Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation
Abstract
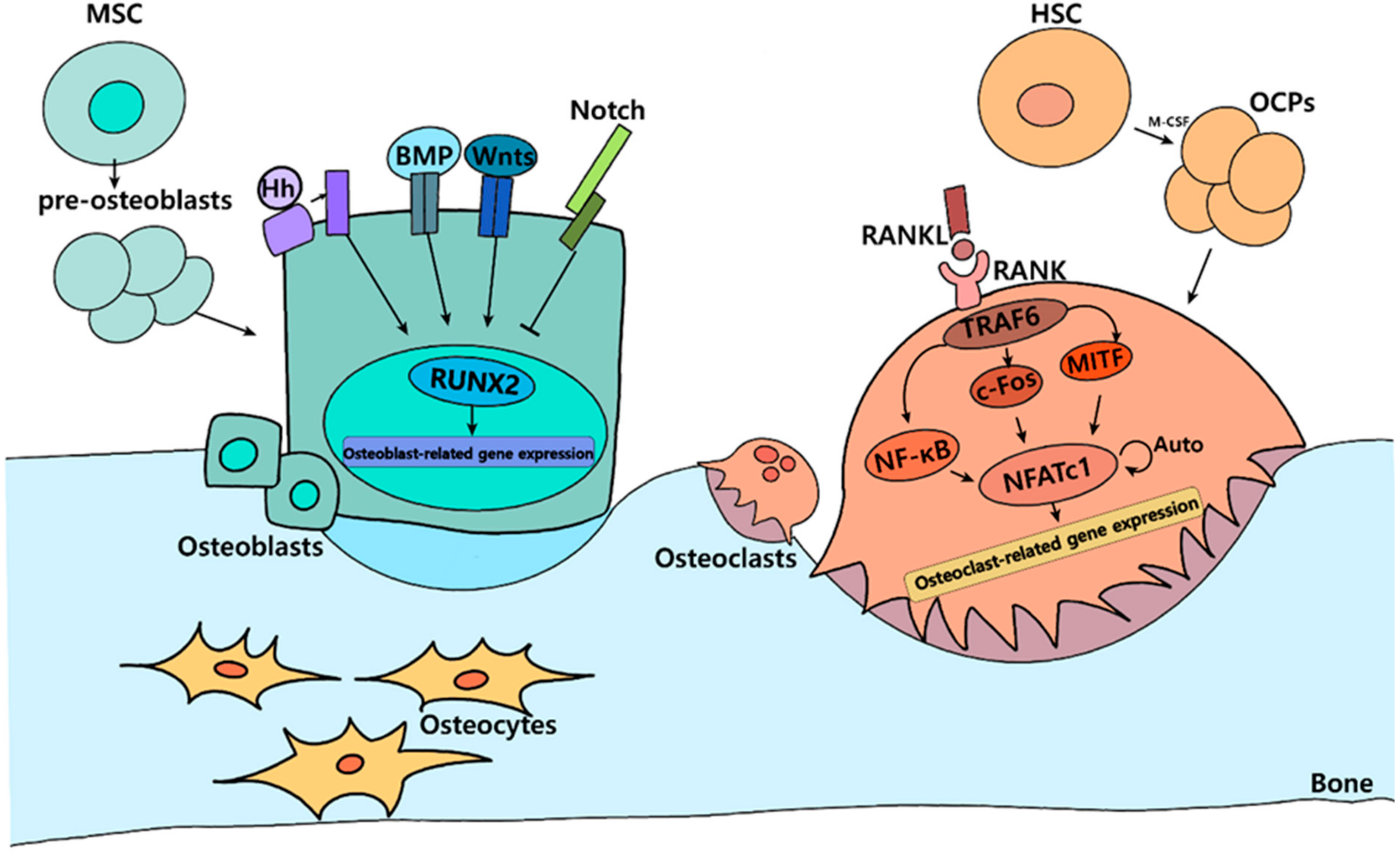
1. Introduction
2. Histone Acetylation in Osteoblastogenesis and Osteoclastogenesis
2.1. Histone Acetylation
2.2. Role of Histone Acetyltransferase in Osteoblastogenesis and Osteoclastogenesis
2.3. Role of Histone Deacetylase in Osteoblastogenesis and Osteoclastogenesis
3. Histone Methylation in Osteoblastogenesis and Osteoclastogenesis
3.1. Histone Methylation
3.2. Role of Histone Methyltransferase in Osteoblastogenesis and Osteoclastogenesis
3.3. Role of Histone Demethylase in Osteoblastogenesis and Osteoclastogenesis
4. Histone Cleavage in Osteoclastogenesis
4.1. Histone Cleavage
4.2. Role of Histone Cleavage in Osteoclastogenesis
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.V.; Dunleavy, E.; Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009, 10, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Gabay, O.; Clouse, K.A. Epigenetics of cartilage diseases. Jt. Bone Spine Rev. Rhum. 2016, 83, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Arnaudo, A.M.; Garcia, B.A. Proteomic characterization of novel histone post-translational modifications. Epigenet. Chromatin 2013, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.C.; Nielsen, E.C.; Matthiesen, R.; Jensen, L.H.; Sehested, M.; Finn, P.; Grauslund, M.; Hansen, A.M.; Jensen, O.N. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteom. 2006, 5, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom. 2007, 6, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Peng, C.; Montellier, E.; Lu, Z.; Chen, Y.; Ishii, H.; Debernardi, A.; Buchou, T.; Rousseaux, S.; Jin, F.; et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014, 10, 365–370. [Google Scholar] [CrossRef]
- Fierz, B.; Muir, T.W. Chromatin as an expansive canvas for chemical biology. Nat. Chem. Biol. 2012, 8, 417–427. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Azad, G.K.; Tomar, R.S. Proteolytic clipping of histone tails: The emerging role of histone proteases in regulation of various biological processes. Mol. Biol. Rep. 2014, 41, 2717–2730. [Google Scholar] [CrossRef] [PubMed]
- Dhaenens, M.; Glibert, P.; Meert, P.; Vossaert, L.; Deforce, D. Histone proteolysis: A proposal for categorization into ‘clipping’ and ‘degradation’. Bioessays 2015, 37, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Osley, M.A. Epigenetics: How to lose a tail. Nature 2008, 456, 885–886. [Google Scholar] [CrossRef]
- Zhou, P.; Wu, E.; Alam, H.B.; Li, Y. Histone cleavage as a mechanism for epigenetic regulation: Current insights and perspectives. Curr. Mol. Med. 2014, 14, 1164–1172. [Google Scholar] [CrossRef]
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simoes, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. BioMed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef]
- Sobacchi, C.; Schulz, A.; Coxon, F.P.; Villa, A.; Helfrich, M.H. Osteopetrosis: Genetics, treatment and new insights into osteoclast function. Nat. Rev. Endocrinol. 2013, 9, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Galindo, M.; Zaidi, S.K.; Vradii, D.; Bhat, B.M.; Robinson, J.A.; Choi, J.Y.; Komori, T.; Stein, J.L.; Lian, J.B.; et al. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003, 63, 5357–5362. [Google Scholar] [PubMed]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Rutkovskiy, A.; Stenslokken, K.O.; Vaage, I.J. Osteoblast Differentiation at a Glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yang, S.; Shao, J.; Li, Y.P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. J. Virtual Libr. 2007, 12, 3068–3092. [Google Scholar] [CrossRef]
