Decreased Autophagy Impairs Decidualization of Human Endometrial Stromal Cells: A Role for ATG Proteins in Endometrial Physiology
Abstract
1. Introduction
2. Results
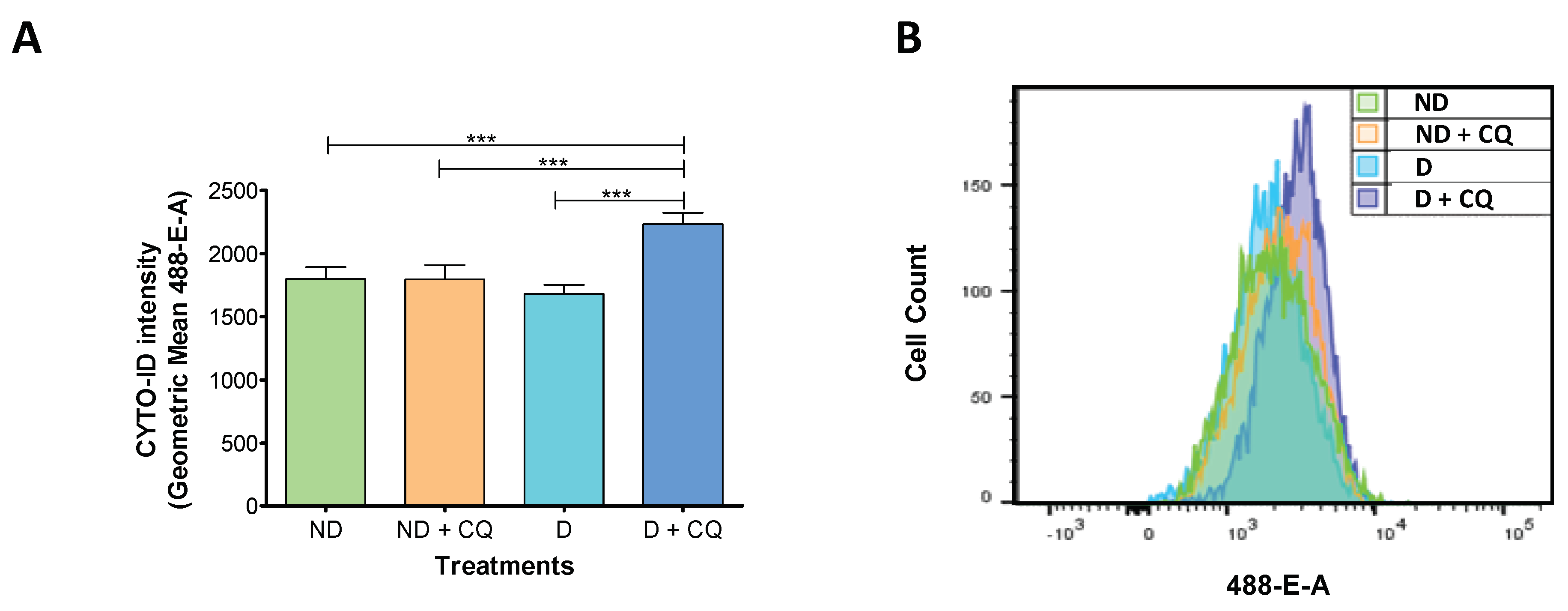
2.1. Autophagic Flux is Increased During t-HESC Decidualization
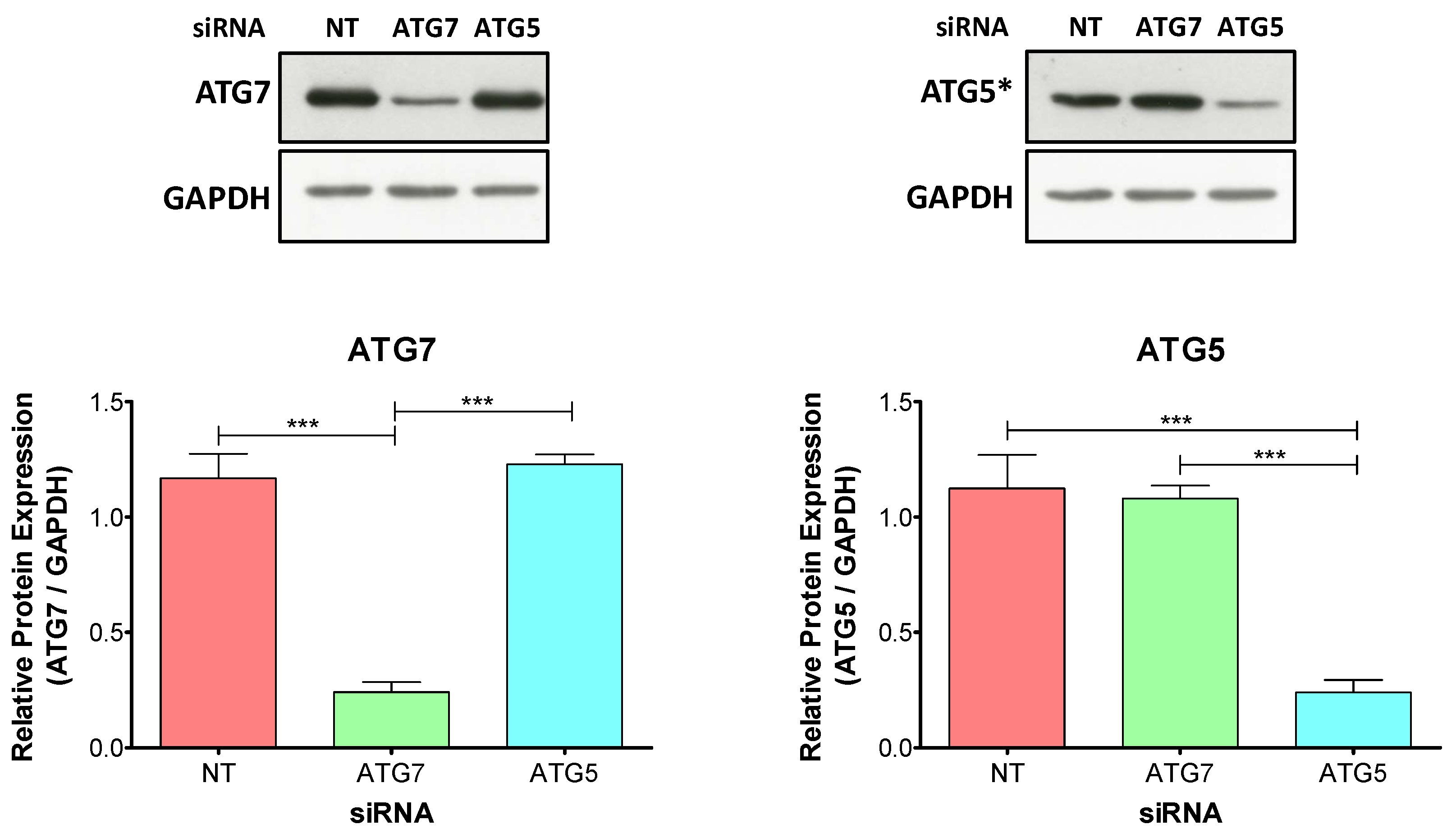
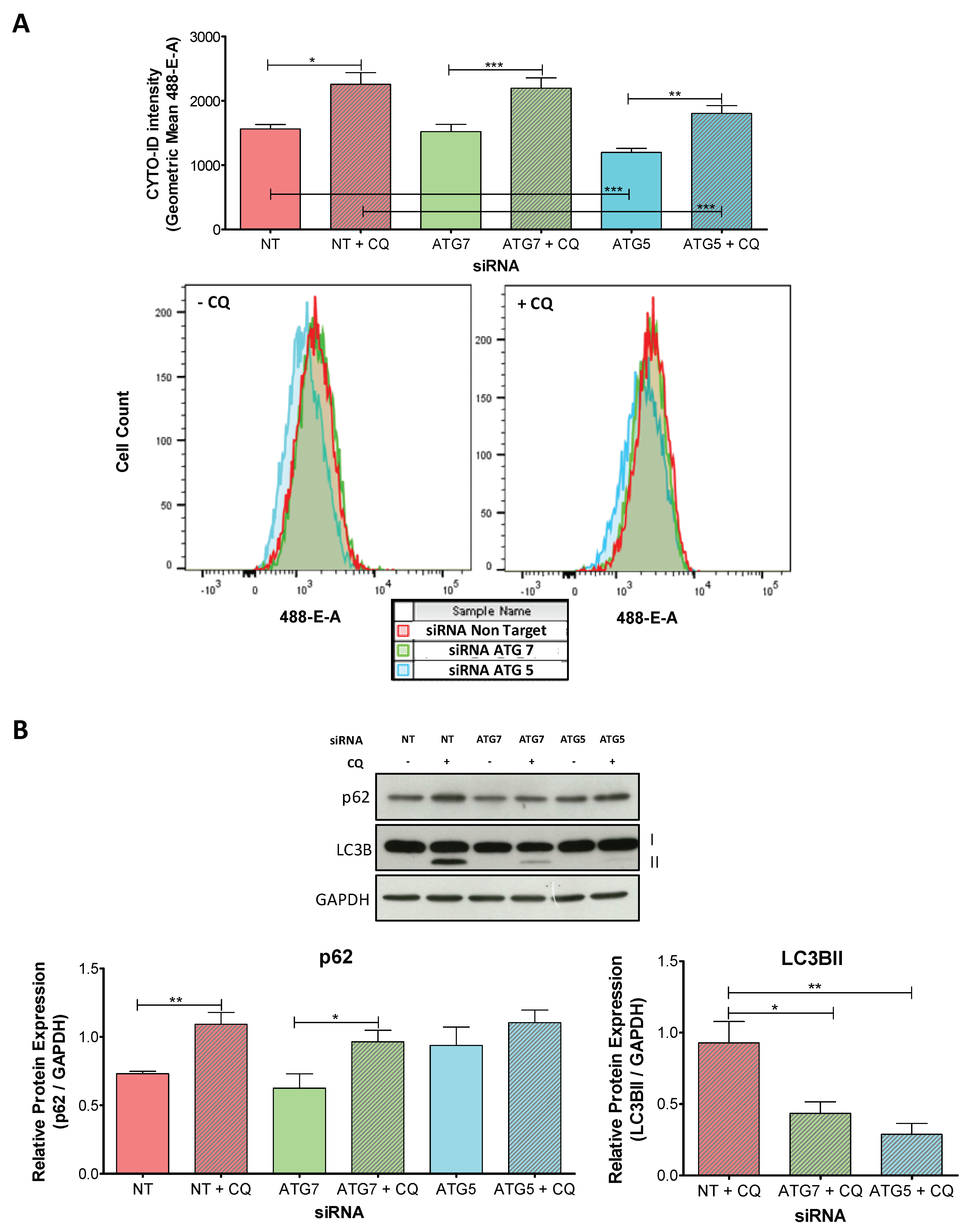
2.2. The Knockdown of ATG5 Is More Effective Than the Knockdown of ATG7 to Impair Autophagy During Decidualization
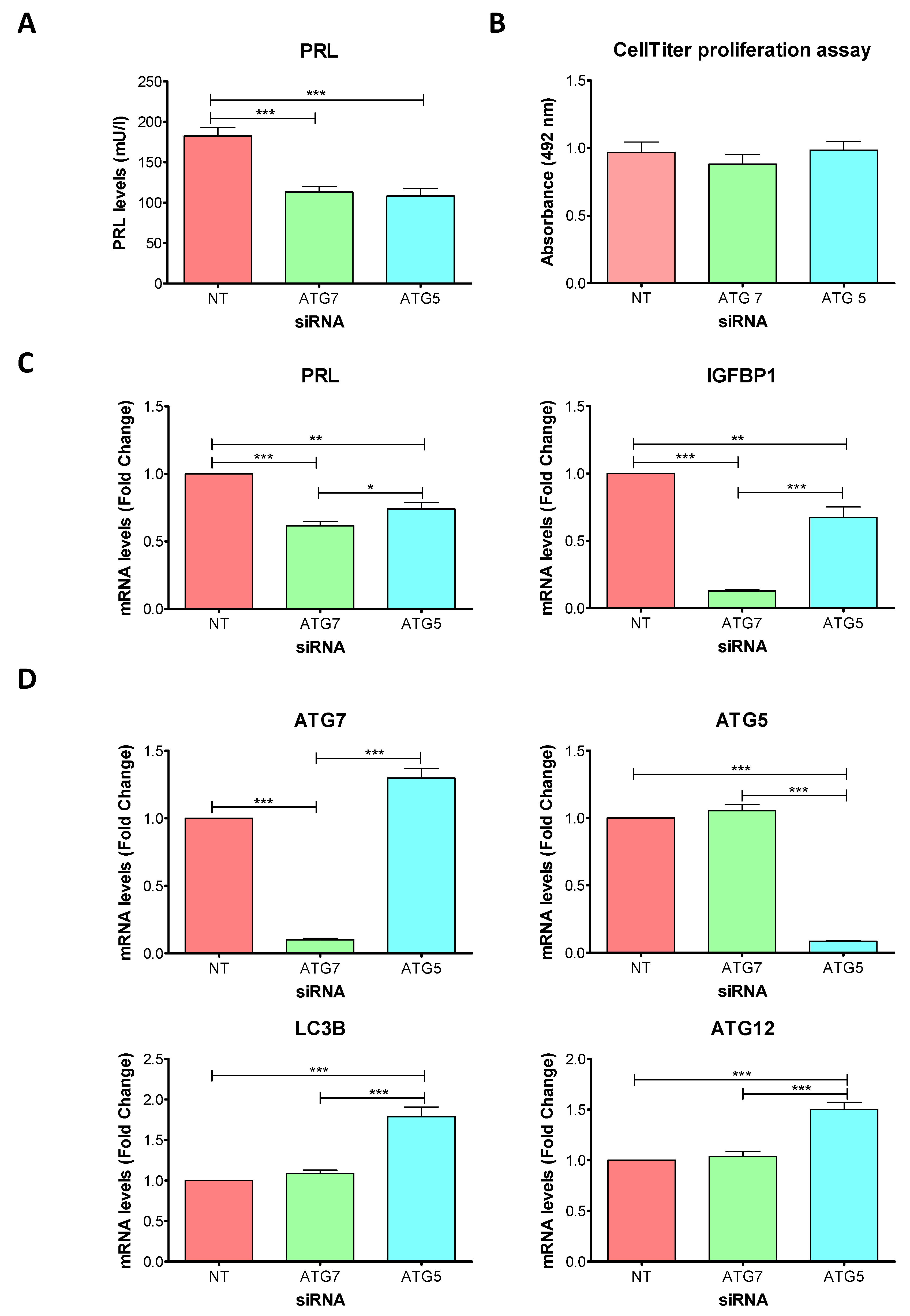
2.3. Knockdowns of ATG7 and ATG5 Impair Decidualization
3. Discussion
3.1. Possible Roles of Autophagy during Decidualization
3.2. Knockdowns of ATG7 and ATG5: Autophagy-Dependent and Autophagy-Independent Roles During Decidualization?
3.3. Autophagy and Endometrial Physiology
4. Materials and Methods
4.1. Cell Culture and in Vitro Decidualization Experiments
4.1.1. Cell Culture
4.1.2. Decidualization Experiments
4.2. Knockdown Experiments: Small Interfering RNA (siRNA) Transfection Followed by in Vitro Decidualization Experiments
4.2.1. siRNA Transfection
4.2.2. Decidualization Experiments Following siRNA Transfection
4.3. Flow Cytometry Autophagic Flux Measurement
4.4. Western Blot Analysis for Protein Detection
4.5. Prolactin Quantification
4.6. Gene Expression Assays
4.7. Proliferation Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 8-Br-cAMP | 8-Bromo-cyclic adenosine monophosphate |
| AF | Autophagic flux |
| ATG | Autophagy-related |
| CQ | Chloroquine |
