Phytosterols Inhibit Side-Chain Oxysterol Mediated Activation of LXR in Breast Cancer Cells
Abstract
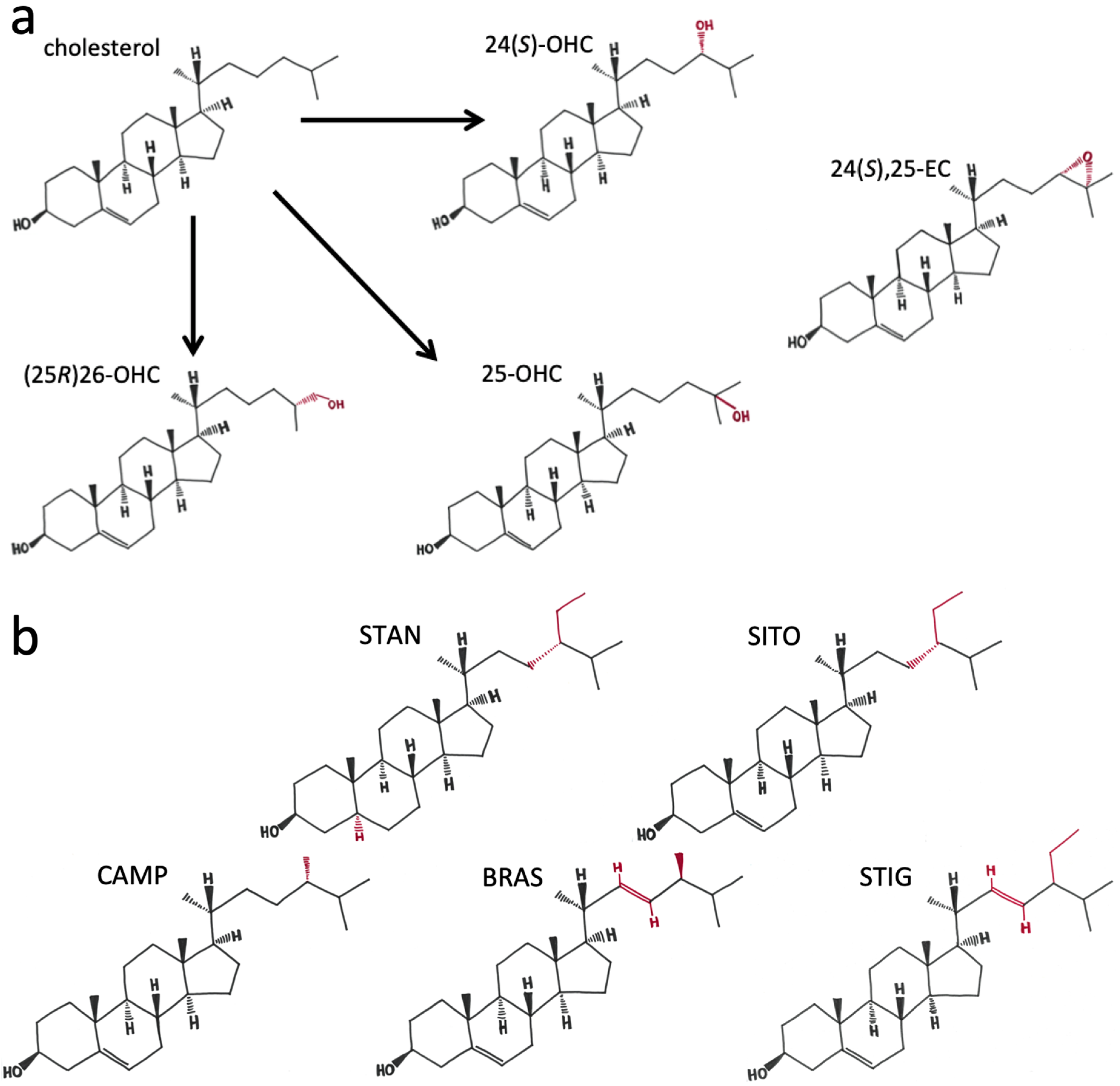
1. Introduction
2. Results
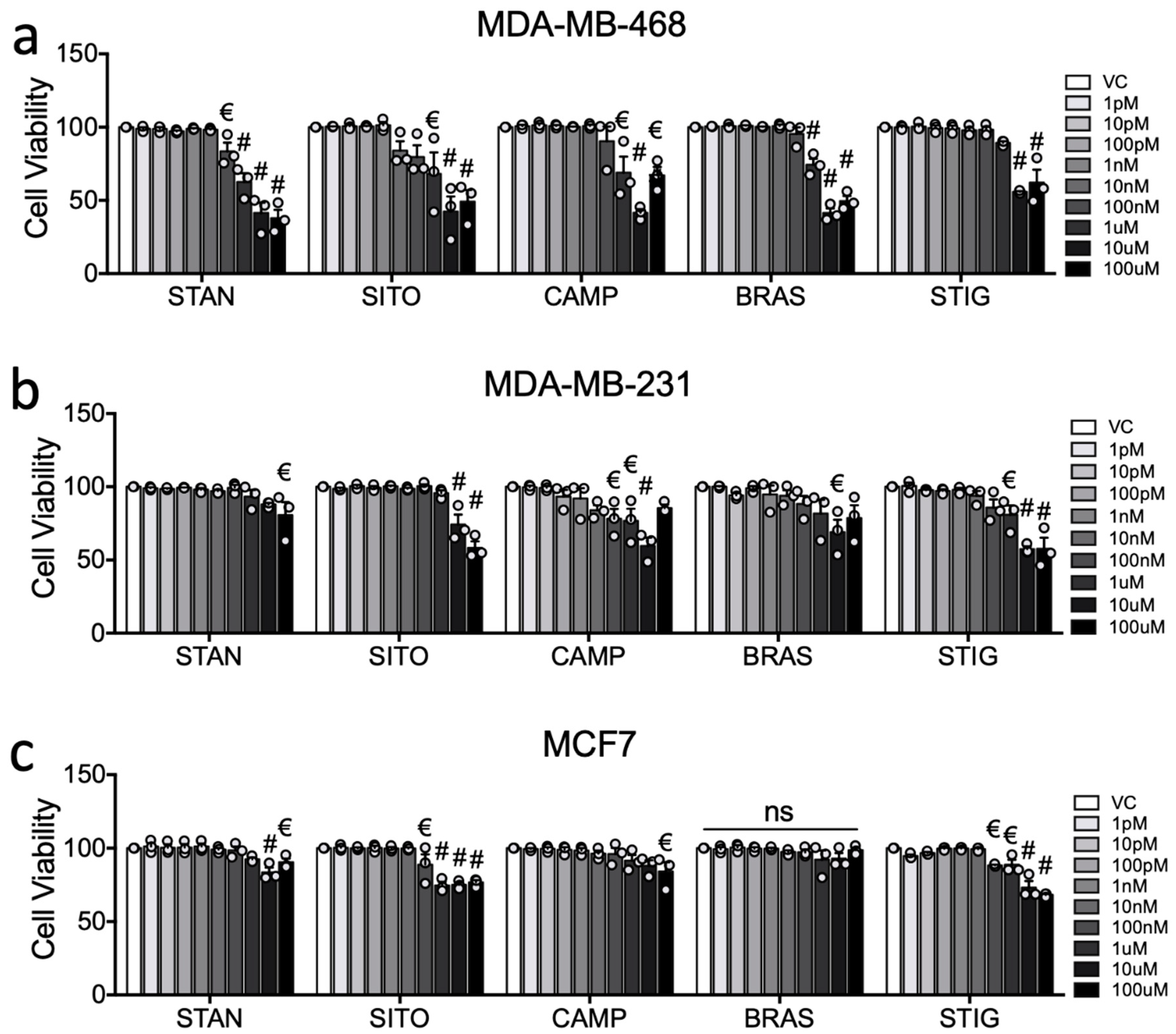
2.1. PSSs Are Poor Transcriptional Activators of LXRA in Breast Cancer Cell Cultures
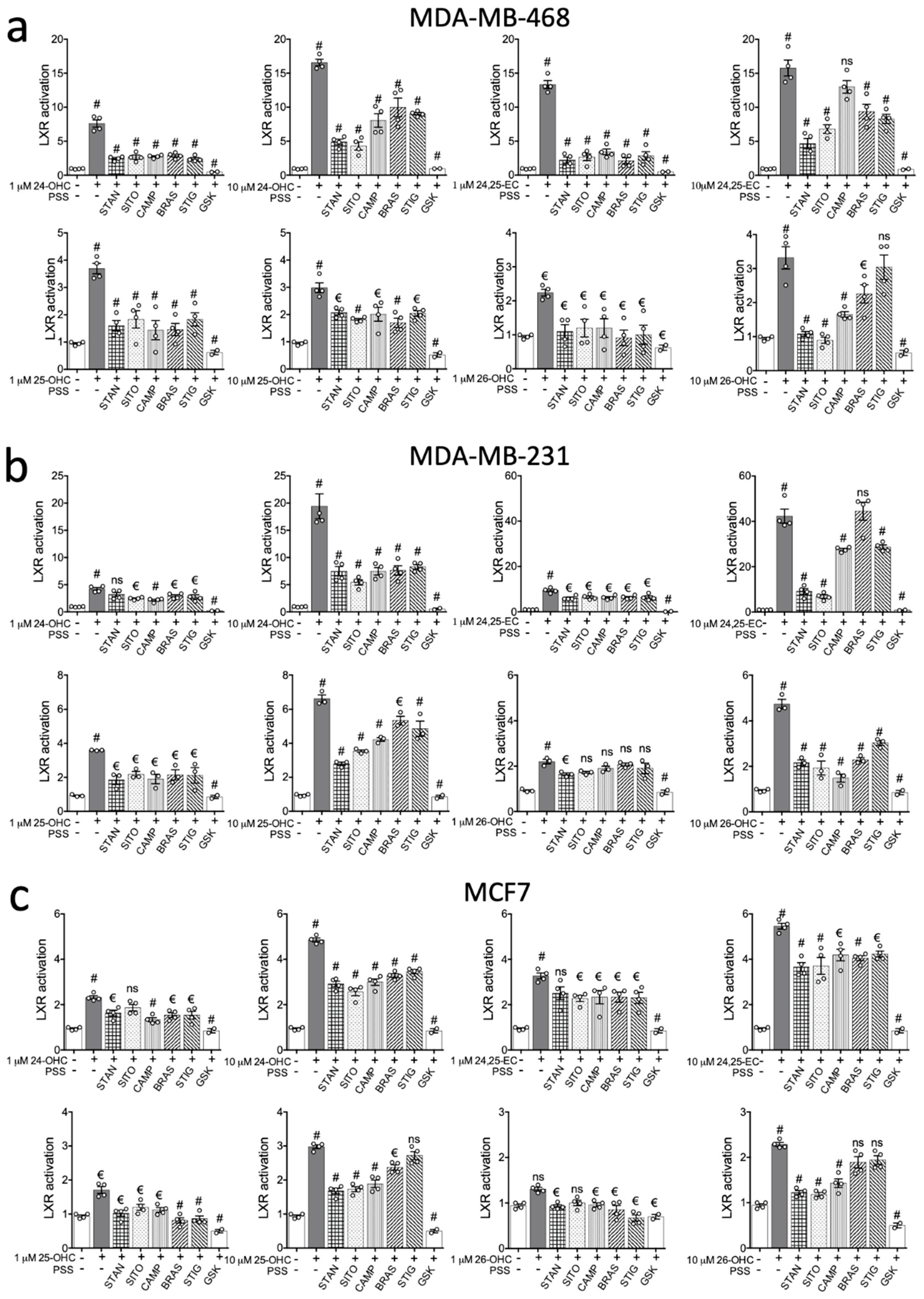
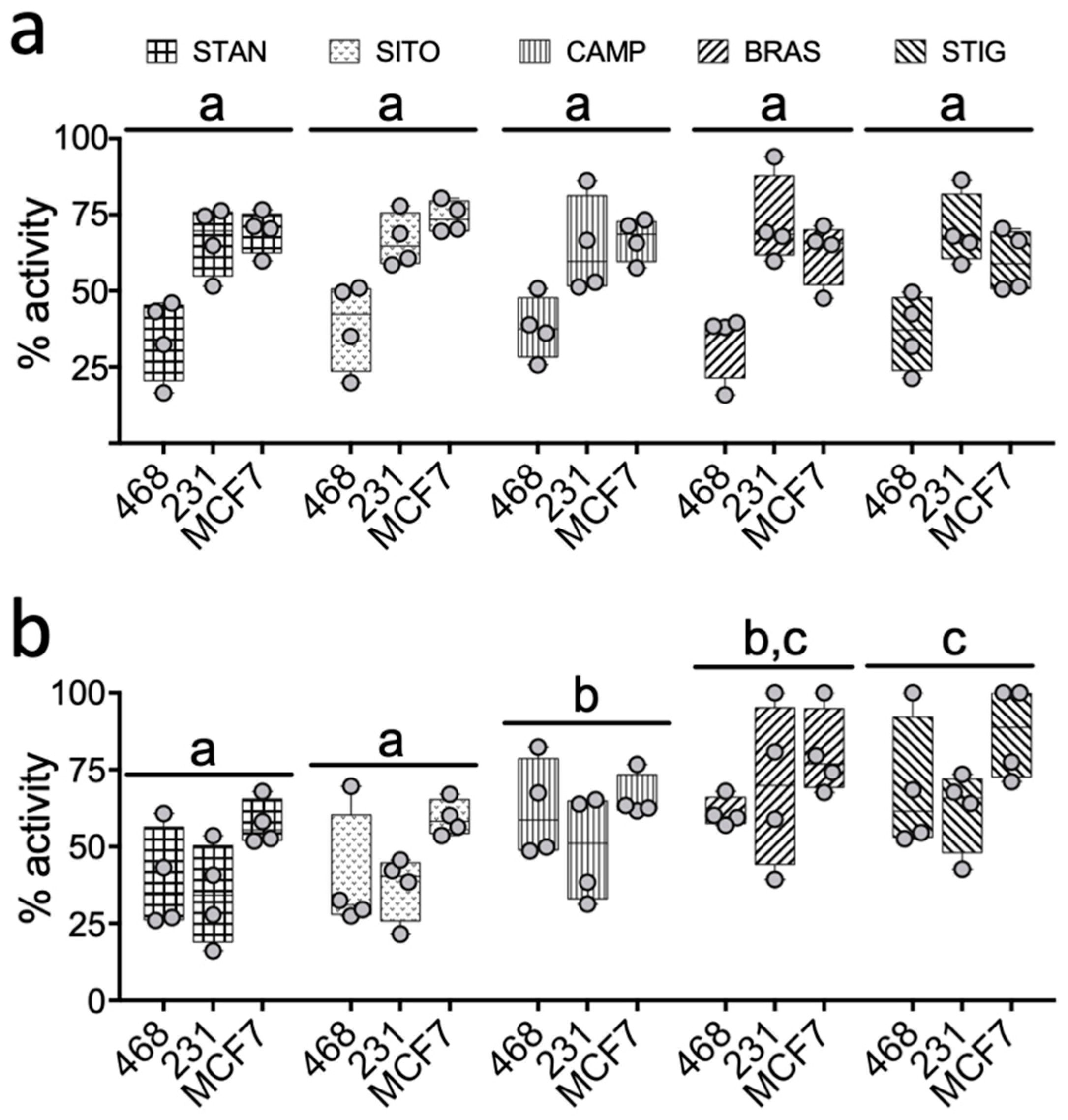
2.2. PSSs Impair Side-Chain Oxysterol Mediated Activation of LXRA
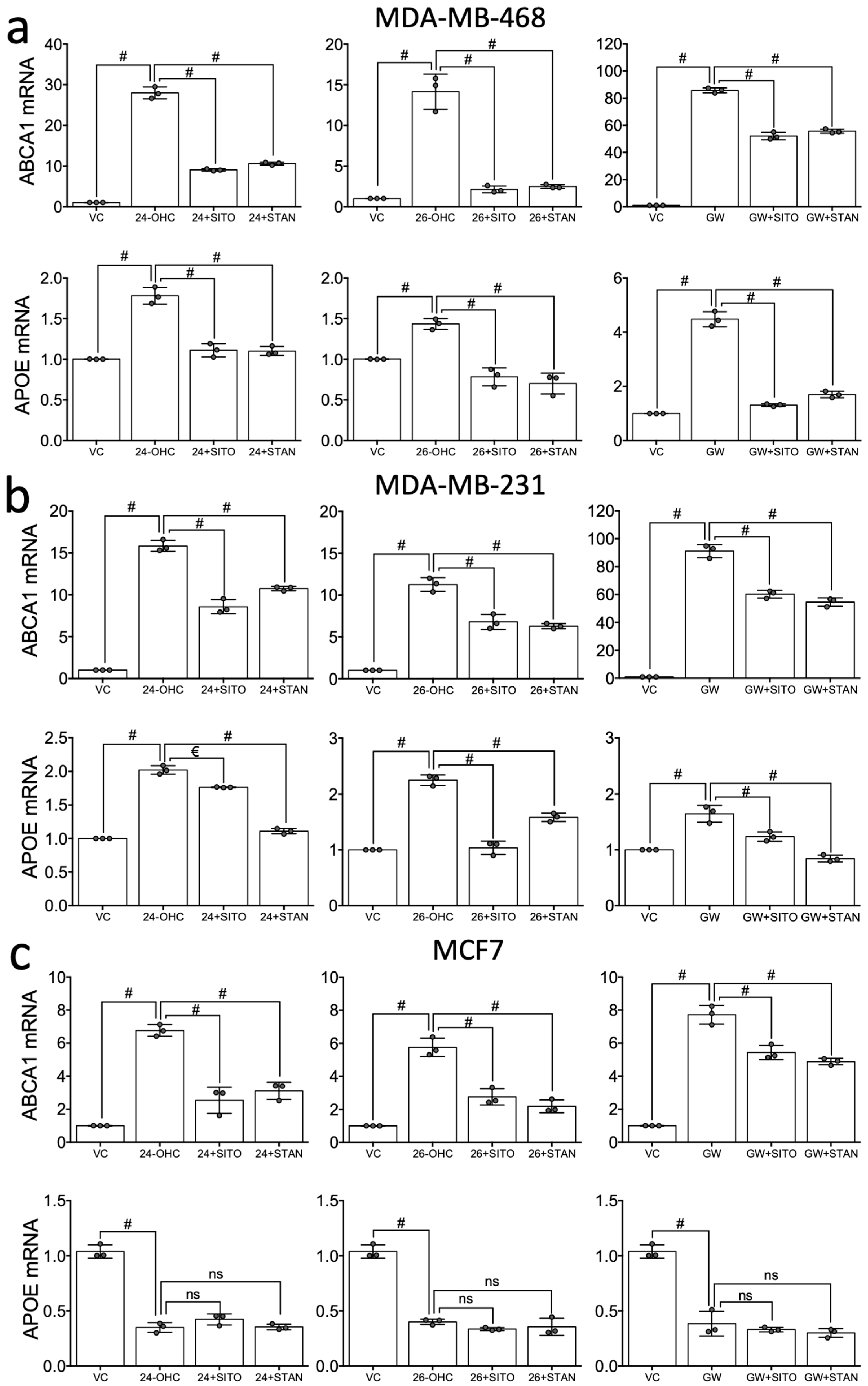
2.3. STAN and SITO Inhibit Oxysterol Mediated Activation of the LXR Target Genes ABCA1 and APOE
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell Lines
4.2. Drugs and Reagents
4.3. MTT Assays
4.4. Reporter Cell Lines and Luciferase Assays
4.5. mRNA Isolation, Reverse Transcription and qPCR
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 24(S)-OHC | 24(S)-hydroxycholesterol |
| 24,25-EC | 24,25-epoxycholesterol |
| 25-OHC | 25-hydroxycholesterol |
| (25R)26-OHC | (25R),26-hydroxycholesterol |
| BRAS | brassicasterol |
| BCa | breast cancer |
| CAMP | campesterol |
| ER | oestrogen receptor |
| LXR | liver X receptor |
| PSS | phytosterol/phytostanol |
| STIG | stigmasterol |
| STAN | sitostanol |
| SITO | β-sitosterol |
References
- Bonsang-Kitzis, H.; Chaltier, L.; Belin, L.; Savignoni, A.; Rouzier, R.; Sablin, M.P.; Lerebours, F.; Bidard, F.C.; Cottu, P.; Sastre-Garau, X.; et al. Beyond Axillary Lymph Node Metastasis, BMI and Menopausal Status Are Prognostic Determinants for Triple-Negative Breast Cancer Treated by Neoadjuvant Chemotherapy. PLoS ONE 2015, 10, e0144359. [Google Scholar] [CrossRef]