- Park-Min, K.H. Epigenetic regulation of bone cells. Connect. Tissue Res. 2017, 58, 76–89. [Google Scholar] [CrossRef]
- Han, Y.; You, X.; Xing, W.; Zhang, Z.; Zou, W. Paracrine and endocrine actions of bone-the functions of secretory proteins from osteoblasts, osteocytes, and osteoclasts. Bone Res. 2018, 6, 16. [Google Scholar] [CrossRef]
- Tanaka, S.; Takahashi, N.; Udagawa, N.; Tamura, T.; Akatsu, T.; Stanley, E.R.; Kurokawa, T.; Suda, T. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J. Clin. Investig. 1993, 91, 257–263. [Google Scholar] [CrossRef]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.W.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef]
- Faulkner, B.; Astleford, K.; Mansky, K.C. Regulation of Osteoclast Differentiation and Skeletal Maintenance by Histone Deacetylases. Molecules 2019, 24, 1355. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Jeffries, M.A. Epigenetics and Bone Remodeling. Curr. Osteoporos. Rep. 2017, 15, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.A.R.; Stein, J.L.; Westendorf, J.J.; van Wijnen, A.J. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone 2015, 81, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Parthun, M.R.; Widom, J.; Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell 1996, 87, 85–94. [Google Scholar] [CrossRef]
- Kleff, S.; Andrulis, E.D.; Anderson, C.W.; Sternglanz, R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 1995, 270, 24674–24677. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Allis, C.D. Special HATs for special occasions: Linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 1996, 6, 176–184. [Google Scholar] [CrossRef]
- Marmorstein, R.; Zhou, M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.A.; Hassan, M.Q.; Koss, M.; Montecino, M.; Selleri, L.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 2011, 194, 146–150. [Google Scholar] [CrossRef]
- Gordon, J.A.; Hassan, M.Q.; Saini, S.; Montecino, M.; van Wijnen, A.J.; Stein, G.S.; Stein, J.L.; Lian, J.B. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol. Cell. Biol. 2010, 30, 3531–3541. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, M.B.; Shim, J.H.; Zou, W.; Sitara, D.; Schweitzer, M.; Hu, D.; Lotinun, S.; Sano, Y.; Baron, R.; Park, J.M.; et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J. Clin. Investig. 2010, 120, 2457–2473. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Yoon, W.J.; Seo, S.B.; Woo, K.M.; Kim, G.S.; Ryoo, H.M.; Baek, J.H. BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J. Biol. Chem. 2010, 285, 36410–36419. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Shevde, N.K.; Pike, J.W. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res. 2005, 20, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Partridge, N.C. Parathyroid hormone activation of matrix metalloproteinase-13 transcription requires the histone acetyltransferase activity of p300 and PCAF and p300-dependent acetylation of PCAF. J. Biol. Chem. 2010, 285, 38014–38022. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Carballo, E.; Ulsamer, A.; Susperregui, A.R.; Manzanares-Cespedes, C.; Sanchez-Garcia, E.; Bartrons, R.; Rosa, J.L.; Ventura, F. Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. J. Bone Miner. Res. 2011, 26, 718–729. [Google Scholar] [CrossRef]
