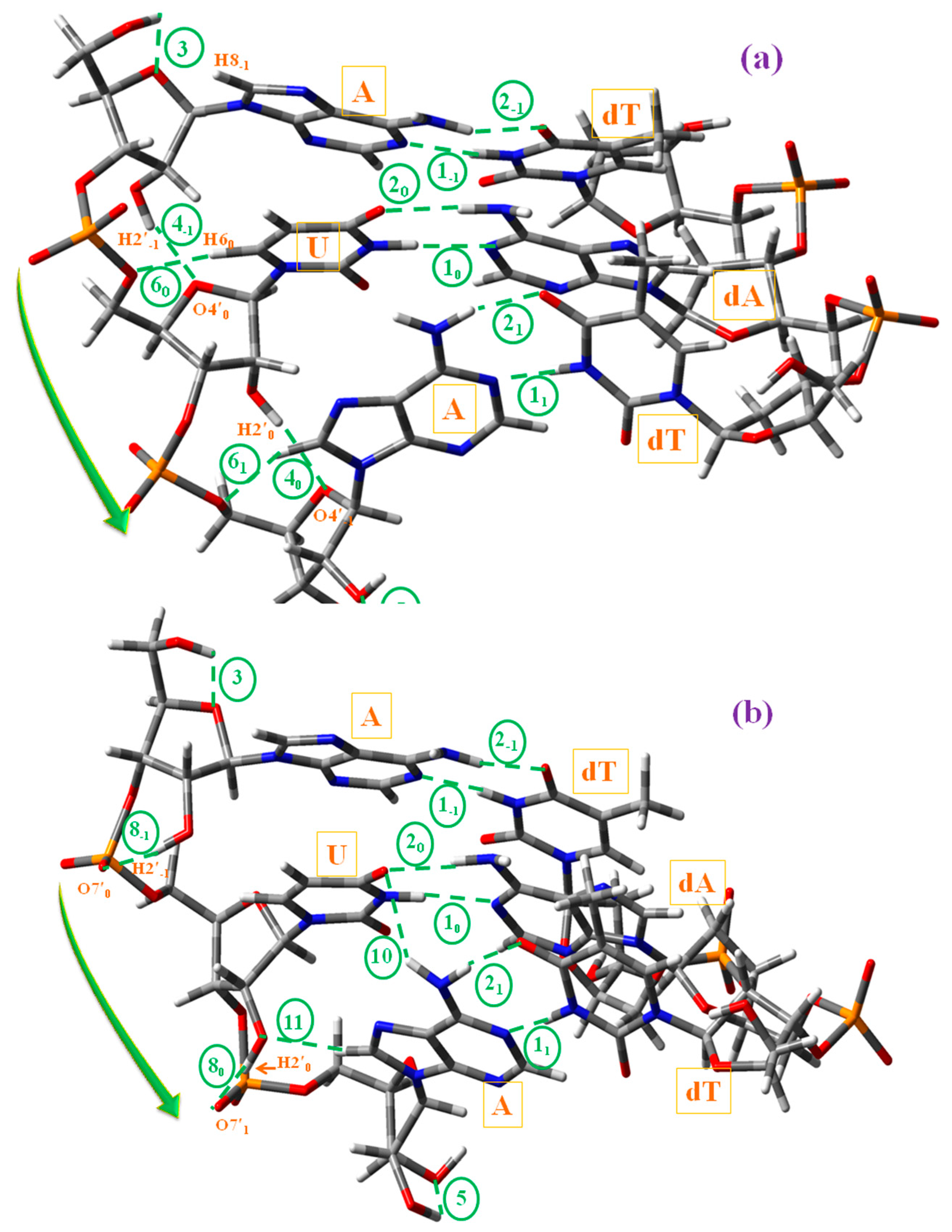
- (i)
- With uridine/s2U/s4U/s2,4U and adenosine H-bond ① corresponds to N3-H3···N1A and H-bond ② to O4···H6A-N6A. Both H-bonds were the most important and responsible for the double helix maintenance. H-bond ① was found slightly longer than H-bond ②. With the cytidine (C) and guanosine (G) nucleosides appeared a new H-bond ⑫ (not included in Scheme 2), N4C–H4C···O6G between NH2 of C and O6 oxygen of G.
- (ii)
- H-bond ③ refers to O5′−1–H5′−1···O4′−1 in the plane n − 1, and it appeared by rotation of the O5′H group to the most stable orientation β+. This H-bond will not be observed in the helix.
- (iii)
- H-bonds ④, ⑧ and ⑭ were responsible for the formation of the A-, B-and C-types microhelixes, respectively. These are very important H-bonds and appear as a consequence of the high reactivity of the H2′n hydroxyl hydrogen bonded to O4′n+1 (A-type), to O7’n+1 (B-type), or to O5′n+1 (C-type). Of these H-bonds, the strongest one corresponds to H-bond ⑧.
- (iv)
- H-bond ⑤ refers to O2′1–H2′1···O3′1 of the primer (plane n + 1) and it appeared only on the RNA strand.
- (v)
- H-bond ⑥ corresponded to C6–H6···O5′ in pyrimidines and C8A–H8A···O5′A in purines. It is a weak H-bond/contact that was found only in A-type microhelixes, which increased the stabilization of this microhelix together with H-bond ④.
- (vi)
- The weak H-bond ⑦ referred to C–H···O, and it was found only in the plane n − 1 and in the 5′-UUU-3′ microhelix of A-type (not included in Scheme 2). This H-bond would not be observed with the rising of the microhelix.
- (vii)
- H-bond ⑩ corresponded to N-H···O between the NH2 amino hydrogen of adenosine in plane n + 1 and the oxygen atom of uridine in plane n. It appeared only in the 5′-AUA-3′ microhelix of B-type.
- (viii)
- The weak H-bond ⑪ referred to C-H···O between the hydrogen H8 of adenosine of the plane n + 1 and O2′ of the ribose ring of plane n. It is found in the RNA strand of 5′-AUA-3′microhelixes of B-type.
- (ix)