| D | Decidualized |
| E | Estradiol |
| ESC | Endometrial stromal cells |
| IGFBP1 | Insulin-like growth factor binding protein 1 |
| MPA | Medroxyprogesterone |
| ND | Non decidualized |
| NT | Non-target |
| PRL | Prolactin |
| siRNA | Small interfering RNA |
| SN | Supernatant |
| t-HESC | Immortalized human endometrial stromal cells |
References
- Strowitzki, T.; Germeyer, A.; Popovici, R.; von Wolff, M. The human endometrium as a fertility-determining factor. Hum. Reprod. 2006, 12, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef] [PubMed]
- Ramathal, C.Y.; Bagchi, I.C.; Taylor, R.N.; Bagchi, M.K. Endometrial decidualization: Of mice and men. Semin. Reprod. Med. 2010, 28, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.K.; Lim, H.; Das, S.K.; Reese, J.; Paria, B.C.; Daikoku, T.; Wang, H. Molecular cues to implantation. Endocr. Rev. 2004, 25, 341–373. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, T.; Tanaka, K.; Oguro, T.; Tochigi, H.; Prechapanich, J.; Uchino, S.; Itakura, A.; Šućurović, S.; Murakami, K.; Brosens, J.J.; et al. Androgens modulate the morphological characteristics of human endometrial stromal cells decidualized in vitro. Reprod. Sci. 2014, 21, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Lane, B.; Oxberry, W.; Mazella, J.; Tseng, L. Decidualization of human endometrial stromal cells in vitro: Effects of progestin and relaxin on the ultrastructure and production of decidual secretory proteins. Hum. Reprod. 