- Boumah, C.E.; Lee, M.; Selvamurugan, N.; Shimizu, E.; Partridge, N.C. Runx2 recruits p300 to mediate parathyroid hormone’s effects on histone acetylation and transcriptional activation of the matrix metalloproteinase-13 gene. Mol. Endocrinol. 2009, 23, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, Y.; Jin, C.; Zhang, M.; Lv, L.; Zhang, X.; Liu, H.; Zhou, Y. Histone H3K9 Acetyltransferase PCAF Is Essential for Osteogenic Differentiation Through Bone Morphogenetic Protein Signaling and May Be Involved in Osteoporosis. Stem Cells 2016, 34, 2332–2341. [Google Scholar] [CrossRef]
- Pelletier, N.; Champagne, N.; Stifani, S.; Yang, X.J. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene 2002, 21, 2729–2740. [Google Scholar] [CrossRef]
- Hakelien, A.M.; Bryne, J.C.; Harstad, K.G.; Lorenz, S.; Paulsen, J.; Sun, J.; Mikkelsen, T.S.; Myklebost, O.; Meza-Zepeda, L.A. The regulatory landscape of osteogenic differentiation. Stem Cells 2014, 32, 2780–2793. [Google Scholar] [CrossRef]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-derived Mesenchymal Stem Cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef] [PubMed]
- Weilbaecher, K.N.; Motyckova, G.; Huber, W.E.; Takemoto, C.M.; Hemesath, T.J.; Xu, Y.; Hershey, C.L.; Dowland, N.R.; Wells, A.G.; Fisher, D.E. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol. Cell 2001, 8, 749–758. [Google Scholar] [CrossRef]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Punj, V.; Kim, J.M.; Lee, S.; Ulmer, T.S.; Lu, W.; Rice, J.C.; An, W. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016, 30, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xing, G.; Zhang, L.; Shang, J.; Li, Y.; Li, C.; Tian, F.; Yang, X. Satb1 promotes osteoclastogenesis by recruiting CBP to upregulate miR-223 expression in chronic kidney disease-mineral and bone disorder. Die Pharm. 2017, 72, 680–686. [Google Scholar] [CrossRef]
- Meier, J.C.; Tallant, C.; Fedorov, O.; Witwicka, H.; Hwang, S.Y.; van Stiphout, R.G.; Lambert, J.P.; Rogers, C.; Yapp, C.; Gerstenberger, B.S.; et al. Selective Targeting of Bromodomains of the Bromodomain-PHD Fingers Family Impairs Osteoclast Differentiation. ACS Chem. Biol. 2017, 12, 2619–2630. [Google Scholar] [CrossRef] [PubMed]
- Caron, C.; Boyault, C.; Khochbin, S. Regulatory cross-talk between lysine acetylation and ubiquitination: Role in the control of protein stability. Bioessays 2005, 27, 408–415. [Google Scholar] [CrossRef]
- Li, M.; Luo, J.; Brooks, C.L.; Gu, W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002, 277, 50607–50611. [Google Scholar] [CrossRef]
- Lu, J.; Qu, S.; Yao, B.; Xu, Y.; Jin, Y.; Shi, K.; Shui, Y.; Pan, S.; Chen, L.; Ma, C. Osterix acetylation at K307 and K312 enhances its transcriptional activity and is required for osteoblast differentiation. Oncotarget 2016, 7, 37471–37486. [Google Scholar] [CrossRef]
- Wang, C.Y.; Yang, S.F.; Wang, Z.; Tan, J.M.; Xing, S.M.; Chen, D.C.; Xu, S.M.; Yuan, W. PCAF acetylates Runx2 and promotes osteoblast differentiation. J. Bone Miner. Metab. 2013, 31, 381–389. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Youn, B.U.; Jin, H.M.; Kim, J.Y.; Moon, J.B.; Ko, A.; Seo, S.B.; Lee, K.Y.; Kim, N. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 2011, 436, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Feigenson, M.; Shull, L.C.; Taylor, E.L.; Camilleri, E.T.; Riester, S.M.; van Wijnen, A.J.; Bradley, E.W.; Westendorf, J.J. Histone Deacetylase 3 Deletion in Mesenchymal Progenitor Cells Hinders Long Bone Development. J. Bone Miner. Res. 2017, 32, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, P.; Ge, J.; Cheng, J.; Dong, W.; Yuan, H.; Du, Y.; Yang, M.; Sun, R.; Jiang, H. Histone deacetylase 8 suppresses osteogenic differentiation of bone marrow stromal cells by inhibiting histone H3K9 acetylation and RUNX2 activity. Int. J. Biochem. Cell Biol. 2014, 54, 68–77. [Google Scholar] [CrossRef] [PubMed]