- The intra-strand H-bond ⑬ corresponds to N-H···N between the NH2 amino group of cytosine (plane n − 1) and adenine (plane n; not included in Scheme 2), which was found in 5′-CUC-3′microhelixes.
3.5.2. Types of DNA:RNA Hybrid Microhelixes Optimized
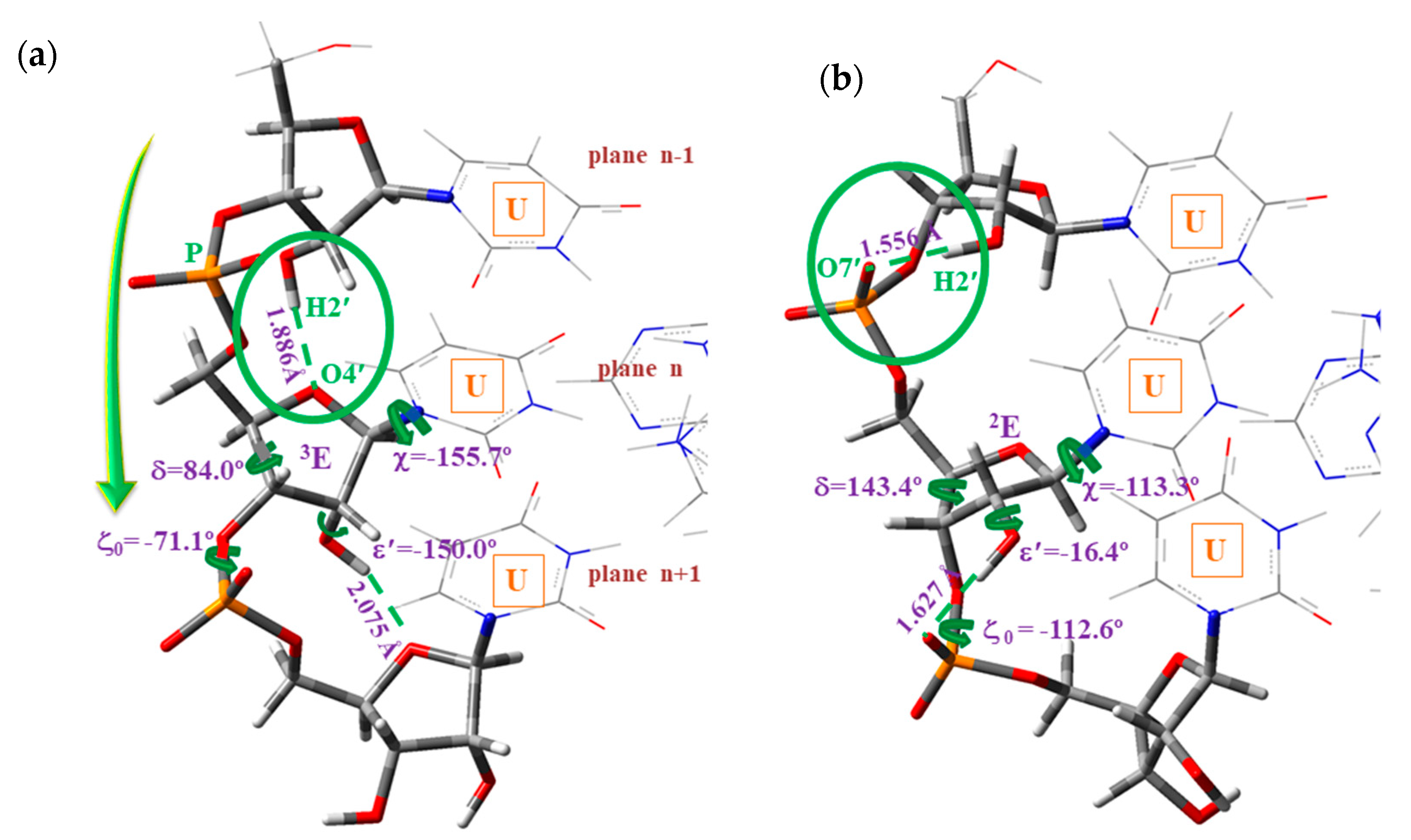
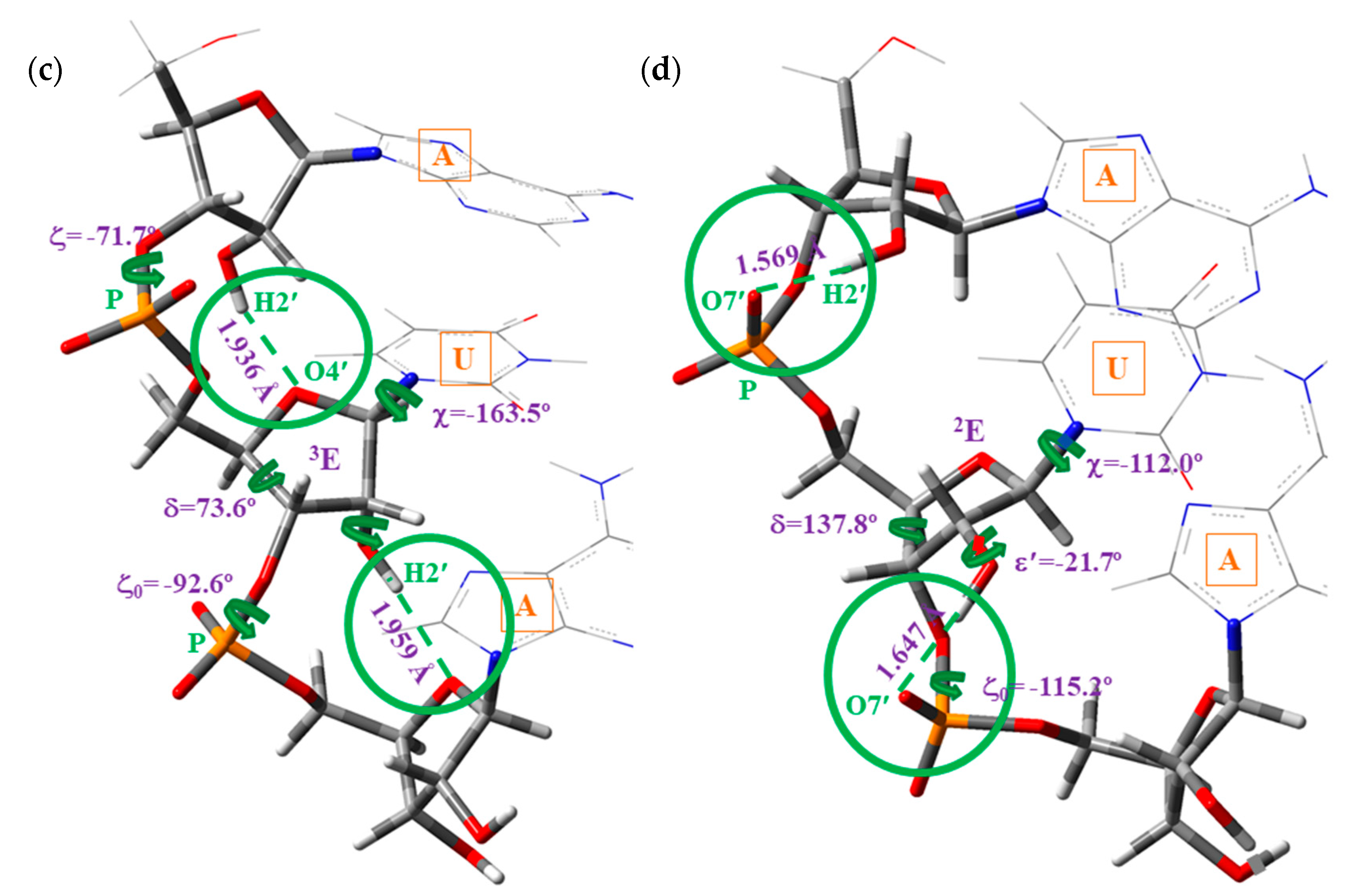
- (i)
- A-type: Corresponds to the O2′–H2′(n)···O4′(n+1) H-bond ④ between contiguous nucleotides of the same strand, Scheme 3a,c, and with the pyrimidine ring in high-anti orientation relative to the furanose ring.
- (ii)
- B-type: Corresponds to the O2′–H2′(n)···O7′(n+1) H-bond ⑧ between contiguous nucleotides of the same strand, Scheme 3b,d, and with the pyrimidine ring appearing in anti orientation related to the furanose ring. These microhelixes always appear remarkably more stable than A-type microhelix, mainly because H-bond ⑧ is noticeably stronger than H-bond ④.
- (iii)
- C-type: Corresponds to the intra-strand O2′–H2′(n)···O5′(n+1) H-bond ⑬ between adjacent nucleotides, Scheme S1a. This type of microhelix was only optimized in 5′-AUA-3′, and because it was significantly less stable than both A-type and B-type microhelixes, therefore its values were omitted in the present manuscript.
- (iv)
- D-type: Corresponds to the O2′–H2′(n)···O3′(n) H-bond/interaction in the same nucleotide. This type of microhelix was found only stable when it was mixed with C-type in 5′-AUA-3′, Scheme S1b. Because it was the least stable type of the simulated microhelixes its values were also omitted in the present manuscript.
3.5.3. Effect on the Helical Parameters of the Mixed Purine–Pyrimidine Stacks Versus Purine–Purine or Pyrimidine–Pyrimidine Stacks
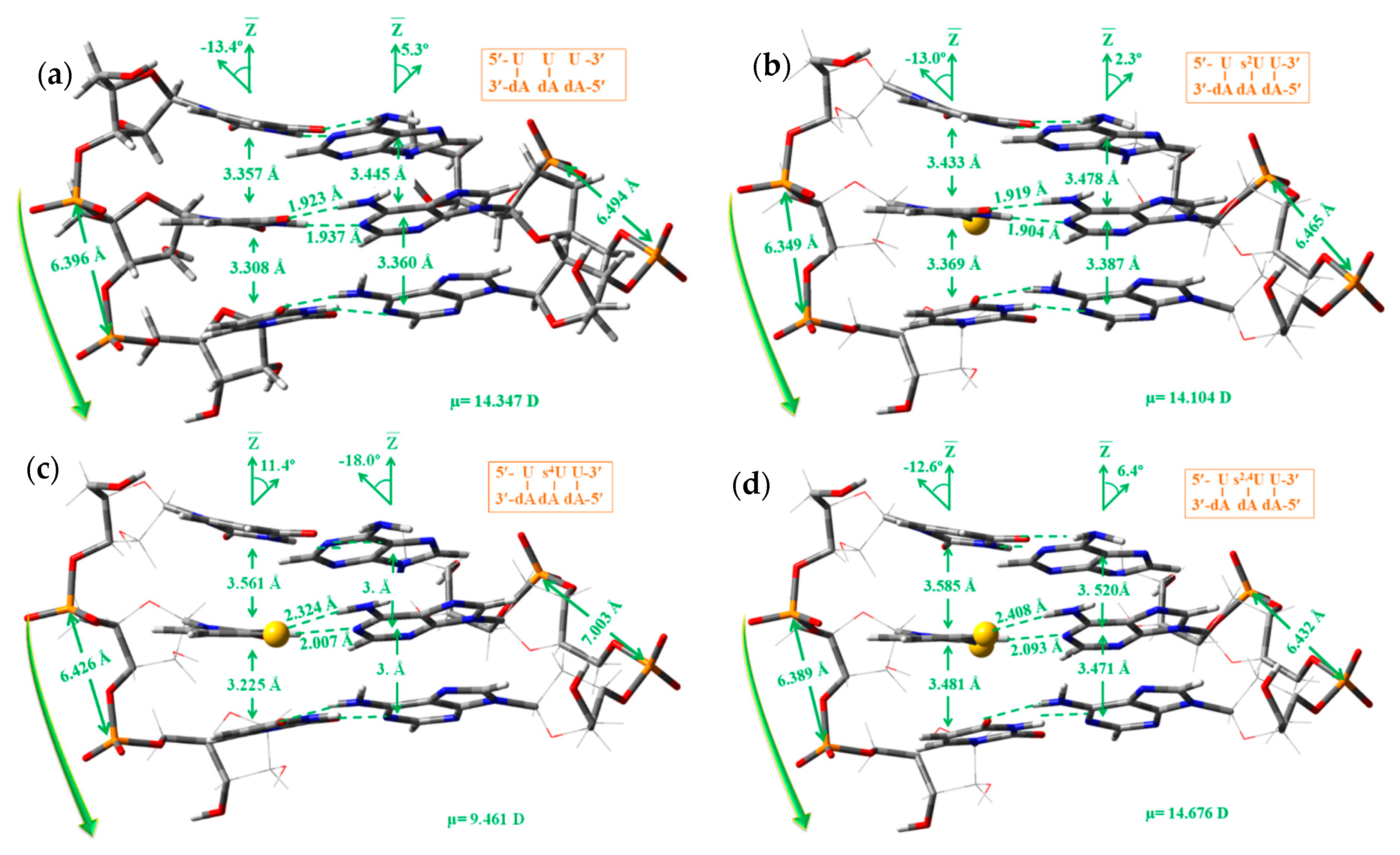
- (i)
- The microhelix 5′-AUA-3′ was remarkably more stable than 5′-UUU-3′ in both the A-type and B-type, i.e., helixes in which the nucleotides appeared noticeably mixed in each strand were more stable than those in which the mixing was small. It was due to the highest H-bond interactions and stacking interactions between the different planes of nucleotides in mixed strands.
