Identification of QTLs for Stripe Rust Resistance in a Recombinant Inbred Line Population
Abstract
1. Introduction
2. Results
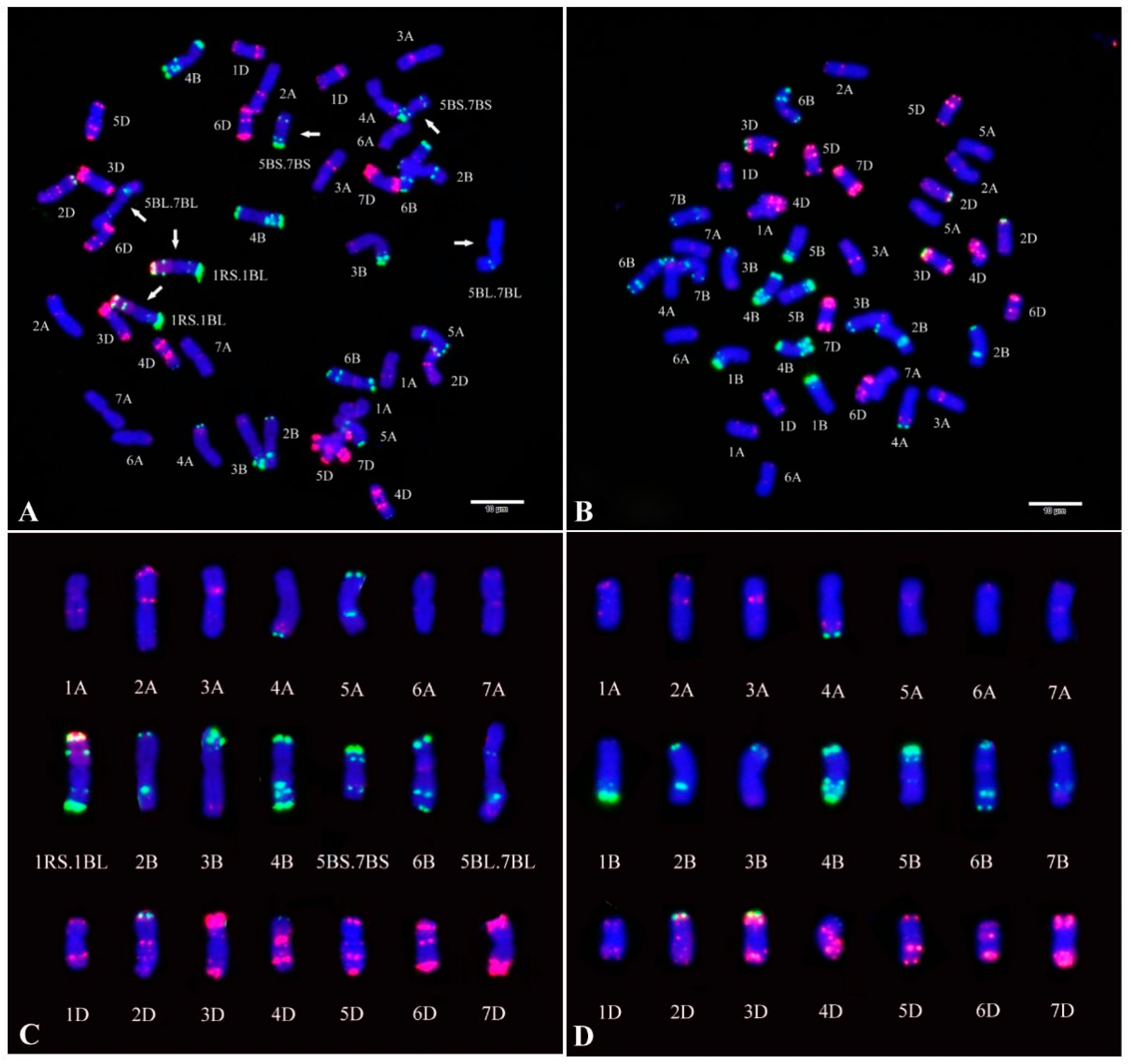
2.1. Fluorescence In Situ Hybridization (FISH) Analysis of CH55 and CH42
2.2. Phenotypic Analysis
2.3. Analysis of SLAF-Seq Data and SNP Markers
2.4. Genetic Map Construction and Consistency Analysis
2.5. QTL Mapping of Stripe Rust Resistance of the RILs
2.6. Additive Effects of QTLs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Field Trials
4.2. Broadsense Heritability
4.3. DNA Extractions
4.4. SLAF Library Construction and High-Throughput Sequencing
4.5. Analysis of SLAF-Seq Data and Genetic Map Construction
4.6. QTL Analysis
4.7. FISH Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Pst | Puccinia striiformis f. sp. tritici |
| APR | Adult-Plant Resistance |
| CYR | Chinese Yellow Rust |
| QTL | Quantitative Trait Loci |
| SNP | Single Nucleotide Polymorphism |
| CH42 | Chuanmai 42 |
| CH55 | Chuanmai 55 |
| SLAF-seq | Specific Locus Amplified Fragment Sequencing |
| NGS | Next Generation Sequencing |
| TGW | Thousand Grain Weight |
| RILs | Recombinant Inbred Lines |
| BSA | Bulked Segregant Analysis |
| FISH | Fluorescence In Situ Hybridization |
| ND-FISH | Non-denaturing FISH |
| DAPI | 4’,6-diamidino-2-phenylindole |
| GC | Guanine Cytosine |
| ICIM | Inclusive Composite Interval Mapping |
| LOD | Logarithm of Odds |
| IWGSC | International Wheat Genome Sequencing Consortium |
| CTAB | cetyltrimethylammonium ammonium bromide |
| GATK | Genome Analysis Toolkit |
References
- Beddow, J.M.; Pardey, P.G.; Chai, Y.; Hurley, T.M.; Kriticos, D.J.; Braun, H.J.; Park, R.F.; Cuddy, W.S.; Yonow, T. Research investment implications of shifts in the global geography of wheat stripe rust. Nat. Plants 2015, 1, 15132. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M. High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 2013, 4, 608–627. [Google Scholar] [CrossRef]
- Chen, X.M. Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 2014, 36, 311–326. [Google Scholar] [CrossRef]
- He, Z.H.; Lan, C.X.; Chen, X.M.; Zou, Y.C.; Zhuang, Q.S.; Xia, X.C. Progress and perspective in research of adult-plant resistance to stripe rust and powdery mildew in wheat. Sci. Agric. Sin. 2011, 44, 2193–2215. [Google Scholar] [CrossRef]
- Wang, M.N.; Chen, X.M. Stripe rust resistance. In Stripe Rust, 5th ed.; Chen, X.M., Kang, Z.S., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 353–558. [Google Scholar]
- Feng, J.Y.; Wang, M.N.; See, D.R.; Chao, S.M.; Zheng, Y.L.; Chen, X.M. Characterization of novel gene Yr79 and four additional quantitative trait loci for all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat PI182103. Phytopathology 2018, 108, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, S.R.; Xia, X.C.; Zhou, Q.; He, Y.J.; Wei, Y.M.; Zheng, Y.Y.; He, Z.H. Molecular mapping of a recessive stripe rust resistance gene yrMY37 in Chinese wheat cultivar Mianmai37. Mol. Breed. 2015, 35, 97. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2015-2016 Supplement. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf (accessed on 2 April 2019).