- La Noce, M.; Mele, L.; Laino, L.; Iolascon, G.; Pieretti, G.; Papaccio, G.; Desiderio, V.; Tirino, V.; Paino, F. Cytoplasmic Interactions between the Glucocorticoid Receptor and HDAC2 Regulate Osteocalcin Expression in VPA-Treated MSCs. Cells 2019, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Suh, J.H.; Kim, A.Y.; Lee, Y.S.; Park, S.Y.; Kim, J.B. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006, 20, 2432–2443. [Google Scholar] [CrossRef]
- Paino, F.; La Noce, M.; Tirino, V.; Naddeo, P.; Desiderio, V.; Pirozzi, G.; De Rosa, A.; Laino, L.; Altucci, L.; Papaccio, G. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: Evidence for HDAC2 involvement. Stem Cells 2014, 32, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.M.; Kahler, R.A.; Li, X.; Westendorf, J.J. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 2004, 279, 41998–42007. [Google Scholar] [CrossRef]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef]
- Jeon, E.J.; Lee, K.Y.; Choi, N.S.; Lee, M.H.; Kim, H.N.; Jin, Y.H.; Ryoo, H.M.; Choi, J.Y.; Yoshida, M.; Nishino, N.; et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006, 281, 16502–16511. [Google Scholar] [CrossRef]
- Jensen, E.D.; Schroeder, T.M.; Bailey, J.; Gopalakrishnan, R.; Westendorf, J.J. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J. Bone Miner. Res. 2008, 23, 361–372. [Google Scholar] [CrossRef]
- Tseng, P.C.; Hou, S.M.; Chen, R.J.; Peng, H.W.; Hsieh, C.F.; Kuo, M.L.; Yen, M.L. Resveratrol promotes osteogenesis of human mesenchymal stem cells by upregulating RUNX2 gene expression via the SIRT1/FOXO3A axis. J. Bone Miner. Res. 2011, 26, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Simic, P.; Zainabadi, K.; Bell, E.; Sykes, D.B.; Saez, B.; Lotinun, S.; Baron, R.; Scadden, D.; Schipani, E.; Guarente, L. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating beta-catenin. EMBO Mol. Med. 2013, 5, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Feng, Z.; Wang, X.; Zeng, M.; Liu, J.; Han, S.; Xu, J.; Chen, L.; Cao, K.; Long, J.; et al. SIRT3/SOD2 maintains osteoblast differentiation and bone formation by regulating mitochondrial stress. Cell Death Differ. 2018, 25, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Sharma, S.M.; Bronisz, A.; Srinivasan, R.; Sankar, U.; Ostrowski, M.C. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol. Cell. Biol. 2007, 27, 4018–4027. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, J.; Kim, J.H.; Jin, H.M.; Zhou, B.; Lee, S.Y.; Kim, N. Protein inhibitor of activated STAT 3 modulates osteoclastogenesis by down-regulation of NFATc1 and osteoclast-associated receptor. J. Immunol. 2007, 178, 5588–5594. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Li, N.; Ding, N.; Liu, C.; Yang, X.; Kang, F.; Cao, Z.; Quan, H.; Hou, T.; Xu, J.; et al. HDAC2 regulates FoxO1 during RANKL-induced osteoclastogenesis. Am. J. Physiol. Cell Physiol. 2016, 310, C780–C787. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.; Kaiser, B.; Romsa, A.; Schwarz, T.; Gopalakrishnan, R.; Jensen, E.D.; Mansky, K.C. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J. Biol. Chem. 2011, 286, 12056–12065. [Google Scholar] [CrossRef] [PubMed]
- Blixt, N.C.; Faulkner, B.K.; Astleford, K.; Lelich, R.; Schering, J.; Spencer, E.; Gopalakrishnan, R.; Jensen, E.D.; Mansky, K.C. Class II and IV HDACs function as inhibitors of osteoclast differentiation. PLoS ONE 2017, 12, e0185441. [Google Scholar] [CrossRef]