- (ii)
- The planarity of the pyrimidine ring in strand I was noticeably reduced in the A-type and increased in the B-type as compared to in 5′-UUU-3′. This increment in planarity in the B-type was because of the H-bond ⑩ which deformed the planarity of the uracil ring and weakened the H-bonds ① and ②.
- (iii)
- The β and ε′ exocyclic torsional angles in the B-type helix were slightly incremented in ca. 5°, and α and δ slightly decreased by a similar amount ca. 5°, as compared to in 5′-UUU-3′. The differences in these parameters were small in the A-type.
- (iv)
- The exocyclic torsional angle ζ was noticeably reduced in B-type, 19°, but only very small, 1°, in the A-type. The highest change in A-type corresponded to the ε torsional angle, 50.2° in 5′-AUA-3′ vs. −160.0° in 5′-UUU-3′. The pseudorotation phase angle P was also noticeably changed, with a change in the symmetry of the furanose ring, from 2E to in B-type and from 3E to in A-type.
- (v)
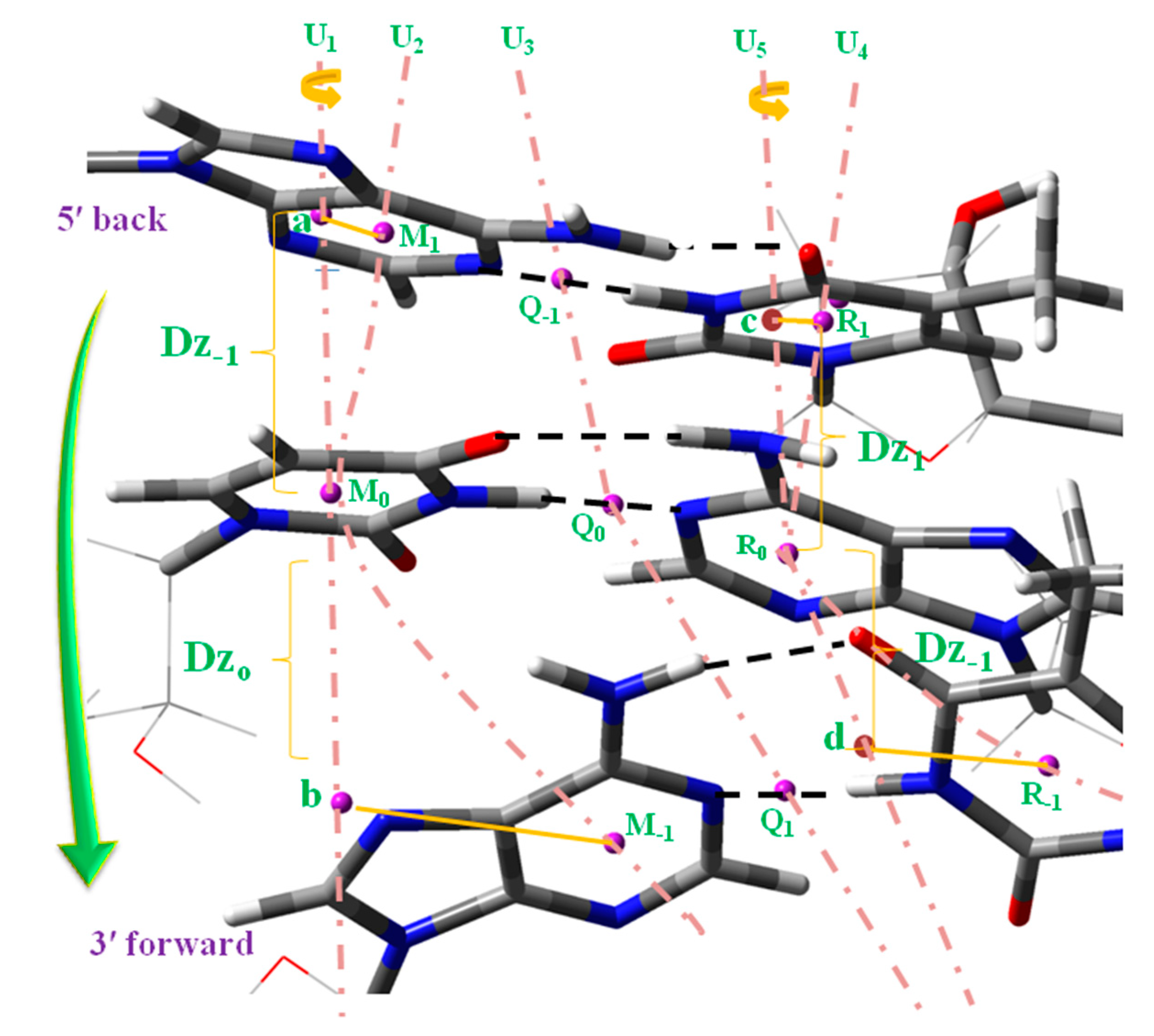
- The rise parameter Dz between the base pair planes (Scheme 4), as well as the diameter of the helix d and the helical radius R, were noticeably increased in the B-type helix 5′-AUA-3′. This feature was also observed in the A-type but in less quantity, i.e., the regions of the ADN:RNA helix not mixed in each strand would appear more compressed.
- (vi)
- The torsional angle N–C···C–N of the base pair appeared significantly reduced, as well as the propeller twist θp dihedral angle, the inclination parameter INC of the base pair related to the origin of the microhelix axis O0, the torsional angle aM0O0Z between the helical axis through O0 and the orthogonal axis to the uracil ring plane n, and the torsional angle cR0O0Z between the helical axis through O0 and the orthogonal axis to the adenine ring plane n.
- (vii)
- The dipole moment was remarkably reduced in the A-type, 14.347 D in 5′-UUU-3′ vs. 6.358 D in 5′-AUA-3′, i.e., the regions of the ADN:RNA helix not mixed (with the same nucleoside) in each strand appeared more available for the water molecules to enter inside of the helix, and therefore these regions appeared more easy to interact with pharmacological compounds soluble in water.
3.5.4. Effect of the Sulphur Atom in the Microhelix
- (i)
- A slight increment in the non-planarity of the uracil ring, especially in the torsional angles involving N1 atom in the microhelixes A-type and N3 atom in B-type. The highest effect was observed in the 5′-A s2,4U A-3′ microhelix of B-type.
- (ii)
- A slight increase of the 3E pucker, especially in microhelixes 5′-U s2UU-3′ of A-type, while it was decreased with s4U. This feature is in accordance to the increment observed in the 3E pucker abundance when s2U is substituted in the anticodon loop of tRNA [66]. This s2U substitution appeared to produce a small increase (2.9 kJ/mol) in the stability of s2U:A base pair as compared to U:A in RNA duplexes [7].
- (iii)
- A small change of ca. 2° in the exocyclic torsional angles of the microhelix A-type, and a large change in B-type, in especial ca. 8° in ζ with the S2 substitution in 5′-UUU-3′, and ca. 13° with s2,4. These changes were smaller when the sulphur atom was inserted into 5′-AUA-3′. However, these changes were not significant for a very flexible helix, because we have found [28] that the highest changes appear with the substitution of uridine by cytidine, such as in the 5′-CUC-3′ microhelix of A-type, with changes of ca. 30° in ζ and ca. 40° in β, as compared to changes in 5′-UUU-3′, and with values slightly lower in the B-type.
- (iv)
- With the s2U nucleoside, a lengthening of the intermolecular N3–H3···N1AH-bond ①, and a small shortening of the O4···H6A–N6A H-bond ② was observed in 5′-AUA-3′ of the A-type, and shortening of both H-bonds in 5′-UUU-3′. However, with the s4U and s2,4U nucleosides both H-bonds were always remarkably lengthened. In microhelixes of B-type the changes with s2U were negligible, but similar with s4U and s2,4U, Table 6.
- (v)
- A slight increment of the rise parameter Dz (base stacking) in the 5′-UUU-3′ microhelixes, especially with s4U and s2,4U nucleotides.