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2017 Supplement. Available online: http://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf (accessed on 2 April 2019).
- Dong, Z.; Hegarty, J.M.; Zhang, J.; Zhang, W.J.; Chao, S.M.; Chen, X.M.; Zhou, Y.H. Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor. Appl. Genet. 2017, 130, 2127–2137. [Google Scholar] [CrossRef]
- Wan, A.M.; Zhao, Z.H.; Chen, X.M.; He, Z.H.; Jin, S.L.; Jia, Q.Z.; Yao, G.; Yang, J.X.; Wang, B.T.; Li, G.B.; et al. Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 2004, 88, 896–904. [Google Scholar] [CrossRef]
- Li, G.Q.; Li, Z.F.; Yang, W.Y.; Zhang, Y.; He, Z.H.; Xu, S.C.; Singh, R.P.; Qu, Y.Y.; Xia, X.C. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai42 and its allelism with Yr24 and Yr26. Theor. Appl. Genet. 2006, 112, 1434–1440. [Google Scholar] [CrossRef]
- Liu, T.G.; Peng, Y.L.; Chen, W.Q.; Zhang, Z.Y. First detection of virulence in Puccinia striiformis f. sp. tritici in China to resistance genes Yr24 (= Yr26) present in wheat cultivar Chuanmai42. Plant Dis. 2010, 94, 1163. [Google Scholar] [CrossRef]
- Sun, X.W.; Liu, D.Y.; Zhang, X.F.; Li, W.B.; Liu, H.; Hong, W.G.; Jiang, C.B.; Guan, N.; Ma, C.X.; Zeng, H.P.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Xu, R.X.; Zhu, B.Y.; Yu, T.; Qu, W.Q.; Lu, L.; Xu, Q.; Qi, X.H.; Chen, X.H. A high-density genetic map of cucumber derived from Specific Length Amplified Fragment sequencing (SLAF-seq). Front. Plant Sci. 2015, 5, 768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.P.; Huang, L.; Gao, A.N.; Zhang, J.; Yang, X.M.; Liu, W.H.; Li, X.Q.; Li, L.H. A high-density genetic map for P genome of Agropyron Gaertn. based on specific-locus amplified fragment sequencing (SLAF-seq). Planta 2015, 242, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.J.; Zhang, H.P.; Liu, K.; Cao, J.J.; Wang, S.X.; Jiang, H.; Wu, Z.Y.; Lu, J.; Zhu, X.F.; Xia, X.C.; et al. Cloning and characterization of TaTGW-7A gene associated with grain weight in wheat via SLAF-seq-BSA. Front. Plant Sci. 2016, 7, 1902. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.L.; Fang, Z.W.; Sun, C.; Zhang, P.; Zhang, X.; Lu, C.; Wang, S.P.; Ma, D.F.; Zhu, Y.X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018, 8, 3086. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119. 2, pTa-535, pTa71, CCS1, and pAWRC. 1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Pahalawatta, V.; Chen, X.M. Genetic analysis and molecular mapping of wheat genes conferring resistance to the wheat stripe rust and barley stripe rust pathogens. Phytopathology 2005, 95, 427–432. [Google Scholar] [CrossRef]
- Wu, J.H.; Zeng, Q.D.; Wang, Q.L.; Liu, S.J.; Yu, S.Z.; Mu, J.M.; Huang, S.; Sela, H.; Distelfeld, A.; Huang, L.L.; et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018, 131, 1481–1496. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Morris, C.; Appels, R.; Lan, C.X. Catalogue of Gene Symbols for Wheat: 2013–2014 Supplement. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2013.pdf (accessed on 2 April 2019).