- Jin, Z.; Wei, W.; Huynh, H.; Wan, Y. HDAC9 Inhibits Osteoclastogenesis via Mutual Suppression of PPARgamma/RANKL Signaling. Mol. Endocrinol. 2015, 29, 730–738. [Google Scholar] [CrossRef]
- Stemig, M.; Astelford, K.; Emery, A.; Cho, J.J.; Allen, B.; Huang, T.H.; Gopalakrishnan, R.; Mansky, K.C.; Jensen, E.D. Deletion of histone deacetylase 7 in osteoclasts decreases bone mass in mice by interactions with MITF. PLoS ONE 2015, 10, e0123843. [Google Scholar] [CrossRef]
- Huh, J.E.; Shin, J.H.; Jang, E.S.; Park, S.J.; Park, D.R.; Ko, R.; Seo, D.H.; Kim, H.S.; Lee, S.H.; Choi, Y.; et al. Sirtuin 3 (SIRT3) maintains bone homeostasis by regulating AMPK-PGC-1beta axis in mice. Sci. Rep. 2016, 6, 22511. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, Y.D.; Kim, H.J.; Lee, Z.H.; Kim, H.H. SOD2 and Sirt3 Control Osteoclastogenesis by Regulating Mitochondrial ROS. J. Bone Miner. Res. 2017, 32, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Han, L.; Iyer, S.; de Cabo, R.; Zhao, H.; O’Brien, C.A.; Manolagas, S.C.; Almeida, M. Sirtuin1 Suppresses Osteoclastogenesis by Deacetylating FoxOs. Mol. Endocrinol. 2015, 29, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Ka, S.O.; Lee, S.M.; Lee, S.I.; Park, J.W.; Park, B.H. Overexpression of sirtuin 6 suppresses inflammatory responses and bone destruction in mice with collagen-induced arthritis. Arthritis Rheum. 2013, 65, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Miao, L.; Lu, Y.; Wang, L. Sirtuin 1 inhibits TNF-alpha-mediated osteoclastogenesis of bone marrow-derived macrophages through both ROS generation and TRPV1 activation. Mol. Cell. Biochem. 2019, 455, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Huh, J.E.; Shin, J.; Park, D.R.; Ko, R.; Jin, G.R.; Seo, D.H.; Kim, H.S.; Shin, H.I.; Oh, G.T.; et al. Sirt6 cooperates with Blimp1 to positively regulate osteoclast differentiation. Sci. Rep. 2016, 6, 26186. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Lachner, M.; Jenuwein, T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002, 14, 286–298. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Cheng, X. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Volkel, P.; Angrand, P.O. The control of histone lysine methylation in epigenetic regulation. Biochimie 2007, 89, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kooistra, S.M.; Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012, 13, 297–311. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.F.; Nimura, K.; Lo, W.N.; Saga, K.; Kaneda, Y. Histone H3 lysine 36 methyltransferase Whsc1 promotes the association of Runx2 and p300 in the activation of bone-related genes. PLoS ONE 2014, 9, e106661. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Niu, N.; Li, L.; Shao, R.; Ouyang, H.; Zou, W. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol. 2018, 16, e2006522. [Google Scholar] [CrossRef] [PubMed]
- Hemming, S.; Cakouros, D.; Isenmann, S.; Cooper, L.; Menicanin, D.; Zannettino, A.; Gronthos, S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014, 32, 802–815. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, Y.H.; Li, L.Y.; Lang, J.; Yeh, S.P.; Shi, B.; Yang, C.C.; Yang, J.Y.; Lin, C.Y.; Lai, C.C.; et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 2011, 13, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, P.C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem. Biophys. Res. Commun. 2013, 432, 612–617. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chung, C.C.; Liu, Y.C.; Yeh, S.P.; Hsu, J.L.; Hung, M.C.; Su, H.L.; Li, L.Y. Enhancer of Zeste Homolog 2 and Histone Deacetylase 9c Regulate Age-Dependent Mesenchymal Stem Cell Differentiation into Osteoblasts and Adipocytes. Stem Cells 2016, 34, 2183–2193. [Google Scholar] [CrossRef]