- Crispo, A.; Grimaldi, M.; D’Aiuto, M.; Rinaldo, M.; Capasso, I.; Amore, A.; D’Aiuto, G.; Giudice, A.; Ciliberto, G.; Montella, M. BMI and breast cancer prognosis benefit: Mammography screening reveals differences between normal weight and overweight women. Breast 2015, 24, 86–89. [Google Scholar] [CrossRef]
- Dos Santos, C.R.; Fonseca, I.; Dias, S.; de Almeida, J.C.M. Plasma level of LDL-cholesterol at diagnosis is a predictor factor of breast tumor progression. BMC Cancer 2014, 14, 132. [Google Scholar]
- Brennan, S.F.; Woodside, J.V.; Lunny, P.M.; Cardwell, C.R.; Cantwell, M.M. Dietary fat and breast cancer mortality: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2017, 57, 1999–2008. [Google Scholar] [CrossRef]
- Chlebowski, R.T.; Blackburn, G.L.; Thomson, C.A.; Nixon, D.W.; Shapiro, A.; Hoy, M.K.; Goodman, M.T.; Giuliano, A.E.; Karanja, N.; McAndrew, P.; et al. Dietary fat reduction and breast cancer outcome: Interim efficacy results from the Women’s Intervention Nutrition Study. J. Natl. Cancer Inst. 2006, 98, 1767–1776. [Google Scholar] [CrossRef]
- Manthravadi, S.; Shrestha, A.; Madhusudhana, S. Impact of statin use on cancer recurrence and mortality in breast cancer: A systematic review and meta-analysis. Int. J. Cancer 2016, 139, 1281–1288. [Google Scholar] [CrossRef]
- Liu, B.; Yi, Z.; Guan, X.; Zeng, Y.X.; Ma, F. The relationship between statins and breast cancer prognosis varies by statin type and exposure time: A meta-analysis. Breast Cancer Res. Treat. 2017, 164, 1–11. [Google Scholar] [CrossRef]
- Janowski, B.A.; Willy, P.J.; Devi, T.R.; Falck, J.R.; Mangelsdorf, D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 1996, 383, 728–731. [Google Scholar] [CrossRef]