- Cobo, N.; Wanjugi, H.; Lagudah, E.; Dubcovsky, J. A high-resolution map of wheat QYr.ucw-1BL, an adult plant stripe rust resistance locus in the same chromosomal region as Yr29. Plant Genome 2019, 12, 180055. [Google Scholar] [CrossRef]
- Lan, C.X.; Rosewarne, G.M.; Singh, R.P.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Basnet, B.R.; Zhang, Y.L.; Yang, E.N. QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin#1. Mol. Breed. 2014, 34, 789–803. [Google Scholar] [CrossRef]
- Pretorius, Z.A.; Lan, C.X.; Prins, R.; Knight, V.; McLaren, N.W.; Singh, R.P.; Bender, C.M.; Kloppers, F.J. Application of remote sensing to identify adult plant resistance loci to stripe rust in two bread wheat mapping populations. Precis. Agric. 2017, 18, 411–428. [Google Scholar] [CrossRef]
- Bokore, F.E.; Cuthbert, R.D.; Knox, R.E.; Randhawa, H.S.; Hiebert, C.W.; DePauw, R.M.; Singh, A.K.; Singh, A.; Sharpe, A.G.; Diaye, A.N.; et al. Quantitative trait loci for resistance to stripe rust of wheat revealed using global field nurseries and opportunities for stacking resistance genes. Theor. Appl. Genet. 2017, 130, 2617–2635. [Google Scholar] [CrossRef] [PubMed]
- Cobo, N.; Pflüger, L.; Chen, X.M.; Dubcovsky, J. Mapping QTL for resistance to new virulent races of wheat stripe rust from Two Argentinean wheat cultivars. Crop Sci. 2018, 58, 1–14. [Google Scholar] [CrossRef]
- Ma, J.; Qin, N.; Cai, B.; Chen, G.Y.; Ding, P.Y.; Zhang, H.; Yang, C.C.; Huang, L.; Mu, Y.; Tang, H.P.; et al. Identification and validation of a novel major QTL for all-stage stripe rust resistance on 1BL in the winter wheat line 20828. Theor. Appl. Genet. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- William, M.; Singh, R.P.; Huerta-Espino, J.; Ortiz Islas, S.; Hoisington, D. Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 2003, 93, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Rosewarne, G.M.; Singh, R.P.; Huerta-Espino, J.; Herrera-Foessel, S.A.; Forrest, K.L.; Hayden, M.J.; Rebetzke, G.J. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet × Pastor wheat population. Theor. Appl. Genet. 2012, 124, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Xu, S.C.; Zhang, L.J.; Miao, Q.; Zhai, Q.; Li, N. SSR marker of wheat stripe rust resistance gene Yr2. J. Triticeae Crop. 2005, 25, 17–19. [Google Scholar]
- Lin, F.; Chen, X.M. Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 2007, 114, 1277–1287. [Google Scholar] [CrossRef]
- Ren, R.S.; Wang, M.N.; Chen, X.M.; Zhang, Z.J. Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI183527. Theor. Appl. Genet. 2012, 125, 847–857. [Google Scholar] [CrossRef]
- Zhou, X.L.; Wang, M.N.; Chen, X.M.; Lu, Y.; Kang, Z.S.; Jing, J.X. Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI178759. Theor. Appl. Genet. 2014, 127, 935–945. [Google Scholar] [CrossRef]