- Dudakovic, A.; Camilleri, E.T.; Paradise, C.R.; Samsonraj, R.M.; Gluscevic, M.; Paggi, C.A.; Begun, D.L.; Khani, F.; Pichurin, O.; Ahmed, F.S.; et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J. Biol. Chem. 2018, 293, 12894–12907. [Google Scholar] [CrossRef]
- Hemming, S.; Cakouros, D.; Vandyke, K.; Davis, M.J.; Zannettino, A.C.; Gronthos, S. Identification of Novel EZH2 Targets Regulating Osteogenic Differentiation in Mesenchymal Stem Cells. Stem Cells Dev. 2016, 25, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Khani, F.; Thaler, R.; Paradise, C.R.; Deyle, D.R.; Kruijthof-de Julio, M.; Galindo, M.; Gordon, J.A.; Stein, G.S.; Dudakovic, A.; van Wijnen, A.J. Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation. J. Cell. Biochem. 2017, 118, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Kota, S.K.; Roening, C.; Patel, N.; Kota, S.B.; Baron, R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone 2018, 117, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Jang, A.R.; Kim, D.I.; Park, M.J.; Lim, S.K.; Kim, M.S.; Park, J.H. PRMT1 mediates RANKL-induced osteoclastogenesis and contributes to bone loss in ovariectomized mice. Exp. Mol. Med. 2018, 50, 111. [Google Scholar] [CrossRef] [PubMed]
- Higashihori, N.; Lehnertz, B.; Sampaio, A.; Underhill, T.M.; Rossi, F.; Richman, J.M. Methyltransferase G9A Regulates Osteogenesis via Twist Gene Repression. J. Dent. Res. 2017, 96, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Shin, Y.; Kim, J.; Ulmer, T.S.; An, W. H3K27me1 is essential for MMP-9-dependent H3N-terminal tail proteolysis during osteoclastogenesis. Epigenet. Chromatin 2018, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Qiao, Y.; Mun, S.H.; Lee, M.J.; Murata, K.; Bae, S.; Zhao, B.; Park-Min, K.H.; Ivashkiv, L.B. Cutting Edge: EZH2 Promotes Osteoclastogenesis by Epigenetic Silencing of the Negative Regulator IRF8. J. Immunol. 2016, 196, 4452–4456. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ge, W. The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis. Cell Death Dis. 2018, 9, 33. [Google Scholar] [CrossRef]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.H.; Wang, C.Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012, 11, 50–61. [Google Scholar] [CrossRef]
- Yang, D.; Okamura, H.; Nakashima, Y.; Haneji, T. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J. Biol. Chem. 2013, 288, 33530–33541. [Google Scholar] [CrossRef]
- Yang, D.; Okamura, H.; Teramachi, J.; Haneji, T. Histone demethylase Utx regulates differentiation and mineralization in osteoblasts. J. Cell. Biochem. 2015, 116, 2628–2636. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Aguilar, R.; Henriquez, B.; Lian, J.B.; Stein, J.L.; Stein, G.S.; van Wijnen, A.J.; van Zundert, B.; Allende, M.L.; Montecino, M. Epigenetic Control of the Bone-master Runx2 Gene during Osteoblast-lineage Commitment by the Histone Demethylase JARID1B/KDM5B. J. Biol. Chem. 2015, 290, 28329–28342. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sinha, K.; Deng, J.M.; Yasuda, H.; Krahe, R.; Behringer, R.R.; de Crombrugghe, B. Mesenchymal Deletion of Histone Demethylase NO66 in Mice Promotes Bone Formation. J. Bone Miner. Res. 2015, 30, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.M.; Yasuda, H.; Coombes, M.M.; Dent, S.Y.; de Crombrugghe, B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010, 29, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.M.; Yasuda, H.; Zhou, X.; de Crombrugghe, B. Osterix and NO66 histone demethylase control the chromatin of Osterix target genes during osteoblast differentiation. J. Bone Miner. Res. 2014, 29, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ermann, J.; Niu, N.; Yan, G.; Yang, Y.; Shi, Y.; Zou, W. Histone demethylase LSD1 regulates bone mass by controlling WNT7B and BMP2 signaling in osteoblasts. Bone Res. 2018, 6, 14. [Google Scholar] [CrossRef]