- (vi)
- A slight lengthening of the helix diameter d in A-type microhelixes (with the exception of s4U) and shortening in B-type, with the exception of s2U. The helical radius R always increased with the exception of s4 in 5′-AUA-3′ of the B-type. A general slight deformation in the helical parameters INC and θp was also observed.
- (vii)
- A slight increment in the dipole moment (μ) in the B-type, in accordance to a higher stability of the RNA helixes with sulphur was observed in water solution [26,66]. However, in the A-type a trend was not observed, and thus in this case the effect of sulphur could not be considered of special interest because of the very large range of observed values of μ, from 17.4 D in 5′-UUU-3′ to 6.4 D in 5′-AUA-3′.
3.5.5. Hydration of Few Microhelixes
- (i)
- (ii)
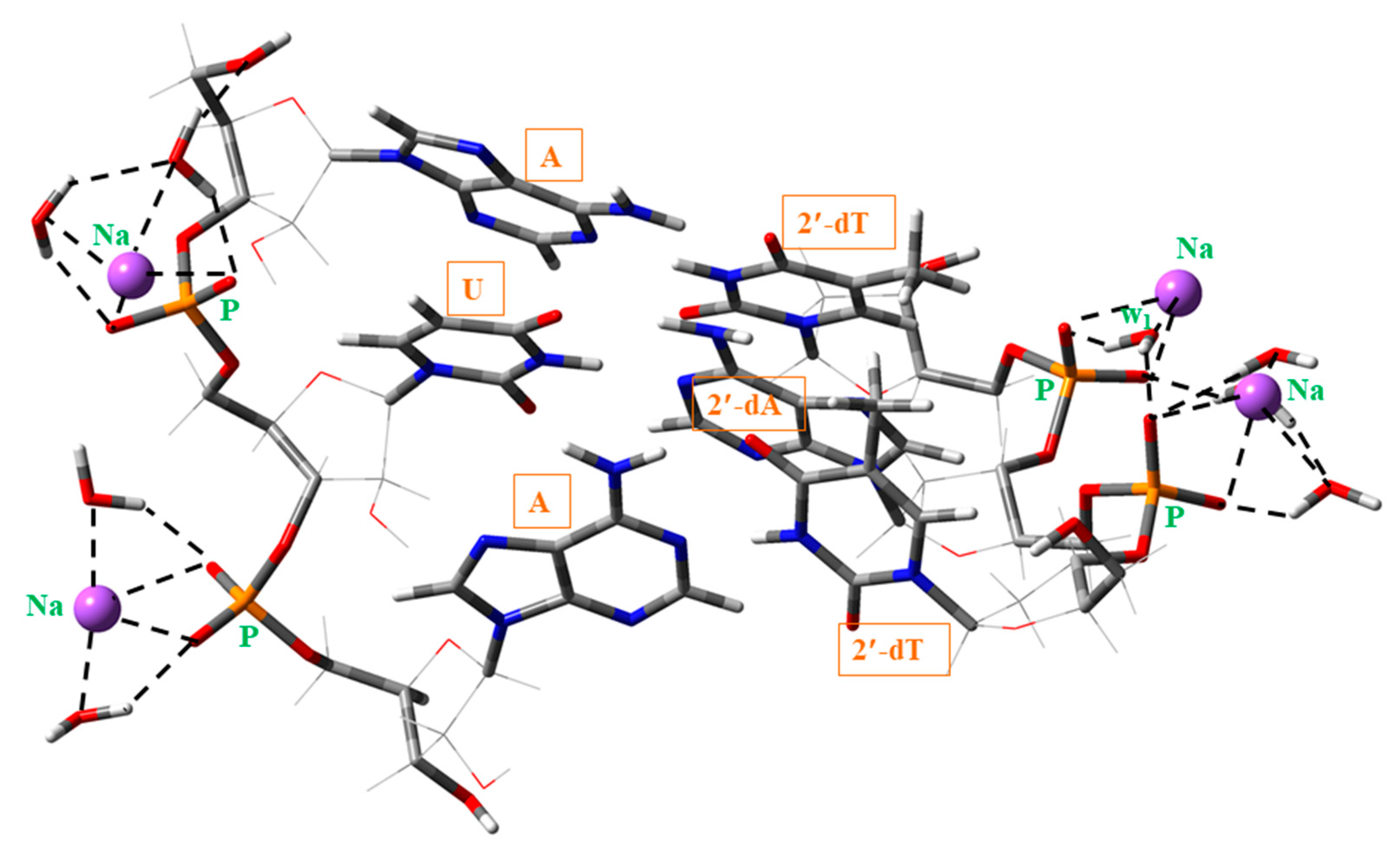
- A slight compression of the microhelix by the water molecules, with a shortening of the P1···P2 (~0.15 Å) and P3···P4 (~1 Å) distances, the helix diameter d (~0.4 Å) and the helical radius R (~0.1 Å). The very large shortening in the P3···P4 distance (strand II) was due to the strong H-bond of the water molecule w1 (Figure 10), which acts as a bridge between the oxygen atoms of the neighboring phosphate groups, and therefore approaching both phosphate groups. This compression of the helix also led to a noticeable modification in other backbone parameters, Table S2, especially those in strand II.
- (iii)
- The compression of the microhelix also affected the rise parameter Dz, Table S3, with a noticeable decrease in its value, 0.03 Å in Dz−1 and 0.24 Å in Dz0.
- (iv)
- A noteworthy change in the propeller twist θp (~9°) and a small increment in the aMoOoZ (~2°) and cMoOoZ (~3°) helical parameters.
- (v)
- A small increment of ca. 2° in the furanose pucker P, but it was enough to change its symmetry to 3E.
- (vi)
- A strong increment of the dipole moment μ, ca. 3 D, by the water molecules.
4. Summary and Conclusions
- The ring in the uracil molecule appeared easier to be deformed than when it was thio-substituted. This high flexibility of the ring in uracil molecule could explain the great biological importance of this molecule, in special for the genetic code. In contrast, a lower flexibility of the uracil ring with thio-substitution could explain the less biological importance of these thio-molecules.
- The water molecules slightly affected the planarity of the uracil ring, and modified its stability. In the isolated state 2TU was slightly less stable than 4TU, but with the hydration it was the reverse. With the progress of the hydration, the trend in the different geometrical parameters appeared in general similar among uracil and its thio-substituted derivatives.
- Molecular docking of 2-thiouracil against three different pathogens such as Bacillus subtilis, Escherichia coli and Candida albicans revealed a strong intermolecular H-bonded interaction between the N–H site of ASN33 and O atom of 2TU and with the N–H site of GLU32 and O atom of 2TU. Thus, 2TU could be used to be more effective on gram negative strains.
- With the sulphur-substitution the H-bonds of the WC pairs appeared somewhat weakened, especially on O4 position, slightly destabilizing the pair, i.e., the pairs with the sulphur atom inserted in the uracil ring were less stable than those in the canonical form.
- Depending on where the 2′-OH group was H-bonded, four types of microhelixes were optimized. However, here only the A-type and B-type were analyzed in detail because of their higher stability. A-type helixes are characterized by the intra-strand H-bond 2′-OH(n)···O4′(n + 1) with the furanose ring atom O4′, while B-type helixes are described by the H-bond 2′-OH(n)···O7′(n) with the adjacent O7′ oxygen atom of the phosphate group.
- The A-type microhelix with only uridine in strand I (RNA) appeared to modify/prefer the sugar ring orientation of dA in strand II (DNA) as (C-type) instead of as 3E (A-type). Although both microhelixes were stable, however with C-type in strand II was more stable than with A-type, and it led to a lower deformation of the helix. However, with the C-type in strand I, the microhelix was noticeably less stable than with A-type.
- Purine–pyrimidine stacks such as in 5′-AUA-3′ appeared remarkably more stable than purine–purine or pyrimidine–pyrimidine stacks as in 5′-UUU-3′. That is, ADN:RNA not mixed helix regions were more compressed (higher Dz, d, R) and strongly rotated each base in the base-pair (higher θp, INC, aM0O0Z, cR0O0Z). In addition, these regions could be more available for water molecules and soluble pharmacological compounds to enter and interact with the helix.
- The base pairs with sulphur led to a slight increment in the non-planarity of the uracil ring, and to a small change in most of the exocyclic torsional angles. However, the change was noticeably large in the torsional angle ζ of B-type microhelixes. These effects did not appear to be significant in a very flexible helix with very large changes of ca. 40° when uridine was substituted by cytidine.
- The sulphur atom inserted in 5′-UUU-3′microhelixes, especially in s4 and s2,4 positions, led to a slight increment of the rise parameter Dz (base stacking), and to a slight lengthening of the helix diameter d in A-type microhelixes and shortening in B-type, with the exception of s2U. The helical radius R usually increased with the sulphur substitution. The sulphur atom also affected the helical parameters INC and θp, with a slight modification.