- Xu, H.X.; Zhang, J.; Zhang, P.; Qie, Y.M.; Niu, Y.C.; Li, H.J.; Ma, P.T.; Xu, Y.F.; An, D.G. Development and validation of molecular markers closely linked to the wheat stripe rust resistance gene YrC591 for marker-assisted selection. Euphytica 2014, 198, 317–323. [Google Scholar] [CrossRef]
- Li, Z.F.; Zheng, T.C.; He, Z.H.; Li, G.Q.; Xu, S.C.; Li, X.P.; Yang, G.Y.; Singh, R.P.; Xia, X.C. Molecular tagging of stripe rust resistance gene YrZH84 in Chinese wheat line Zhou 8425B. Theor. Appl. Genet. 2006, 112, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Muleta, K.T.; Bulli, P.; Rynearson, S.; Chen, X.M.; Pumphrey, M. Loci associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a core collection of spring wheat (Triticum aestivum). PLoS ONE 2017, 12, e0179087. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, K.; Singh, R.P.; Huerta-Espino, J.; William, H.M. Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 2003, 93, 881–890. [Google Scholar] [CrossRef]
- Muleta, K.T.; Rouse, M.N.; Rynearson, S.; Chen, X.M.; Buta, B.G.; Pumphrey, M.O. Characterization of molecular diversity and genome-wide mapping of loci associated with resistance to stripe rust and stem rust in Ethiopian bread wheat accessions. BMC Plant Biol. 2017, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Imtiaz, M.; Ahmad, M.; Cromey, M.G.; Griffin, W.B.; Hampton, J.G. Detection of molecular markers linked to the durable adult plant stripe rust resistance gene Yr18 in bread wheat (Triticum aestivum L.). Plant Breed. 2004, 123, 401–404. [Google Scholar] [CrossRef]
- Bansal, U.K.; Hayden, M.J.; Keller, B.; Wellings, C.R.; Park, R.F.; Bariana, H.S. Relationship between wheat rust resistance genes Yr1 and Sr48 and a microsatellite marker. Plant Pathol. 2009, 58, 1039–1043. [Google Scholar] [CrossRef]
- Eriksen, L.; Afshari, F.; Christiansen, M.J.; McIntosh, R.A.; Jahoor, A.; Wellings, C.R. Yr32 for resistance to stripe (yellow) rust present in the wheat cultivar Carstens V. Theor. Appl. Genet. 2004, 108, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; He, Z.H.; Lu, J.L.; Li, J.; Ren, Y.; Ma, C.X.; Xia, X.C. Molecular mapping of stripe rust resistance gene YrJ22 in Chinese wheat cultivar Jimai 22. Mol. Breed. 2016, 36, 118. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Rajaram, S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol. Entomol. Hung. 2000, 35, 133–139. [Google Scholar]
- Yang, E.N.; Rosewarne, G.M.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Tang, Z.X.; Sun, C.F.; Ren, Z.L.; Singh, R.P. QTL analysis of the spring wheat “Chapio” identifies stable stripe rust resistance despite inter-continental genotype × environment interactions. Theor. Appl. Genet. 2013, 126, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Rosewarne, G.M.; Li, Z.F.; Singh, R.P.; Yang, E.N.; Herrera-Foessel, S.A.; Huerta-Espino, J. Different QTLs are associated with leaf rust resistance in wheat between China and Mexico. Mol. Breed. 2015, 35, 127. [Google Scholar] [CrossRef]