1994, 9, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Telgmann, R.; Gellersen, B. Marker genes of decidualization: Activation of the decidual prolactin gene. Hum. Reprod. Update 1998, 4, 472–479. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef]
- Reggiori, F.; Komatsu, M.; Finley, K.; Simonsen, A. Autophagy: More Than a Nonselective Pathway. Int. J. Cell Biol. 2012, 2012, 1–18. [Google Scholar] [CrossRef]
- Burman, C.; Ktistakis, N.T. Autophagosome formation in mammalian cells. Semin. Immunopathol. 2010, 32, 397–413. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Mariño, G.; Kroemer, G. Autophagy and aging. Cell 2015, 146, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Aoki, A.; Kusabiraki, T.; Shima, T.; Yoshino, O.; Cheng, S.B.; Sharma, S.; Saito, S. Role of autophagy in oocytogenesis, embryogenesis, implantation, and pathophysiology of pre-eclampsia. J. Obstet. Gynaecol. Res. 2017, 43, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jo, M.; Lee, E.; Oh, Y.K.; Choi, D. The Role of Autophagy in Human Endometrium. Biol. Reprod. 2012, 86, 1–10. [Google Scholar] [CrossRef]
- Rhee, J.S.; Saben, J.L.; Mayer, A.L.; Schulte, M.B.; Asghar, Z.; Stephens, C.; Chi, M.M.; Moley, K.H. Diet-induced obesity impairs endometrial stromal cell decidualization: A potential role for impaired autophagy. Hum. Reprod. 2016, 31, 1315–1326. [Google Scholar] [CrossRef]
- Avagliano, L.; Terraneo, L.; Virgili, E.; Martinelli, C.; Doi, P.; Samaja, M.; Bulfamante, G.P.; Marconi, A.M. Autophagy in normal and abnormal early human pregnancies. Reprod. Sci. 2015, 22, 838–844. [Google Scholar] [CrossRef]