- Janowski, B.A.; Grogan, M.J.; Jones, S.A.; Wisely, G.B.; Kliewer, S.A.; Corey, E.J.; Mangelsdorf, D.J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA 1999, 96, 266–271. [Google Scholar] [CrossRef]
- Nelson, E.R.; Wardell, S.E.; Jasper, J.S.; Park, S.; Suchindran, S.; Howe, M.K.; Carver, N.J.; Pillai, R.V.; Sullivan, P.M.; Sondhi, V.; et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 2013, 342, 1094–1098. [Google Scholar] [CrossRef]
- Baek, A.E.; Yu, Y.R.A.; He, S.S.; Wardell, S.E.; Chang, C.Y.; Kwon, S.; Pillai, R.V.; McDowell, H.B.; Thompson, J.W.; Dubois, L.G.; et al. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 2017, 8, 864. [Google Scholar] [CrossRef]
- Wu, Q.; Ishikawa, T.; Sirianni, R.; Tang, H.; McDonald, J.G.; Yuhanna, I.S.; Thompson, B.; Girard, L.; Mineo, C.; Brekken, R.A.; et al. 27-Hydroxycholesterol Promotes Cell-Autonomous, ER-Positive Breast Cancer Growth. Cell Rep. 2013, 5, 637–645. [Google Scholar] [CrossRef]
- Dalenc, F.; Iuliano, L.; Filleron, T.; Zerbinati, C.; Voisin, M.; Arellano, C.; Chatelut, E.; Marquet, P.; Samadi, M.; Roche, H.; et al. Circulating oxysterol metabolites as potential new surrogate markers in patients with hormone receptor-positive breast cancer: Results of the OXYTAM study. J. Steroid Biochem. Mol. Biol. 2016, 169, 210–218. [Google Scholar] [CrossRef]
- Moreau, R.A.; Nystrom, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef]
- Pollak, O.J.; Kritchevsky, D. Sitosterol. Monogr. Atherosc. 1981, 10, 1–219. [Google Scholar]