- The addition of Na atoms and water molecules on the microhelix led to a noticeable shortening of H-bond ① and slight lengthening of H-bond ② in 5′-AUA-3′. It also led to a slight compression of the microhelix with shortening of the P1···P2 and P3···P4 distances, the helix diameter d, the helical radius R and the rise parameter Dz. This compression of the microhelix could be only due to a very large modification of all the backbone parameters. This modification also affected the propeller twist θp (with a large change), and other helical parameters. A strong increment of the dipole moment µ of the cluster was also obtained with the hydration, especially in B-type microhelixes.
- The thio-uridine nucleosides generally had a larger µ than those with uridine. It means that in water solution the bonding of the thio-uridine nucleosides to the primer appeared facilitated over uridine nucleoside. Since the WC pair with the sulphur atom was weaker, it could delay (or deform) the growing of the RNA viral/tumoral, and therefore could explain the use of these thionucleo bases as pharmaceutical drugs.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| WC | Watson–Crick |
| DFT | Density Functional Theory |
| NBO | Natural Bond Orbital |
References
- Lipsett, M.N. Isolation of 4-thiouridylic acid from soluble ribonucleic acid of Escherichia coli. J. Biol. Chem. 1965, 240, 3975–3978. [Google Scholar] [PubMed]
- Dolak, L.; Sokolski, W.T.; Mizsak, S.; Stroman, D.W.; Sebek, O.K. Microbial formation of 4 thiouracil. Antimicrob. Agents Chemother. 1977, 11, 569–570. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ribeiro Da Silva, M.A.V.; Amaral, L.M.P.F.; Szterner, P. Experimental study on the thermochemistry of 2-thiouracil, 5-methyl-2-thiouracil and 6-methyl-2-thiouracil. J. Chem. Thermodyn. 2013, 57, 380–386. [Google Scholar] [CrossRef]
- Al-Masoudi, N.A.; Saleh, B.A.; Abdul Karim, N.; Issa, A.Y.; Pannecouque, C. Synthesis and anti-HIV activity of new 2-thiolumazine and 2-thiouracil metal complexes. Heteroat. Chem. 2011, 22, 44–50. [Google Scholar] [CrossRef]
- Palumbo, A.; d’Ischia, M.; Cioffi, F.A. 2-Thiouracil is a selective inhibitor of neuronal nitric oxide synthase antagonising tetrahydrobiopterin-dependent enzyme activation and dimerization. FEBS Lett. 2000, 485, 109–112. [Google Scholar] [CrossRef]
- Napolitano, A.; Palumbo, A.; d’Ischia, M.; Prote, G. Mechanism of Selective Incorporation of the Melanoma Seeker 2-Thiouracil into Growing Melanin. J. Med. Chem. 1996, 39, 5192–5201. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.T.; Fahrenbach, A.C.; Sheng, J.; Pian, J.L.; Szostak, J.W. Thermodynamic insights into 2-thiouridine-enhanced RNA hybridization. Nucleic Acids Res. 2015, 43, 7675–7687. [Google Scholar] [CrossRef] [PubMed]
- Roche, K.L.; Nukui, M.; Krishna, B.A.; O’Connor, C.M.; Murphy, E.A. Selective 4-thiouracil labeling of RNA transcripts within latently infected cells after infection with human cytomegalovirus expressing functional uracil phosphoribosyltransferase. J. Virol. 2018, 92, e00880. [Google Scholar] [CrossRef] [PubMed]
- Baptista, T.; Devys, D. Saccharomyces cerevisiae Metabolic Labeling with 4-thiouracil and the Quantification of Newly Synthesized mRNA As a Proxy for RNA Polymerase II Activity. J. Vis. Exp. JoVE 2018, 22, e57982. [Google Scholar] [CrossRef] [PubMed]
- Knüppel, R.; Kuttenberger, C.; Ferreira-Cerca, S. Toward time-resolved analysis of RNA metabolism in archaea using 4-thiouracil. Front. Microbiol. 2017, 8, 286. [Google Scholar] [CrossRef]
- Wyrzykiewicz, E.; Nowakowska, Z.; Bartkowiak, G.; Kedzia, B. Synthesis and antimicrobial properties of s-substituted derivatives of 4-thiouracil. Pol. J. Chem. 1997, 71, 201–206. [Google Scholar]
- Alcolea Palafox, M.; Rastogi, V.K.; Tanwar, R.P.; Mittal, L. Vibrational frequencies and structure of 2-thiouracil by Hartree-Fock, post-Hartree-Fock and density functional methods. Spectrochim. Acta A 2003, 59, 2473–2486. [Google Scholar] [CrossRef]
- Alcolea Palafox, M.; Rastogi, V.K.; Singh, S.P. Effect of the Sulphur Atom on Geometry and Spectra of the Biomolecule 2-Thiouracil and in the WC Base Pair 2-Thiouridine-Adenosine. Influence of water in the first hydration Shell. J. Biomol. Struct. Dyn. 2018, 36, 1225–1254. [Google Scholar] [CrossRef] [PubMed]
- Yekeler, H. Ab initio study on tautomerism of 2-thiouracil in the gas phase and in solution. J. Comput. Aided Mol. Des. 2000, 14, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Marino, T.; Russo, N.; Sicilia, E.; Toscano, M. Tautomeric equilibria of 2- and 4-thiouracil in gas phase and in solvent: A density functional study. Int. J. Quantum Chem. 2001, 82, 44–52. [Google Scholar] [CrossRef]
- Yang, W.; Hu, Y. Conformations of 2-thiouracil in the aqueous solution and its adsorption behavior on the gold substrates explored by DFT calculations and experimental methods. Spectrochim. Acta A 2015, 134, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Rubin, Y.V.; Morozov, Y.; Venkateswarlu, D.; Leszczynski, J. Prototropic equilibria in 4-thiouracil: A combined spectroscopic and ab initio SCF-MO investigation. J. Phys. Chem. A 1998, 102, 2194–2200. [Google Scholar] [CrossRef]
- Zou, X.; Dai, X.; Liu, K.; Zhao, H.; Song, D.; Su, H. Photophysical and photochemical properties of 4-thiouracil: Time-resolved IR spectroscopy and DFT studies. J. Phys. Chem. B 2014, 118, 5864–5872. [Google Scholar] [CrossRef]
- Qiu, Z.; Xia, Y.; Wang, H.; Diao, K. MP2 study on the hydrogen-bonding interactions between 4-thiouracil and four RNA bases. Struct. Chem. 2010, 21, 99–105. [Google Scholar] [CrossRef]
- Prasanthkumar, K.P.; Suresh, C.H.; Aravindakumar, C.T. Dimer radical cation of 4-thiouracil: A pulse radiolysis and theoretical study. J. Phys. Org. Chem. 2013, 26, 510–516. [Google Scholar] [CrossRef]
- Shukla, M.K.; Leszczynski, J. A theoretical study of hydration of 4-thiouracil in the electronic singlet excited state. J. Mol. Struct. 2006, 771, 149–155. [Google Scholar] [CrossRef]
- Khvorostov, A.; Lapinski, L.; Rostkowska, H.; Nowak, M.J. Unimolecular photochemistry of 4-thiouracils. Photochem. Photobiol. 2005, 81, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Jaiswal, S.; Kumar, M.; Singh, P.; Srivastav, G.; Yadav, R.A. DFT study of molecular geometries and vibrational characteristics of uracil and its thio-derivatives and their radical cations. Spectrochim. Acta A 2010, 75, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, F.; Xu, W.; Liu, C. The effects of incorporating 2,4-dithiouracil into uracil tetrad: A theoretical study. J. Mol. Struct. (Theochem) 2005, 716, 137–141. [Google Scholar] [CrossRef]