- Zhan, L.; Li, J.; Wei, B. Autophagy in endometriosis: Friend or foe? Biochem. Biophys. Res. Commun. 2018, 495, 60–63. [Google Scholar] [CrossRef]
- Krikun, G.; Mor, G.; Alvero, A.; Guller, S.; Schatz, F.; Sapi, E.; Rahman, M.; Caze, R.; Qumsiyeh, M.; Lockwood, C.J. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology 2004, 145, 2291–2296. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Van Grol, J.; Subauste, C.S. Atg5 but not Atg7 in dendritic cells enhances IL-2 and IFN-γ production by Toxoplasma gondii-reactive CD4+ T cells. Microbes Infect. 2015, 17, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.O.; Yoo, S.M.; Ahn, H.H.; Nah, J.; Hong, S.H.; Kam, T.I.; Jung, S.; Jung, Y.K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aoki, A.; Nakashima, A.; Kusabiraki, T.; Ono, Y.; Yoshino, O.; Muto, M.; Kumasawa, K.; Yoshimori, T.; Ikawa, M.; Saito, S. Trophoblast-Specific Conditional Atg7 Knockout Mice Develop Gestational Hypertension. Am. J. Pathol. 2018, 188, 2474–2486. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef]
- Riffelmacher, T.; Simon, A.K. Mechanistic roles of autophagy in hematopoietic differentiation. FEBS J. 2017, 284, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Tian, F.J.; Lin, Y. Oxidative Stress in Placenta: Health and Diseases. Biomed Res. Int. 2015, 2015, 1–15. [Google Scholar] [CrossRef]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef]