- Olkkonen, V.M.; Gylling, H.; Ikonen, E. Plant sterols, cholesterol precursors and oxysterols: Minute concentrations-Major physiological effects. J. Steroid Biochem. Mol. Biol. 2017, 169, 4–9. [Google Scholar] [CrossRef]
- Fakih, O.; Sanver, D.; Kane, D.; Thorne, J.L. Exploring the biophysical properties of phytosterols in the plasma membrane for novel cancer prevention strategies. Biochimie 2018, 153, 150–161. [Google Scholar] [CrossRef]
- Yang, C.; Yu, L.; Li, W.; Xu, F.; Cohen, J.C.; Hobbs, H.H. Disruption of cholesterol homeostasis by plant sterols. J. Clin. Investig. 2004, 114, 813–822. [Google Scholar] [CrossRef]
- Lutjohann, D.; Bjorkhem, I.; Beil, U.F.; von Bergmann, K. Sterol absorption and sterol balance in phytosterolemia evaluated by deuterium-labeled sterols: Effect of sitostanol treatment. J. Lipid Res. 1995, 36, 1763–1773. [Google Scholar]
- Brauner, R.; Johannes, C.; Ploessl, F.; Bracher, F.; Lorenz, R.L. Phytosterols reduce cholesterol absorption by inhibition of 27-hydroxycholesterol generation, liver X receptor alpha activation, and expression of the basolateral sterol exporter ATP-binding cassette A1 in Caco-2 enterocytes. J. Nutr. 2012, 142, 981–989. [Google Scholar] [CrossRef]
- Kaneko, E.; Matsuda, M.; Yamada, Y.; Tachibana, Y.; Shimomura, I.; Makishima, M. Induction of intestinal ATP-binding cassette transporters by a phytosterol-derived liver X receptor agonist. J. Biol. Chem. 2003, 278, 36091–36098. [Google Scholar] [CrossRef]
- Plat, J.; Nichols, J.A.; Mensink, R.P. Plant sterols and stanols: Effects on mixed micellar composition and LXR (target gene) activation. J. Lipid Res. 2005, 46, 2468–2476. [Google Scholar] [CrossRef]