- Rastogi, V.K.; Alcolea Palafox, M.; Guerrero-Martínez, A.; Tardajos, G.; Vats, J.K.; Tomer, R. Raman and infrared spectra of hydrated 2,4-dithiouracil molecule. AIP Conf. Proc. 2010, 1267, 629–630. [Google Scholar]
- Testa, S.M.; Disney, M.D.; Turner, D.H.; Kierzek, R. Thermodynamics of RNA−RNA duplexes with 2- or 4-thiouridines: Implications for antisense design and targeting a group I intron. Biochemistry 1999, 38, 16655–16662. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.K.; Davis, D.R. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997, 25, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Alcolea Palafox, M. Effect of the sulfur atom on S2 and S4 positions of the uracil ring in different DNA:RNA hybrid microhelixes with three nucleotide base pairs. Biopolymers 2019, 110, e23247. [Google Scholar] [CrossRef]
- Gyi, J.I.; Lane, A.N.; Conn, G.L.; Brown, T. The orientation and dynamics of the C2′-OH and hydration of RNA and DNA·RNA hybrids. Nucleic Acids Res. 1998, 26, 3104–3110. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, D.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Ortiz, S.; Alvarez-Ros, M.C.; Alcolea Palafox, M.; Rastogi, V.K.; Balachandran, V.; Rathor, S.K. FT-IR and FT-Raman spectra of 6-chlorouracil: Molecular structure, tautomerism and solid state simulation. A comparison between 5-chlorouracil and 6-chlorouracil. Spectrochim. Acta A 2014, 130, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Cuellar, A.; Alcolea Palafox, M.; Rastogi, V.K.; Kiefer, W.; Schlücker, S.; Rathor, S.K. FT-IR and FT-Raman spectra of 5-fluoroorotic acid with solid state simulation by DFT methods. Spectrochim. Acta A 2014, 132, 430–445. [Google Scholar] [CrossRef] [PubMed]
- Ponomareva, A.G.; Yurenko, Y.P.; Zhurakivsky, R.O.; van Mourik, T.; Hovorun, D.M. Structural and energetic properties of the potential HIV-1 reverse transcriptase inhibitors d4A and d4G: A comprehensive theoretical investigation. J. Biomol. Struct. Dyn. 2014, 32, 730–740. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.A.; Tamara Molina, A.; Álvarez-Ros, M.C.; Alcolea Palafox, M. Conformational analysis of the anti-HIV Nikavir prodrug: Comparisons with AZT and Thymidine, and establishment of structure-activity relationships/tendencies in other 6′-derivatives. J. Biomol. Struct. Dyn. 2015, 33, 723–748. [Google Scholar] [CrossRef] [PubMed]
- Alcolea Palafox, M. Molecular structure differences between the antiviral Nucleoside Analogue 5-iodo-2′-deoxyuridine and the natural nucleoside 2′-deoxythymidine using MP2 and DFT methods: Conformational analysis, crystal simulations, DNA pairs and possible behaviour. J. Biomol. Struct. Dyn. 2014, 32, 831–851. [Google Scholar] [CrossRef] [PubMed]
- Danilov, V.I.; Anisimov, V.M.; Kurita, N.; Hovorun, D.M. MP2 and DFT studies of the DNA rare base pairs: The molecular mechanism of the spontaneous substitution mutations conditioned by tautomerism of bases. Chem. Phys. Lett. 2005, 412, 285–293. [Google Scholar] [CrossRef]
- Danilov, V.I.; van Mourik, T.; Kurita, N.; Wakabayashi, H.; Tsukamoto, T.; Hovorun, D.M. On the Mechanism of the Mutagenic Action of 5-Bromouracil: A DFT Study of Uracil and 5-Bromouracil in a Water Cluster. J. Phys. Chem. A 2009, 113, 2233–2235. [Google Scholar] [CrossRef] [PubMed]
- Alcolea Palafox, M.; Posada-Moreno, P.; Villarino-Marín, A.L.; Martinez-Rincon, C.; Ortuño-Soriano, I.; Zaragoza-García, I. DFT Calculation of four new potential agents muscarinic of bispyridinium type: Structure, synthesis, biological activity, hydration, and relations with the potents W84 and DUO-3O. J. Comput. Aided Mol. Des. 2011, 25, 145–161. [Google Scholar] [CrossRef]
- Pullman, B. Quantum Theory of Chemical Reactions; Daudel, R., Ed.; Reidel Publishing Co.: Amsterdam, The Netherlands, 1980. [Google Scholar]
- Alcolea Palafox, M.; Rastogi, V.K.; Kumar, H.; Kostova, I.; Vats, J.K. Tautomerism in 5-bromouracil: Relationships with other 5-haloderivatives and effect of the microhydration. Spectrosc. Lett. 2011, 44, 300–306. [Google Scholar] [CrossRef]
- Muñoz-Freán, S.; Alcolea Palafox, M.; Rastogi, V.K. Effect of the microhydration on the tautomerism in the anticarcinogenic drug 5-Fluorouracil and relationships with other 5-haloderivatives. J. Mol. Struct. 2013, 1054–1055, 32–46. [Google Scholar] [CrossRef]
- Alcolea Palafox, M.; Iza, N.; de la Fuente, M.; Navarro, R. Simulation of the first hydration shell of nucleosides D4T and Thymidine: Structures obtained using MP2 and DFT methods. J. Phys. Chem. B 2009, 113, 2458–2476. [Google Scholar] [CrossRef] [PubMed]
- Alcolea Palafox, M. Anticancer drug IUdR and other 5-halogen derivatives of 2′-deoxyuridine: Conformers, hydrates and structure-activity relationships. Struct. Chem. 2014, 25, 53–69. [Google Scholar] [CrossRef]
- Yang, J.M.; Chen, C.C. GEMDOCK: A generic evolutionary method for molecular docking. Proteins 2004, 55, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.M.; Shen, T.W. A pharmacophore-based evolutionary approach for screening selective estrogen receptor modulators. Proteins 2005, 59, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- DeLano, W.R. The PyMOL Molecular Graphics System, Version 1.7.4.5; Schrödinger, LLC: New York, NY, USA, 2010. [Google Scholar]
- Chu, C.; Das, K.; Tyminski, J.R.; Bauman, J.D.; Guan, R.; Qiu, W.; Montelione, G.T.; Arnold, E.; Shatkin, A.J. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA 2011, 108, 10104–10108. [Google Scholar] [CrossRef] [PubMed]
- Protein Data Bank. Available online: http://www.rcsb.org/pdb (accessed on 15 February 2019).
- Tiekink, E.R.T. Crystal-structure of 2-thiouracil. Z. Kristallogr. 1989, 187, 79–84. [Google Scholar] [CrossRef]
- Shishkin, O.V.; Gorb, L.; Leszczynski, J. Conformational flexibility of pyrimidine rings of nucleic acid bases in polar environment: PCM study. Struct. Chem. 2009, 20, 743–749. [Google Scholar] [CrossRef]
- Rejnek, J.; Hanus, M.; Kabelác, M.; Ryjácek, F.; Hobza, P. Correlated ab initio study of nucleic acid bases and their tautomers in the gas phase, in a microhydrated environment and in aqueous solution. Part 4. Uracil and thymine. Phys. Chem. Chem. Phys. 2005, 7, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Awad, S.M.; Zohny, Y.M.; Ali, S.A.; Mahgoub, S.; Said, A.M. Design, synthesis, molecular modeling, and biological evaluation of novel thiouracil derivatives as potential antithyroid agents. Molecules 2018, 23, 2913. [Google Scholar] [CrossRef] [PubMed]
- Vassart, G.; Dumont, J.E. The thyrotropin receptor and the regulation of thyrocyte function and growth. Endocr. Rev. 1992, 13, 596–611. [Google Scholar] [PubMed]