- Zhao, C.H.; Cui, F.; Wang, X.Q.; Shan, S.C.; Li, X.F.; Bao, Y.G.; Wang, H.G. Effects of 1BL/1RS translocation in wheat on agronomic performance and quality characteristics. Field Crop. Res. 2012, 127, 79–84. [Google Scholar] [CrossRef]
- Xie, W.L.; Ben-David, R.; Zeng, B.; Dinoor, A.; Xie, C.J.; Sun, Q.X.; Röder, M.S.; Fahoum, A.; Fahima, T. Suppressed recombination rate in 6VS/6AL translocation region carrying the Pm21 locus introgressed from Haynaldia villosa into hexaploid wheat. Mol. Breed. 2012, 29, 399–412. [Google Scholar] [CrossRef]
- Agenbag, G.M.; Pretorius, Z.A.; Boyd, L.A.; Bender, C.M.; Prins, R. Identification of adult plant resistance to stripe rust in the wheat cultivar Cappelle-Desprez. Theor. Appl. Genet. 2012, 125, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.F.; Campbell, A.B.; Hannah, A.E. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948, 26, 496–500. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.X.; Cheng, T.R.; Yang, W.R.; Pan, H.T.; Zhong, J.J.; Huang, L.; Liu, E.Z. High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunus mume Sieb. et Zucc). DNA Res. 2015, 22, 183–191. [Google Scholar] [CrossRef]
- Li, H.H.; Ribaut, J.M.; Li, Z.L.; Wang, J.K. Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet. 2008, 116, 243–260. [Google Scholar] [CrossRef]
- Wang, J.K.; Li, H.H.; Zhang, L.Y.; Meng, L. Users’ Manual of QTL IciMapping; The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS): Beijing, China, 2014; pp. 96–131. [Google Scholar]
- Han, F.P.; Lamb, J.C.; Birchler, J.A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef]
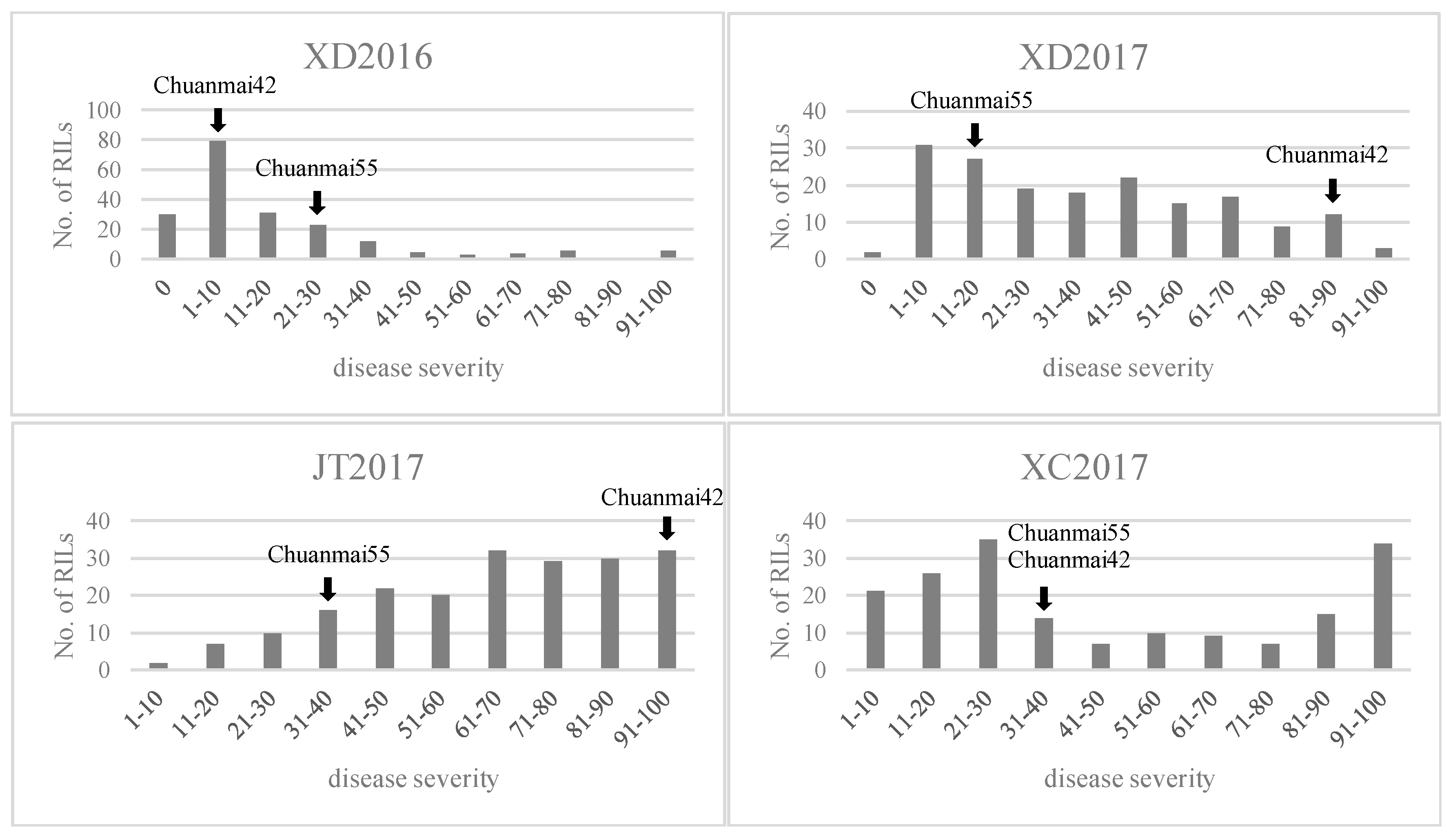

 represents field trials in 2015–2016 growing seasons at Xindu (XD2016);
represents field trials in 2015–2016 growing seasons at Xindu (XD2016);  represents field trials in 2016–2017 growing seasons at Xindu (XD2017);
represents field trials in 2016–2017 growing seasons at Xindu (XD2017);  represents field trials in 2016–2017 growing seasons at Jitian (JT2017);
represents field trials in 2016–2017 growing seasons at Jitian (JT2017);  represents field trials in 2016–2017 growing seasons at Xichong (XC2017).
represents field trials in 2016–2017 growing seasons at Xichong (XC2017).
 represents field trials in 2015–2016 growing seasons at Xindu (XD2016);
represents field trials in 2015–2016 growing seasons at Xindu (XD2016);  represents field trials in 2016–2017 growing seasons at Xindu (XD2017);
represents field trials in 2016–2017 growing seasons at Xindu (XD2017);  represents field trials in 2016–2017 growing seasons at Jitian (JT2017);
represents field trials in 2016–2017 growing seasons at Jitian (JT2017);  represents field trials in 2016–2017 growing seasons at Xichong (XC2017).