- Park, Y.; Carr, T.P. Unsaturated fatty acids and phytosterols regulate cholesterol transporter genes in Caco-2 and HepG2 cell lines. Nutr. Res. 2013, 33, 154–161. [Google Scholar] [CrossRef]
- Alemany, L.; Laparra, J.M.; Barbera, R.; Alegria, A. Relative expression of cholesterol transport-related proteins and inflammation markers through the induction of 7-ketosterol-mediated stress in Caco-2 cells. Food Chem. Toxicol. 2013, 56, 247–253. [Google Scholar] [CrossRef]
- Liang, Y.T.; Wong, W.T.; Guan, L.; Tian, X.Y.; Ma, K.Y.; Huang, Y.; Chen, Z.Y. Effect of phytosterols and their oxidation products on lipoprotein profiles and vascular function in hamster fed a high cholesterol diet. Atherosclerosis 2011, 219, 124–133. [Google Scholar] [CrossRef]
- Awad, A.B.; Williams, H.; Fink, C.S. Effect of phytosterols on cholesterol metabolism and MAP kinase in MDA-MB-231 human breast cancer cells. J. Nutr. Biochem. 2003, 14, 111–119. [Google Scholar] [CrossRef]
- Awad, A.B.; Williams, H.; Fink, C.S. Phytosterols reduce in vitro metastatic ability of MDA-MB-231 human breast cancer cells. Nutr. Cancer 2001, 40, 157–164. [Google Scholar] [CrossRef]
- Awad, A.B.; Fink, C.S.; Williams, H.; Kim, U. In vitro and in vivo (SCID mice) effects of phytosterols on the growth and dissemination of human prostate cancer PC-3 cells. Eur. J. Cancer Prev. 2001, 10, 507–513. [Google Scholar] [CrossRef]
- Link, L.B.; Canchola, A.J.; Bernstein, L.; Clarke, C.A.; Stram, D.O.; Ursin, G.; Horn-Ross, P.L. Dietary patterns and breast cancer risk in the California Teachers Study cohort. Am. J. Clin. Nutr. 2013, 98, 1524–1532. [Google Scholar] [CrossRef]