- Lyshchik, A.; Higashi, T.; Asato, R.; Tanaka, S.; Ito, J.; Mai, J.J.; Pellot-Barakat, C.; Insana, M.F.; Brill, A.B.; Saga, T.; et al. Thyroid gland tumor diagnosis at US elastography. Radiology 2005, 237, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Misra, K. Modeling and simulation analysis of propyl-thiouracil (PTU), an anti-thyroid drug on thyroid peroxidase (TPO), thyroid stimulating hormone receptor (TSHR), and sodium iodide (NIS) symporter based on systems biology approach. Netw. Model. Anal. Health Inform. Bioinform. 2013, 2, 45–57. [Google Scholar] [CrossRef]
- Davies, T.F.; Latif, R.; Yin, X. New genetic insights from autoimmune thyroid disease. J. Thyroid. Res. 2012, 5, 623852. [Google Scholar] [CrossRef]
- Farid, N.R.; Szkudlinski, M.W. Minireview: Structural and functional evolution of the thyrotropin receptor. Endocrinology 2004, 145, 4048–4057. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Raaka, B.M.; Gershengorn, M.C. Human TSH receptor ligands as pharmacological probes with potential clinical application. Expert Rev. Endocrinol. Metab. 2009, 4, 669–679. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruf, J.; Carayon, P. Structural and functional aspects of thyroid peroxidase. Arch. Biochem. Biophys. 2006, 445, 269–277. [Google Scholar] [CrossRef]
- Roy, G.; Mugesh, G. Selenium analogues of antithyroid drugs–recent developments. Chem. Biodivers. 2008, 5, 414–439. [Google Scholar] [CrossRef]
- Dohan, O.; De la Vieja, A.; Paroder, V.; Riedel, C.; Artani, M.; Reed, M.; Ginter, C.S.; Carrasco, N. The sodium/iodide symporter (NIS): Characterization, regulation, and medical significance. Endocr. Rev. 2003, 24, 48–77. [Google Scholar] [CrossRef]
- Riesco-Eizaguirre, G.; Santisteban, P. A perspective view of sodium iodide symporter research and its clinical implications. Eur. J. Endocrinol. 2006, 155, 495–512. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, B.D.; Pal, A.; Del Frate, F.; Topkar, V.V.; Szostak, J.W. Replacing Uridine with 2-Thiouridine Enhances the Rate and Fidelity of Nonenzymatic RNA Primer Extension. J. Am. Chem. Soc. 2015, 137, 2769–2775. [Google Scholar] [CrossRef] [PubMed]
- Sochacka, E.; Szczepanowski, R.H.; Cypryk, M.; Sobczak, M.; Janicka, M.; Kraszewska, K.; Bartos, P.; Chwialkowska, A.; Nawrot, B. 2-thiouracil deprived of thiocarbonyl function preferentially base pairs with guanine rather than adenine in RNA and DNA duplexes. Nucleic Acids Res. 2015, 43, 2499–2512. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Principles in Nucleic Acid Structure; Springer: New York, NY, USA, 1984. [Google Scholar]
- Kozlov, I.A.; Orgel, L.E. Nonenzymatic Template-directed Synthesis of RNA from Monomers. Mol. Biol. 2000, 34, 781–789. [Google Scholar] [CrossRef]
- Fohrer, J.; Hennig, M.; Carlomagno, T. Influence of the 2′-hydroxyl group conformation on the stability of A-form helices in RNA. J. Mol. Biol. 2006, 356, 280–287. [Google Scholar] [CrossRef] [PubMed]
| Parameters | Isolated State | Hydrated with 30 H2O | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| U | 2TU | 4TU | 2,4DTU | U | 2TU | 4TU | 2,4DTU | |||
| B3LYP | B3LYP a | MP2 b | X-ray c | B3LYP | B3LYP | B3LYP | B3LYP | B3LYP | B3LYP | |
| Bond lengths | ||||||||||
| N1–C2 | 1.395 | 1.378 | 1.3769 | 1.338 (4) | 1.393 | 1.377 | 1.355 | 1.345 | 1.356 | 1.346 |
| C2–N3 | 1.384 | 1.368 | 1.3721 | 1.357 (4) | 1.39 | 1.374 | 1.362 | 1.352 | 1.363 | 1.354 |
| N3–C4 | 1.414 | 1.419 | 1.4126 | 1.389 (4) | 1.389 | 1.394 | 1.386 | 1.392 | 1.377 | 1.383 |
| C4–C5 | 1.46 | 1.458 | 1.4541 | 1.432 (5) | 1.443 | 1.443 | 1.426 | 1.427 | 1.415 | 1.416 |
| C5=C6 | 1.35 | 1.351 | 1.3527 | 1.338 (5) | 1.354 | 1.354 | 1.354 | 1.354 | 1.358 | 1.357 |
| N1–C6 | 1.375 | 1.375 | 1.3745 | 1.373 (4) | 1.374 | 1.374 | 1.365 | 1.366 | 1.362 | 1.363 |
| N1–H | 1.009 | 1.01 | 1.0092 | 0.90 (5) | 1.01 | 1.011 | 1.059 | 1.065 | 1.06 | 1.065 |
| C2=S/O | 1.217 | 1.665 | 1.6494 | 1.683 (3) | 1.215 | 1.662 | 1.264 | 1.72 | 1.263 | 1.718 |
| C4=S/O | 1.219 | 1.218 | 1.2264 | 1.227 (4) | 1.663 | 1.66 | 1.252 | 1.249 | 1.683 | 1.679 |
| C5–H | 1.081 | 1.081 | 1.0776 | 0.84 (4) | 1.08 | 1.08 | 1.079 | 1.08 | 1.079 | 1.081 |
| Bond angles | ||||||||||
| N–C2–N | 112.8 | 113.3 | 112.5 | 116.0 (3) | 112.9 | 113.3 | 116.7 | 116.8 | 116.6 | 116.7 |
| C–N3–C | 128.3 | 128.2 | 129 | 125.2 (3) | 128.1 | 128.1 | 124.6 | 124.6 | 124.6 | 124.7 |
| N–C4–C | 113.4 | 113.2 | 113 | 115.4 (3) | 114.1 | 113.9 | 116.1 | 116 | 116.3 | 116.2 |
| C–C5=C | 119.9 | 119.6 | 119.7 | 119.2 (3) | 120.2 | 119.9 | 118.7 | 118,6 | 118.9 | 118.8 |
| C2–N1–H | 114.8 | 115.1 | 115.1 | 118 (3) | 115 | 115.2 | 119.7 | 120.2 | 120.5 | 121.1 |
| N3–C2=S/O | 124.5 | 124.4 | 124.5 | 121.8 (2) | 123.9 | 123.9 | 121.7 | 122.5 | 121.8 | 122.7 |
| N1–C2=S/O | 122.7 | 122.3 | 123 | 122.2 (2) | 123.2 | 122.8 | 121.6 | 120.6 | 121.5 | 120.7 |
| C4–N3–H | 116.1 | 115.5 | 115.1 | 113 (2) | 117 | 116.4 | 117.8 | 116.8 | 118.5 | 117.3 |
| N3–C4=O | 120.3 | 120 | 120.2 | 119.2 (3) | 120.9 | 120.7 | 118.6 | 118.6 | 119.6 | 119.7 |
| C4–C5–H | 118.2 | 118.3 | 118.7 | 124 (3) | 118.5 | 118.7 | 121.2 | 121.5 | 121.5 | 121.6 |
| C5=C6–H | 122.7 | 123 | 123.1 | 126 (2) | 122.9 | 123.3 | 122.1 | 122.4 | 122 | 122.3 |
| Torsional angles | ||||||||||
| N1–C2–N3–C4 | 0 | −0.1 | 0 | −0.1 | −0.1 | 5.4 | 3.4 | 5.2 | 3 | |
| C2–N3–C4–C5 | 0 | 0.1 | 0 | 0.1 | 0.2 | −0.4 | 0 | 0.2 | 1.4 | |
| C6–N1–C2–N3 | 0 | 0.1 | 0 | 0 | 0 | −6.3 | −4.3 | −6.6 | −5 | |
| µ | 4.248 | 4.621 | 5.345 | 5.064 | 5.076 | 4.884 | 3.698 | 4.583. | 3.748 | |
| Atom | Isolated State | Hydrated with 30 H2O | |||||||
|---|---|---|---|---|---|---|---|---|---|
| U | 2TU a | 4TU | 2,4DTU | U | 2TU | 4TU | 2,4DTU | ||
| B3LYP | B3LYP | MP2 | B3LYP | B3LYP | B3LYP | B3LYP | B3LYP | B3LYP | |
| N1 | −0.643 | −0.592 | −0.693 | −0.634 | −0.585 | −0.612 | −0.574 | −0.605 | −0.569 |
| C2 | 0.824 | 0.229 | 0.382 | 0.826 | 0.224 | 0.846 | 0.327 | 0.846 | 0.322 |
| N3 | −0.687 | −0.634 | −0.738 | −0.633 | −0.583 | −0.639 | −0.602 | −0.601 | −0.565 |
| C4 | 0.654 | 0.653 | 0.819 | 0.023 | 0.016 | 0.682 | 0.681 | 0.098 | 0.092 |
| C5 | −0.398 | −0.383 | −0.435 | −0.340 | −0.325 | −0.381 | −0.357 | −0.328 | −0.301 |
| C6 | 0.029 | 0.029 | 0.117 | 0.035 | 0.034 | 0.08 | 0.075 | 0.086 | 0.077 |
| H1 | 0.452 | 0.462 | 0.472 | 0.455 | 0.464 | 0.477 | 0.484 | 0.477 | 0.484 |
| O2 | −0.619 | - | - | −0.609 | - | −0.771 | - | −0.767 | - |
| S2 | - | −0.178 | −0.235 | - | −0.157 | - | −0.358 | - | −0.353 |
| H3 | 0.455 | 0.464 | 0.477 | 0.453 | 0.471 | 0.474 | 0.48 | 0.478 | 0.484 |
| O4 | −0.586 | −0.576 | −0.686 | - | - | −0.719 | −0.710 | - | - |