represents field trials in 2016–2017 growing seasons at Xichong (XC2017).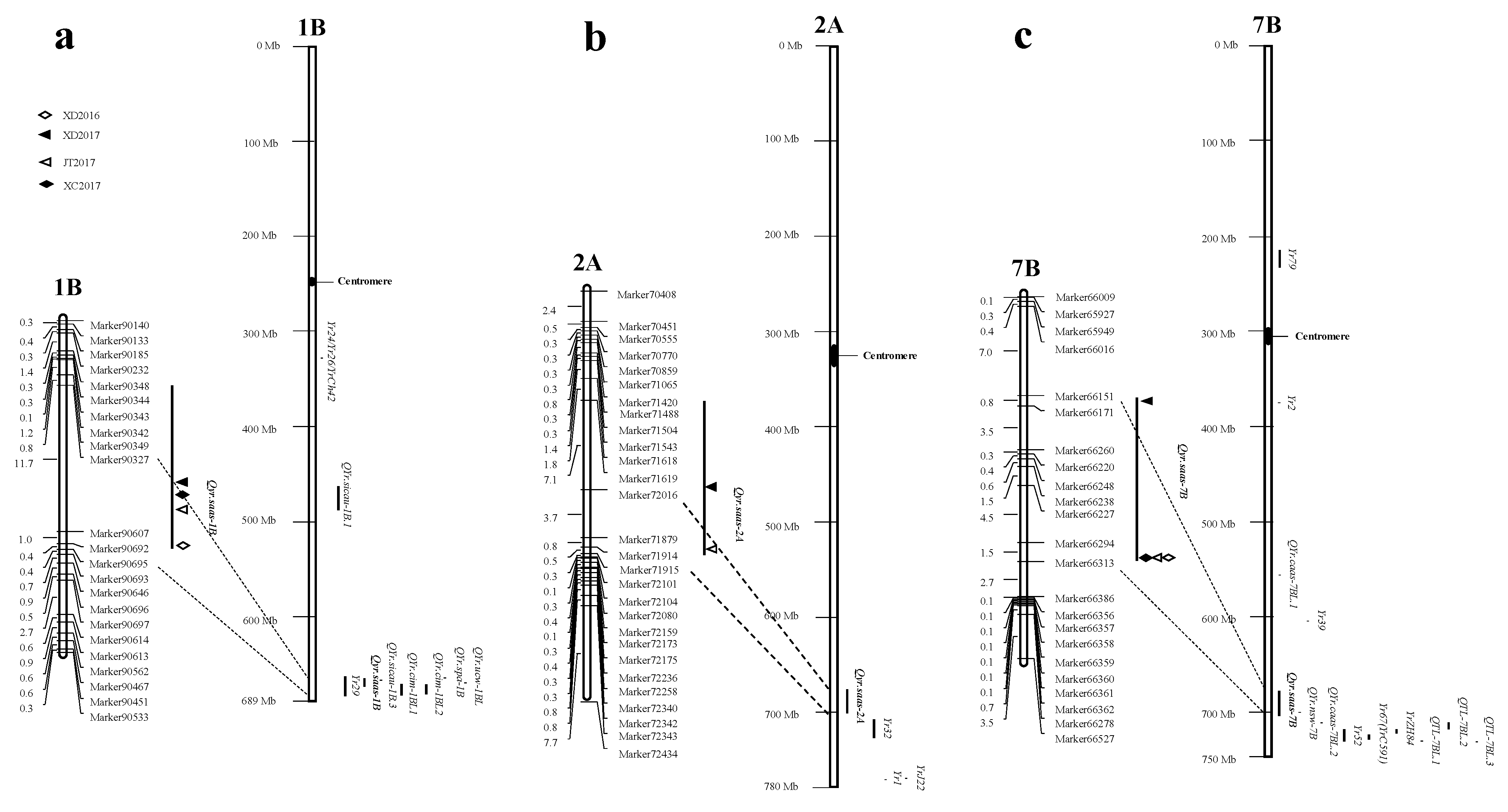
| Environment | Parent Mean | RIL Population Mean | H2 | |||
|---|---|---|---|---|---|---|
| CH55 | CH42 | Min | Max | Mean | ||
| XD2016 | 30 | 10 | 0 | 100 | 20.96 | 0.88 |
| XD2017 | 15 | 85 | 0 | 95 | 39.54 | |
| JT2017 | 35 | 95 | 2.5 | 100 | 66.91 | |
| XC2017 | 40 | 40 | 0.5 | 100 | 48.41 | |
| Sample | Total Reads | Total Bases | Q30 Percentage (%) | GC (%) |
|---|---|---|---|---|
| CH42 | 31,435,118 | 6,285,898,686 | 97.82 | 44.96 |
| CH55 | 33,764,850 | 6,752,071,114 | 97.96 | 44.85 |
| Offspring | 8,531,292 | 1,705,900,571 | 94.82 | 44.79 |
| Total | 1,771,458,398 | 354,218,084,068 | 94.82 | 44.79 |
| Sample | SLAFs Number | Total Depth | Average Depth |
|---|---|---|---|
| CH42 | 862,053 | 23,017,201 | 26.7 |
| CH55 | 863,835 | 20,864,394 | 24.15 |
| Offspring | 493,537 | 5,744,268 | 11.8 |
| Filtering Step | Number of SNPs |
|---|---|
| All reads | 1771.45 M |
| SLAFs in the reads | 2,825,198 |
| SNPs in the SLAFs | 2,507,026 |
| Filtered multiple mutation sites | 2,491,463 |
| SNP without base deletion in the paternal or maternal parents | 640,734 |
| Sequence depth of SNPs > 4 | 446,616 |
| Polymorphic SNPs | 162,394 |
| SNPs of genotype AA × BB | 75,347 |
| SNPs with parental sequence depth >10 and non-significant segregation distortion (p > 0.01) | 6732 |
| Genome | Chromosome | Marker Number | Total Distance | Average Distance | Gap < 5 cM (%) | Largest Gap |
|---|---|---|---|---|---|---|
| A | 1A | 823 | 106.63 | 0.13 | 99.64 | 9.07 |
| 2A | 350 | 99.09 | 0.28 | 99.14 | 16.36 | |
| 3A | 290 | 181.75 | 0.63 | 97.58 | 12.77 | |
| 4A | 321 | 117.36 | 0.37 | 98.75 | 11.72 | |
| 5A | 242 | 170.73 | 0.71 | 97.10 | 12.31 | |
| 6A | 443 | 117.14 | 0.26 | 99.77 | 8.64 | |
| 7A | 142 | 88.56 | 0.62 | 98.58 | 6.85 | |
| Subtotal | 2611 | 881.26 | 0.34 | -- | -- | |
| B | 1B | 491 | 183.21 | 0.37 | 99.59 | 12.92 |
| 2B | 474 | 191.60 | 0.40 | 99.15 | 19.46 | |
| 3B | 565 | 155.65 | 0.28 | 99.11 | 13.94 | |
| 4B | 136 | 134.38 | 0.99 | 97.04 | 14.91 | |
| 5B | 762 | 180.01 | 0.24 | 99.74 | 18.20 | |
| 6B | 322 | 130.55 | 0.41 | 97.82 | 10.64 | |
| 7B | 557 | 215.86 | 0.39 | 99.64 | 7.98 | |
| Subtotal | 3307 | 1191.26 | 0.36 | -- | -- | |
| D | 1D | 159 | 138.40 | 0.87 | 96.20 | 9.35 |
| 2D | 151 | 99.31 | 0.66 | 98.00 | 6.60 | |
| 3D | 104 | 109.02 | 1.05 | 95.15 | 11.72 | |
| 4D | 66 | 83.00 | 1.26 | 93.85 | 13.77 | |
| 5D | 103 | 102.24 | 0.99 | 94.12 | 10.07 | |
| 6D | 83 | 82.98 | 1.00 | 93.90 | 8.52 | |
| 7D | 148 | 141.04 | 0.95 | 95.24 | 14.52 | |