- Weigl, J.; Hauner, H.; Hauner, D. Can Nutrition Lower the Risk of Recurrence in Breast Cancer? Breast Care 2018, 13, 86–91. [Google Scholar]
- Blackburn, G.L.; Wang, K.A. Dietary fat reduction and breast cancer outcome: Results from the Women’s Intervention Nutrition Study (WINS). Am. J. Clin. Nutr. 2007, 86, s878–s881. [Google Scholar] [CrossRef]
- Hutchinson, S.; Thorne, J.L. Lipid-Activated Nuclear Receptors, 1st ed.; Springer: New York, NY, USA, 2019. [Google Scholar]
- Salen, G.; Ahrens, E.H., Jr.; Grundy, S.M. Metabolism of beta-sitosterol in man. J. Clin. Investig. 1970, 49, 952–967. [Google Scholar] [CrossRef]
- Salen, G.; Tint, G.S.; Shefer, S.; Shore, V.; Nguyen, L. Increased sitosterol absorption is offset by rapid elimination to prevent accumulation in heterozygotes with sitosterolemia. Arteriosc. Thromb. 1992, 12, 563–568. [Google Scholar] [CrossRef]
- Shahzad, N.; Khan, W.; Md, S.; Ali, A.; Saluja, S.S.; Sharma, S.; Al-Allaf, F.A.; Abduljaleel, Z.; Ibrahim, I.A.A.; Abdel-Wahab, A.F.; et al. Phytosterols as a natural anticancer agent: Current status and future perspective. Biomed. Pharmacother. 2017, 88, 786–794. [Google Scholar] [CrossRef]
- Stiles, A.R.; Kozlitina, J.; Thompson, B.M.; McDonald, J.G.; King, K.S.; Russell, D.W. Genetic, anatomic, and clinical determinants of human serum sterol and vitamin D levels. Proc. Natl. Acad. Sci. USA 2014, 111, E4006–E4014. [Google Scholar] [CrossRef]
- Ras, R.T.; Geleijnse, J.M.; Trautwein, E.A. LDL-cholesterol-lowering effect of plant sterols and stanols across different dose ranges: A meta-analysis of randomised controlled studies. Br. J. Nutr. 2014, 112, 214–219. [Google Scholar] [CrossRef]