- Yasui, T.; Hirose, J.; Tsutsumi, S.; Nakamura, K.; Aburatani, H.; Tanaka, S. Epigenetic regulation of osteoclast differentiation: Possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J. Bone Miner. Res. 2011, 26, 2665–2671. [Google Scholar] [CrossRef]
- Liu, Y.; Arai, A.; Kim, T.; Kim, S.; Park, N.H.; Kim, R.H. Histone Demethylase Jmjd7 Negatively Regulates Differentiation of Osteoclast. Chin. J. Dent. Res. 2018, 21, 113–118. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, J.W.; Lee, K.H.; Yoon, H.; Shin, D.H.; Ju, U.I.; Seok, S.H.; Lim, S.H.; Lee, Z.H.; Kim, H.H.; et al. Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res. 2014, 24, 1231–1249. [Google Scholar] [CrossRef]
- Youn, M.Y.; Yokoyama, A.; Fujiyama-Nakamura, S.; Ohtake, F.; Minehata, K.; Yasuda, H.; Suzuki, T.; Kato, S.; Imai, Y. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J. Biol. Chem. 2012, 287, 12994–13004. [Google Scholar] [CrossRef]
- Duncan, E.M.; Muratore-Schroeder, T.L.; Cook, R.G.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 2008, 135, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali-Ellis, Z.; Goossens, W.; Margaryan, N.V.; Hendrix, M.J. Cleavage of Histone 3 by Cathepsin D in the involuting mammary gland. PLoS ONE 2014, 9, e103230. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, C.; Lee, S.; Deng, Y.; Wither, M.; Oh, S.; Ning, F.; Dege, C.; Zhang, Q.; Liu, X.; et al. Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc. Natl. Acad. Sci. USA 2017, 114, E7717–E7726. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Verma, N.; Chauhan, S.; Tomar, R.S. Unexpected histone H3 tail-clipping activity of glutamate dehydrogenase. J. Biol. Chem. 2013, 288, 18743–18757. [Google Scholar] [CrossRef] [PubMed]
- Melo, F.R.; Vita, F.; Berent-Maoz, B.; Levi-Schaffer, F.; Zabucchi, G.; Pejler, G. Proteolytic histone modification by mast cell tryptase, a serglycin proteoglycan-dependent secretory granule protease. J. Biol. Chem. 2014, 289, 7682–7690. [Google Scholar] [CrossRef]
- Melo, F.R.; Wallerman, O.; Paivandy, A.; Calounova, G.; Gustafson, A.M.; Sabari, B.R.; Zabucchi, G.; Allis, C.D.; Pejler, G. Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells. J. Allergy Clin. Immunol. 2017, 140, 474–485. [Google Scholar] [CrossRef]
- Shen, J.; Xiang, X.; Chen, L.; Wang, H.; Wu, L.; Sun, Y.; Ma, L.; Gu, X.; Liu, H.; Wang, L.; et al. JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep. 2017, 18, 2131–2143. [Google Scholar] [CrossRef]
- Yi, S.J.; Kim, K. Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep. 2018, 51, 211–218. [Google Scholar] [CrossRef]
- Shin, Y.; Ghate, N.B.; Moon, B.; Park, K.; Lu, W.; An, W. DNMT and HDAC inhibitors modulate MMP-9-dependent H3 N-terminal tail proteolysis and osteoclastogenesis. Epigenet. Chromatin 2019, 12, 25. [Google Scholar] [CrossRef]

| Osteoblstogenesis | Osteoclastogenesis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Synonyms | Function | Histone Modifications Studied | Non-Histone Substrates or Interacting Proteins | Ref. | Function | Histone Modifications Studied | Non-Histone Substrates or Interacting Proteins | Ref. |