- Yamanaka-Tatematsu, M.; Nakashima, A.; Fujita, N.; Shima, T.; Yoshimori, T.; Saito, S. Autophagy Induced by HIF1α Overexpression Supports Trophoblast Invasion by Supplying Cellular Energy. PLoS ONE 2013, 8, 2–10. [Google Scholar] [CrossRef]
- Woods, L.; Perez-Garcia, V.; Kieckbusch, J.; Wang, X.; Demayo, F.; Colucci, F.; Hemberger, M. Decidualisation and placentation defects are a major cause of age-related reproductive decline. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Kumeta, H.; Nakatogawa, H.; Satoo, K.; Adachi, W.; Ishii, J.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 2008, 13, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, S.G.; Yoo, S.M.; Son, J.H.; Jung, Y.K. Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy 2014, 10, 1906–1920. [Google Scholar] [CrossRef] [PubMed]
- Mauthe, M.; Langereis, M.; Jung, J.; Zhou, X.; Jones, A.; Omta, W.; Tooze, S.A.; Stork, B.; Paludan, S.R.; Ahola, T.; et al. An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J. Cell Biol. 2016, 214, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Bestebroer, J.; V’kovski, P.; Mauthe, M.; Reggiori, F. Hidden Behind Autophagy: The Unconventional Roles of ATG Proteins. Traffic 2013, 14, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Kawai, Y.; Fergusson, M.M.; Rovira, I.I.; Bishop, A.J.; Motoyama, N.; Cao, L.; Finkel, T. Atg7 Modulates p53 Activity to Regulate Cell Cycle and Survival During Metabolic Stress. Science 2012, 336, 225–228. [Google Scholar] [CrossRef]