- Marinozzi, M.; Castro Navas, F.F.; Maggioni, D.; Carosati, E.; Bocci, G.; Carloncelli, M.; Giorgi, G.; Cruciani, G.; Fontana, R.; Russo, V. Side-Chain Modified Ergosterol and Stigmasterol Derivatives as Liver X Receptor Agonists. J. Med. Chem. 2017, 60, 6548–6562. [Google Scholar] [CrossRef]
- Navas, F.F.C.; Giorgi, G.; Maggioni, D.; Pacciarini, M.; Russo, V.; Marinozzi, M. C24-hydroxylated stigmastane derivatives as Liver X Receptor agonists. Chem. Phys. Lipids 2018, 212, 44–50. [Google Scholar] [CrossRef]
- Lifsey, H.C. The Impact of Bioactive Phytosterol, Stigmasterol, on Cholesterol Elimination Pathways in Mice. Master’s Thesis, University of Kentucky, Lexington, Kentucky, 2018. [Google Scholar]
- Fouache, A.; Zabaiou, N.; De Joussineau, C.; Morel, L.; Silvente-Poirot, S.; Namsi, A.; Lizard, G.; Poirot, M.; Makishima, M.; Baron, S.; et al. Flavonoids differentially modulate liver X receptors activity-Structure-function relationship analysis. J. Steroid Biochem. Mol. Biol. 2019, 190, 173–182. [Google Scholar] [CrossRef]
- Solheim, S.; Hutchinson, S.A.; Lundanes, E.; Wilson, S.R.; Thorne, J.L.; Roberg-Larsen, H. Fast liquid chromatography-mass spectrometry reveals side chain oxysterol heterogeneity in breast cancer tumour samples. J. Steroid Biochem. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Thorne, J.L.; Battaglia, S.; Baxter, D.E.; Hayes, J.L.; Hutchinson, S.A.; Jana, S.; Millican-Slater, R.A.; Smith, L.; Teske, M.C.; Wastall, L.M.; et al. MiR-19b non-canonical binding is directed by HuR and confers chemosensitivity through regulation of P-glycoprotein in breast cancer. Biochim. Biophys. Acta 2018, 1861, 996–1006. [Google Scholar] [CrossRef]