| Histone acetyltransferases | |||||||||
| PCAF | KAT2B | ↑ | H3K9ac | RUNX2 acetylation | [45,47,48,60] | ↑ | NFATc1 acetylation | [53,61] | |
| P300 | KAT3B | ↑ | H3K18ac, H3K16ac, H4ac | [40,41,43,44,45,46] | ↑ | H3K18ac | Interaction with MITF | [52] | |
| CBP | KAT3A | ↑ | H4ac | Osterix acetylation at K307 and K312 | [42,44,59] | ↑ | H3K18ac | [53,54,55] | |
| MOZ | KAT6A, MYST3 | ↑ | Interaction with RUNX2 | [49] | ↑ | [56] | |||
| MORF | KAT6B, MYST4 | ↑ | Interaction with RUNX2 | [49] | ↑ | [56] | |||
| Histone deacetylases | |||||||||
| HDAC1 | ↓ | [65] | ↓ | [74,75] | |||||
| HDAC2 | ↓ | [64,66] | ↑ | [76] | |||||
| HDAC3 | ↓ | Interaction with RUNX2 | [62,67] | ↑ | [77] | ||||
| HDAC4 | ↓ | RUNX2 deacetylation; Interaction with SMAD3 | [68,69] | ↓ | [78] | ||||
| HDAC5 | ↓ | RUNX2 deacetylation; Interaction with SMAD3 | [68,69] | ↓ | NFATc1 deacetylation | [61,78] | |||
| HDAC7 | ↓ | Interaction with RUNX2 | [70] | ↓ | [77,80] | ||||
| HDAC8 | ↓ | H3K9ac | [63] | ||||||
| HDAC9 | ↓ | [78,79] | |||||||
| HDAC10 | ↓ | [78] | |||||||
| HDAC11 | ↓ | [78] | |||||||
| SIRT1 | ↑ | Beta-catenin deacetylation | [71,72] | ↓ | FoxO deacetylation | [83,85] | |||
| SIRT3 | ↑ | SOD deacetylation | [73] | ↓ | [81,82] | ||||
| SIRT6 | ↓or↑ | Interaction with Blimp1 | [84,86] | ||||||
| Osteoblstogenesis | Osteoclastogenesis | |||||||
|---|---|---|---|---|---|---|---|---|
| Name | Synonyms | Function | Histone Modifications Studied | Non-Histone Substrates or Interacting Proteins | Ref. | Function | Histone Modifications Studied | Ref. |
| Histone metyltransferases | ||||||||
| G9 | EHMT2 | ↑ | H3K9me2 | [105] | ↑ | H3K9me1 | [106] | |
| WHSC1 | NSD2 | ↑ | H3K36me3 | [94] | ||||
| SETD2 | ↑ | H3K36me3 | [95] | |||||
| EZH2 | ↓ | H3K27me3 | [96,97,98,99,100,101] | ↑ | H3K27me3 | [107] | ||
| SUV420H2 | ↓ | H4K20me3 | [102] | |||||
| PRMT1 | ↑ | Direct interaction with p65 | [104] | |||||
| PRMT5 | ↓ | H3R3me2s, H3R8me2s | [103] | |||||
| Dot1 | ↓ | H3K79me2 | [108] | |||||
| Histone demetylases | ||||||||
| LSD1 | KDM1A | ↓ | H3K4me2 | [116] | ||||
| JMJD2B | KDM4B | ↑ | H3K9me3 | [109] | ||||
| JARID1B | KDM5B | ↓ | H3K4me3 | [112] | ||||
| UTX | KDM6A | ↑ | H3K27me3 | [111] | ||||
| JMJD3 | KDM6B | ↑ | H3K27me3 | [109,110] | ↑ | H3K27me3 | [117] | |
| PHF2 | ↑ | RUNX2 demetylation | [119] | |||||
| NO66 | ↓ | H3K4me3, H3K36me3 | Interaction with osterix | [113,114,115] | ||||
| JMJD5 | KDM8 | ↓ | (NFATc1 hydroxylation) | [120] | ||||
| JMJD7 | ↓ | [118] | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, S.-J.; Lee, H.; Lee, J.; Lee, K.; Kim, J.; Kim, Y.; Park, J.-I.; Kim, K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int. J. Mol. Sci. 2019, 20, 3147. https://doi.org/10.3390/ijms20133147
Yi S-J, Lee H, Lee J, Lee K, Kim J, Kim Y, Park J-I, Kim K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. International Journal of Molecular Sciences. 2019; 20(13):3147. https://doi.org/10.3390/ijms20133147
Chicago/Turabian StyleYi, Sun-Ju, Hyerim Lee, Jisu Lee, Kyubin Lee, Junil Kim, Yeojin Kim, Jae-Il Park, and Kyunghwan Kim. 2019. "Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation" International Journal of Molecular Sciences 20, no. 13: 3147. https://doi.org/10.3390/ijms20133147
APA StyleYi, S.-J., Lee, H., Lee, J., Lee, K., Kim, J., Kim, Y., Park, J.-I., & Kim, K. (2019). Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. International Journal of Molecular Sciences, 20(13), 3147. https://doi.org/10.3390/ijms20133147