| S4 | - | - | - | −0.116 | −0.094 | - | - | −0.243 | −0.228 |
| H5 | 0.259 | 0.272 | 0.271 | 0.277 | 0.28 | 0.273 | 0.277 | 0.28 | 0.284 |
| H6 | 0.25 | 0.254 | 0.252 | 0.253 | 0.257 | 0.265 | 0.266 | 0.264 | 0.266 |
| Type | Organisms | Kind | PDB Code | Binding Energy (kcal/mol) | H-Bond (kcal/mol) | VDW (kcal/mol) |
|---|---|---|---|---|---|---|
| Bacteria Fungi | Bacillus subtilis | G+ve | 4OZ5 | −59.59 | −20.52 | −39.078 |
| Escherichia coli | G-ve | 5C59 | −67.53 | −18.89 | −48.648 | |
| Candida albicans | - | 2Y7I | −60.21 | −27.65 | −32.569 |
| Ligand | Target Protein (Receptor) | Docking Parameters Based on the Rank | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Inhibition Constant (mM) | Intermolecular Energy (kcal/mol) | ||||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | ||
| 2,4-DTU | NIS | −4.81 | −4.55 | −3.74 | 0.29 | 0.46 | 1.81 | −4.81 | −4.55 | −3.74 |
| TSHR | −3.15 | −3.12 | −3.01 | 4.90 | 5.15 | 6.20 | −3.15 | −3.12 | −3.01 | |
| TPO | −2.99 | −2.95 | −2.94 | 6.47 | 6.89 | 6.99 | −2.99 | −2.95 | −2.94 | |
| MMI | NIS | −3.37 | −3.33 | −3.11 | 3.38 | 3.62 | 5.22 | −3.37 | −3.33 | −3.11 |
| Base Pair | Interaction s2U/U···(H)dA or H···dA | Modified Base Pair with s2U | Standard Base Pair with U |
|---|---|---|---|
| WC | O4···(H)N6 | 175.2 | 175.3 |
| N3(H)···N | 175.6 | 178.7 | |
| N3–H···N–C(dA) | −172.4 | 115.5 | |
| Reverse WC | O4···H–C(dA) | 140.7 | 134.1 |
| N3–H···N(dA) | 170.6 | 179.5 | |
| N3–H···N–C(dA) | −178.4 | 155.3 |
| Structure | Bond Lengths (Å) | Bond Angles (°) | Torsional Angles (°) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N1–C2 | C2=O/S | C2–N3 | N3–C4 | C4=O/S | N1–C2–N3 | C2–N3–C4 | N–C–N3–C | C–N3–C–C | C–N1–C–N | ||
| Nucleic acid bases···A | Uracil | 1.392 | 1.214 | 1.373 | 1.385 | 1.226 | 113.8 | 127.2 | 0.0 | 0.0 | 0.0 |
| 2TU | 1.374 | 1.662 | 1.360 | 1.395 | 1.222 | 114.4 | 126.5 | 0.0 | 0.0 | 0.0 | |
| 4TU | 1.388 | 1.213 | 1.382 | 1.370 | 1.665 | 114.1 | 126.4 | 0.0 | 0.0 | 0.0 | |
| 2,4DTU | 1.373 | 1.659 | 1.367 | 1.377 | 1.662 | 114.4 | 126.4 | −0.4 | 1.7 | −1.0 | |
| Nucleosides 1 pair···dA | U | 1.403 | 1.225 | 1.376 | 1.393 | 1.236 | 114.9 | 127.1 | 0.7 | −0.5 | −0.2 |
| U rev | 1.388 | 1.230 | 1.360 | 1.397 | 1.218 | 115.7 | 127.2 | 0.8 | −0.5 | −0.6 | |
| s2U | 1.390 | 1.677 | 1.370 | 1.399 | 1.233 | 115.2 | 127.2 | 1.1 | −0.4 | −0.8 | |
| s2U rev | 1.377 | 1.682 | 1.354 | 1.404 | 1.216 | 115.8 | 127.2 | 1.1 | 0.1 | −1.5 | |
| s4U | 1.390 | 1.219 | 1.378 | 1.370 | 1.670 | 115.3 | 126.4 | 0.7 | −0.5 | −0.2 | |
| s2,4U | 1.380 | 1.667 | 1.370 | 1.373 | 1.667 | 115.3 | 126.9 | 1.1 | −1.2 | −0.3 | |
| Nucleotides 3 pairs (A-type) | 5′-U U U-3′ | 1.391 | 1.224 | 1.375 | 1.391 | 1.231 | 115.2 | 126.4 | 4.9 | 2.4 | −9.9 |
| 5′-U U U-3′ (*) | 1.391 | 1.225 | 1.374 | 1.391 | 1.231 | 115.2 | 126.4 | 6.2 | 0.8 | −10.3 | |
| 5′-A U A-3′ | 1.395 | 1.220 | 1.376 | 1.383 | 1.237 | 115.1 | 127.0 | 1.7 | −1.1 | −3.0 | |
| 5′-U s2U U-3′ | 1.372 | 1.684 | 1.361 | 1.396 | 1.228 | 115.8 | 126.6 | 3.2 | 4.4 | −9.6 | |
| 5′-A s2U A-3′ | 1.375 | 1.678 | 1.364 | 1.392 | 1.232 | 115.6 | 127.1 | 3.5 | −2.7 | −3.7 | |
| 5′-U s4U U-3′ | 1.391 | 1.221 | 1.384 | 1.372 | 1.677 | 115.1 | 126.0 | 7.1 | −0.7 | −10.0 | |
| 5′-A s4U A-3′ | 1.393 | 1.217 | 1.387 | 1.366 | 1.688 | 115.2 | 126.2 | 2.7 | −2.3 | −3.2 | |
| 5′-U s2,4U U-3′ | 1.372 | 1.677 | 1.372 | 1.378 | 1.670 | 115.5 | 126.3 | 4.5 | 2.3 | −9.0 | |
| 5′-A s2,4U A-3′ | 1.376 | 1.669 | 1.377 | 1.373 | 1.680 | 115.4 | 126.4 | 4.8 | −4.9 | −3.4 | |
| Nucleotides 3 pairs (B-type) | 5′-U U U-3′ | 1.400 | 1.214 | 1.384 | 1.385 | 1.233 | 114.4 | 127.1 | 0.0 | −4.4 | 6.3 |
| 5′-A U A-3′ | 1.402 | 1.213 | 1.384 | 1.380 | 1.238 | 114.2 | 127.1 | 4.4 | −9.8 | 4.8 | |
| 5′-U s2U U-3′ | 1.384 | 1.666 | 1.370 | 1.390 | 1.230 | 114.5 | 127.4 | −0.7 | −4.9 | 8.1 | |
| 5′-A s2U A-3′ | 1.383 | 1.667 | 1.369 | 1.383 | 1.236 | 114.5 | 127.5 | 2.0 | −8.3 | 6.6 | |
| 5′-s2U s2U s2U-3′ | 1.385 | 1.665 | 1.372 | 1.390 | 1.229 | 114.4 | 127.3 | −0.4 | −5.8 | 8.6 | |
| 5′-U s4U U-3′ | 1.397 | 1.213 | 1.393 | 1.363 | 1.683 | 114.3 | 126.6 | −0.8 | −4.5 | 7.8 | |
| 5′-A s4U A-3′ | 1.398 | 1.212 | 1.394 | 1.362 | 1.688 | 114.2 | 126.7 | −0.3 | −7.9 | 8.8 | |
| 5′-U s2,4U U-3′ | 1.383 | 1.660 | 1.382 | 1.368 | 1.678 | 114.3 | 126.8 | −2.4 | −5.2 | 10.7 | |
| 5′-A s2,4U A-3′ | 1.385 | 1.657 | 1.382 | 1.366 | 1.683 | 114.0 | 127.0 | −1.7 | −7.8 | 11.2 | |
| 5′-s2,4Us2,4U s2,4U-3′ | 1.385 | 1.659 | 1.382 | 1.370 | 1.675 | 114.3 | 126.9 | −1.2 | −6.3 | 9.6 | |
| + 4 Na | 5′-s2Us2U s2U-3′ | 1.390 | 1.662 | 1.366 | 1.389 | 1.228 | 114.3 | 127.5 | 0.2 | −4.1 | 6.1 |
| 5′-s4Us4U s4U-3′ | 1.400 | 1.214 | 1.383 | 1.362 | 1.676 | 114.8 | 126.8 | 5.0 | −4.6 | −0.1 | |
| 5′-s2,4Us2,4U s2,4U-3′ | 1.383 | 1.656 | 1.373 | 1.367 | 1.674 | 114.7 | 127.1 | 8.3 | -4.7 | −6.2 | |
| + 4 Na +20 H2O | 5′-A U A-3′ (**) | 1.394 | 1.222 | 1.367 | 1.382 | 1.231 | 115.4 | 127.2 | 2.6 | −0.7 | −4.4 |
| 5′-U U U-3′ | 1.398 | 1.218 | 1.366 | 1.382 | 1.229 | 114.9 | 127.9 | −0.1 | 3.7 | −3.0 | |
| 5′-s2U s2U s2U-3′ | 1.389 | 1.663 | 1.364 | 1.390 | 1.226 | 114.6 | 127.7 | −0.9 | 0.8 | 0.9 | |
| 5′-s4U s4U s4U-3′ | 1.394 | 1.216 | 1.380 | 1.366 | 1.669 | 115.1 | 126.8 | −1.6 | 6.0 | −3.5 | |
| 5′-s2,4Us2,4U s2,4U-3′ | 1.387 | 1.655 | 1.374 | 1.373 | 1.662 | 114.7 | 127.5 | −2.8 | 1.4 | 2.3 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcolea Palafox, M.; Franklin Benial, A.M.; K. Rastogi, V. Biomolecules of 2-Thiouracil, 4-Thiouracil and 2,4-Dithiouracil: A DFT Study of the Hydration, Molecular Docking and Effect in DNA:RNAMicrohelixes. Int. J. Mol. Sci. 2019, 20, 3477. https://doi.org/10.3390/ijms20143477
Alcolea Palafox M, Franklin Benial AM, K. Rastogi V. Biomolecules of 2-Thiouracil, 4-Thiouracil and 2,4-Dithiouracil: A DFT Study of the Hydration, Molecular Docking and Effect in DNA:RNAMicrohelixes. International Journal of Molecular Sciences. 2019; 20(14):3477. https://doi.org/10.3390/ijms20143477
Chicago/Turabian StyleAlcolea Palafox, M., A. Milton Franklin Benial, and V. K. Rastogi. 2019. "Biomolecules of 2-Thiouracil, 4-Thiouracil and 2,4-Dithiouracil: A DFT Study of the Hydration, Molecular Docking and Effect in DNA:RNAMicrohelixes" International Journal of Molecular Sciences 20, no. 14: 3477. https://doi.org/10.3390/ijms20143477
APA StyleAlcolea Palafox, M., Franklin Benial, A. M., & K. Rastogi, V. (2019). Biomolecules of 2-Thiouracil, 4-Thiouracil and 2,4-Dithiouracil: A DFT Study of the Hydration, Molecular Docking and Effect in DNA:RNAMicrohelixes. International Journal of Molecular Sciences, 20(14), 3477. https://doi.org/10.3390/ijms20143477