- Ra, E.A.; Lee, T.A.; Won Kim, S.; Park, A.; Choi, H.J.; Jang, I.; Kang, S.; Hee Cheon, J.; Cho, J.W.; Eun Lee, J.; et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Honda, S.; Arakawa, S.; Nishida, Y.; Yamaguchi, H.; Ishii, E.; Shimizu, S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mestre Citrinovitz, A.C.; Strowitzki, T.; Germeyer, A. Decreased Autophagy Impairs Decidualization of Human Endometrial Stromal Cells: A Role for ATG Proteins in Endometrial Physiology. Int. J. Mol. Sci. 2019, 20, 3066. https://doi.org/10.3390/ijms20123066
Mestre Citrinovitz AC, Strowitzki T, Germeyer A. Decreased Autophagy Impairs Decidualization of Human Endometrial Stromal Cells: A Role for ATG Proteins in Endometrial Physiology. International Journal of Molecular Sciences. 2019; 20(12):3066. https://doi.org/10.3390/ijms20123066
Chicago/Turabian StyleMestre Citrinovitz, Ana Cecilia, Thomas Strowitzki, and Ariane Germeyer. 2019. "Decreased Autophagy Impairs Decidualization of Human Endometrial Stromal Cells: A Role for ATG Proteins in Endometrial Physiology" International Journal of Molecular Sciences 20, no. 12: 3066. https://doi.org/10.3390/ijms20123066
APA StyleMestre Citrinovitz, A. C., Strowitzki, T., & Germeyer, A. (2019). Decreased Autophagy Impairs Decidualization of Human Endometrial Stromal Cells: A Role for ATG Proteins in Endometrial Physiology. International Journal of Molecular Sciences, 20(12), 3066. https://doi.org/10.3390/ijms20123066