| Subtotal | 814 | 755.99 | 0.93 | -- | -- | |
| Total | 6732 | 2828.51 | 0.42 | -- | -- | |
| Chromosome | Spearman 1 |
|---|---|
| 1A | 0.886 |
| 2A | 0.999 |
| 3A | 0.821 |
| 4A | 1.000 |
| 5A | 0.975 |
| 6A | 0.807 |
| 7A | 0.875 |
| 1B | 0.961 |
| 2B | 0.967 |
| 3B | 1.000 |
| 4B | 0.846 |
| 5B | 0.873 |
| 6B | 0.989 |
| 7B | 0.939 |
| 1D | 0.970 |
| 2D | 0.857 |
| 3D | 0.989 |
| 4D | 0.805 |
| 5D | 0.892 |
| 6D | 0.812 |
| 7D | 1.000 |
| QTL. | Trial | Position | Left Marker | Right Marker | LOD | PVE (%) | Add a |
|---|---|---|---|---|---|---|---|
| Qyr.saas-1B | XD2016 | 172 | Marker90692 | Marker90695 | 3.28 | 6.24 | 6.07 |
| XD2017 | 167 | Marker90327 | Marker90607 | 21.34 | 34.22 | 17.34 | |
| JT2017 | 169 | Marker90327 | Marker90607 | 12.47 | 22.07 | 11.73 | |
| XC2017 | 168 | Marker90327 | Marker90607 | 12.00 | 22.19 | 16.88 | |
| Qyr.saas-2A | XD2017 | 67 | Marker71619 | Marker72016 | 3.42 | 3.77 | 5.75 |
| JT2017 | 72 | Marker71914 | Marker71915 | 3.74 | 5.29 | 5.73 | |
| Qyr.saas-7B | XD2016 | 205 | Marker66294 | Marker66313 | 8.29 | 16.82 | −9.94 |
| XD2017 | 192 | Marker66151 | Marker66171 | 3.05 | 3.27 | −5.35 | |
| JT2017 | 205 | Marker66294 | Marker66313 | 3.22 | 4.45 | −5.28 | |
| XC2017 | 205 | Marker66294 | Marker66313 | 13.19 | 20.64 | −16.42 |
| QTLs | No. of RILs with Corresponding QTL or QTL Combination | Mean Disease Severity | |||||
|---|---|---|---|---|---|---|---|
| 1BL | 2AL | 7BL | JT2017 | XD2016 | XD2017 | XC2017 | |
| + | + | + | 23 | 38.22 a | 3.95 a | 15.78 a | 12.09 a |
| + | + | - | 14 | 52.32 ab | 18.93 b | 24.64 ab | 45.89 b |
| + | - | + | 17 | 59.74 bc | 7.71 ab | 23.62 ab | 17.59 a |
| - | + | + | 14 | 68.93 cd | 11.79 ab | 42.18 cd | 37.68 b |
| + | - | - | 12 | 71.25 cd | 16.25 ab | 33.54 bc | 45.63 b |
| - | - | + | 21 | 72.62 cd | 14.52 ab | 56.90 de | 46.55 b |
| - | + | - | 19 | 77.50 de | 44.21 c | 54.21 d | 80.13 c |
| - | - | - | 14 | 90.00 e | 43.93 c | 70.36 e | 86.96 c |
| Chromosome Arm | Genes/QTLs | Left Marker | Right Marker | Left Physical Position | Right Physical Position | Reference |
|---|---|---|---|---|---|---|
| 1BL | Qyr.saas-1B | Marker90327 | Marker90695 | 664079816 | 673644678 | This study |
| Yr21 | M1(Pto kin2/S2) | M2(Pto kin3/PtoFen-S) | None | None | [20] | |
| Yr24/Yr26/YrCh42 | WRS467 | CM1641 | 328642215 | 328642801 | [21] | |
| Yr29 | Xgwm44 | Xgwm140 | 662195228 | 684861809 | [22,23] | |
| QYr.sicau-1B.1 | Xwmc156 | Xwmc216 | 461685422 | 487427087 | [28] | |
| QYr.sicau-1B.3 | AX-108726041 | AX-111056129 | 667604743 | 667641255 | [28] | |
| QYr.cim-1BL1 | Xgwm259 | Xgwm140 | 672333339 | 684861809 | [24] | |