- Kim, B.; Stephen, S.; Hanby, A.; Horgan, K.; Perry, S.; Richardson, J.; Roundhill, E.; Valleley, E.; Verghese, E.; Williams, B.; et al. Chemotherapy induces Notch1-dependent MRP1 up-regulation, inhibition of which sensitizes breast cancer cells to chemotherapy. BMC Cancer 2015, 15, 634. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hutchinson, S.A.; Lianto, P.; Moore, J.B.; Hughes, T.A.; Thorne, J.L. Phytosterols Inhibit Side-Chain Oxysterol Mediated Activation of LXR in Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3241. https://doi.org/10.3390/ijms20133241
Hutchinson SA, Lianto P, Moore JB, Hughes TA, Thorne JL. Phytosterols Inhibit Side-Chain Oxysterol Mediated Activation of LXR in Breast Cancer Cells. International Journal of Molecular Sciences. 2019; 20(13):3241. https://doi.org/10.3390/ijms20133241
Chicago/Turabian StyleHutchinson, Samantha A., Priscilia Lianto, J. Bernadette Moore, Thomas A. Hughes, and James L. Thorne. 2019. "Phytosterols Inhibit Side-Chain Oxysterol Mediated Activation of LXR in Breast Cancer Cells" International Journal of Molecular Sciences 20, no. 13: 3241. https://doi.org/10.3390/ijms20133241
APA StyleHutchinson, S. A., Lianto, P., Moore, J. B., Hughes, T. A., & Thorne, J. L. (2019). Phytosterols Inhibit Side-Chain Oxysterol Mediated Activation of LXR in Breast Cancer Cells. International Journal of Molecular Sciences, 20(13), 3241. https://doi.org/10.3390/ijms20133241