| QYr.cim-1BL2 | WPt-1770 | WPt-9028 | 671741402 | 681848783 | [25] | |
| QYr.spa-1B | Wsnp_Ra_c53181_56932563 | Wsnp_Ra_c53181_56932563 | 664804354 | 664804467 | [26] | |
| QYr.ucw-1BL | IWA8581 | csLV46G22 | 670389674 | None | [23,27] | |
| 7BL | Qyr.saas-7B | Marker66151 | Marker66313 | 678635912 | 706808017 | This study |
| Yr2 | WMC364 | WMC364 | 375022989 | 375023010 | [31] | |
| Yr39 | Xgwm131 | Xgwm43 | 604774088 | None | [32] | |
| Yr52 | Xcfa2040 | Xbarc182 | 718432553 | 732366237 | [33] | |
| Yr59 | Xbarc32 | Xwmc557 | 723876921 | 728084216 | [34] | |
| Yr67 (YrC591) | Xbarc32 | Xbarc182 | 723876921 | 732366237 | [35] | |
| Yr79 | Xbarc72 | Xwmc335 | 214059722 | 233160839 | [6] | |
| YrZH84 | Xcfa2040 | Xbarc32 | 718432553 | 723876921 | [36] | |
| YrMY37 | Xgwm297 | Xbarc267 | 237502276 | 377136685 | [7] | |
| QTL-7BL.1 | IWA3155 | IWA3416 | 732651049 | 732651181 | [37] | |
| QTL-7BL.2 | Xgwm577 | Xwmc166 | 711234115 | 719852469 | [38] | |
| QTL-7BL.3 | IWB58601 | IWB58601 | 732651454 | 732651554 | [39] | |
| QYr.nsw-7B | Xgwm611 | Xgwm611 | 700632085 | 700632104 | [40] | |
| QYr.caas-7BL.1 | Xbarc176 | XwPt8106 | 557048410 | None | [33] | |
| QYr.caas-7BL.2 | Xgwm577 | XwPt4300 | 711234115 | None | [33] | |
| 2AL | Qyr.saas-2A | Marker72016 | Marker71915 | 677899968 | 701739954 | This study |
| YR1 | Xgwm311 | None | 772967422 | None | [41] | |
| Yr32 | Xwmc198 | Xwmc181 | 707741852 | 728609562 | [42] | |
| YrJ22 | Xwmc658 | IWA1348 | 771166682 | None | [43] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Li, G.; Wan, H.; Li, L.; Li, J.; Yang, W.; Pu, Z.; Yang, Z.; Yang, E. Identification of QTLs for Stripe Rust Resistance in a Recombinant Inbred Line Population. Int. J. Mol. Sci. 2019, 20, 3410. https://doi.org/10.3390/ijms20143410
Yang M, Li G, Wan H, Li L, Li J, Yang W, Pu Z, Yang Z, Yang E. Identification of QTLs for Stripe Rust Resistance in a Recombinant Inbred Line Population. International Journal of Molecular Sciences. 2019; 20(14):3410. https://doi.org/10.3390/ijms20143410
Chicago/Turabian StyleYang, Manyu, Guangrong Li, Hongshen Wan, Liping Li, Jun Li, Wuyun Yang, Zongjun Pu, Zujun Yang, and Ennian Yang. 2019. "Identification of QTLs for Stripe Rust Resistance in a Recombinant Inbred Line Population" International Journal of Molecular Sciences 20, no. 14: 3410. https://doi.org/10.3390/ijms20143410
APA StyleYang, M., Li, G., Wan, H., Li, L., Li, J., Yang, W., Pu, Z., Yang, Z., & Yang, E. (2019). Identification of QTLs for Stripe Rust Resistance in a Recombinant Inbred Line Population. International Journal of Molecular Sciences, 20(14), 3410. https://doi.org/10.3390/ijms20143410






