Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach
Abstract
1. Introduction
2. Results
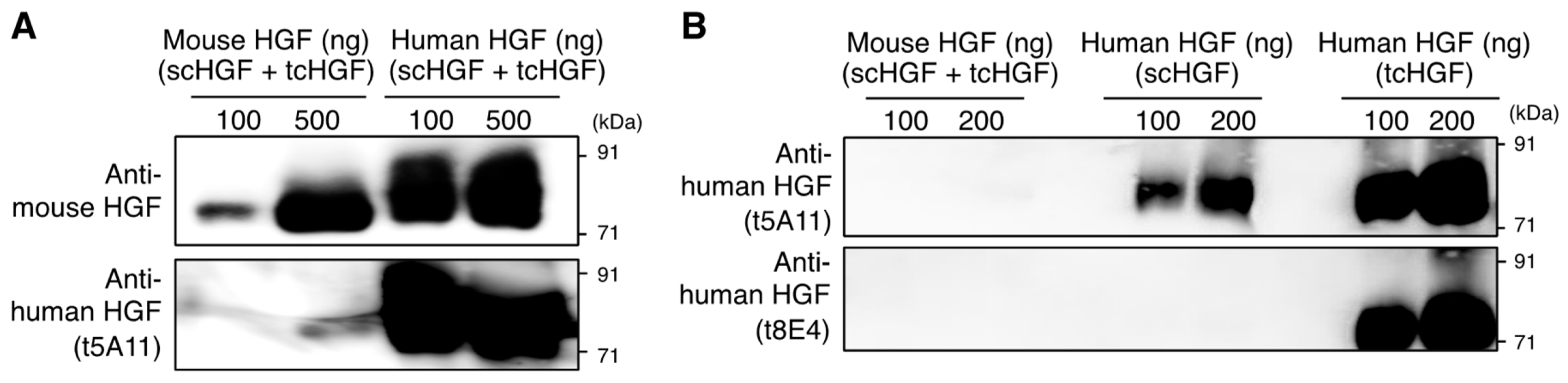
2.1. Specificity of Antibodies
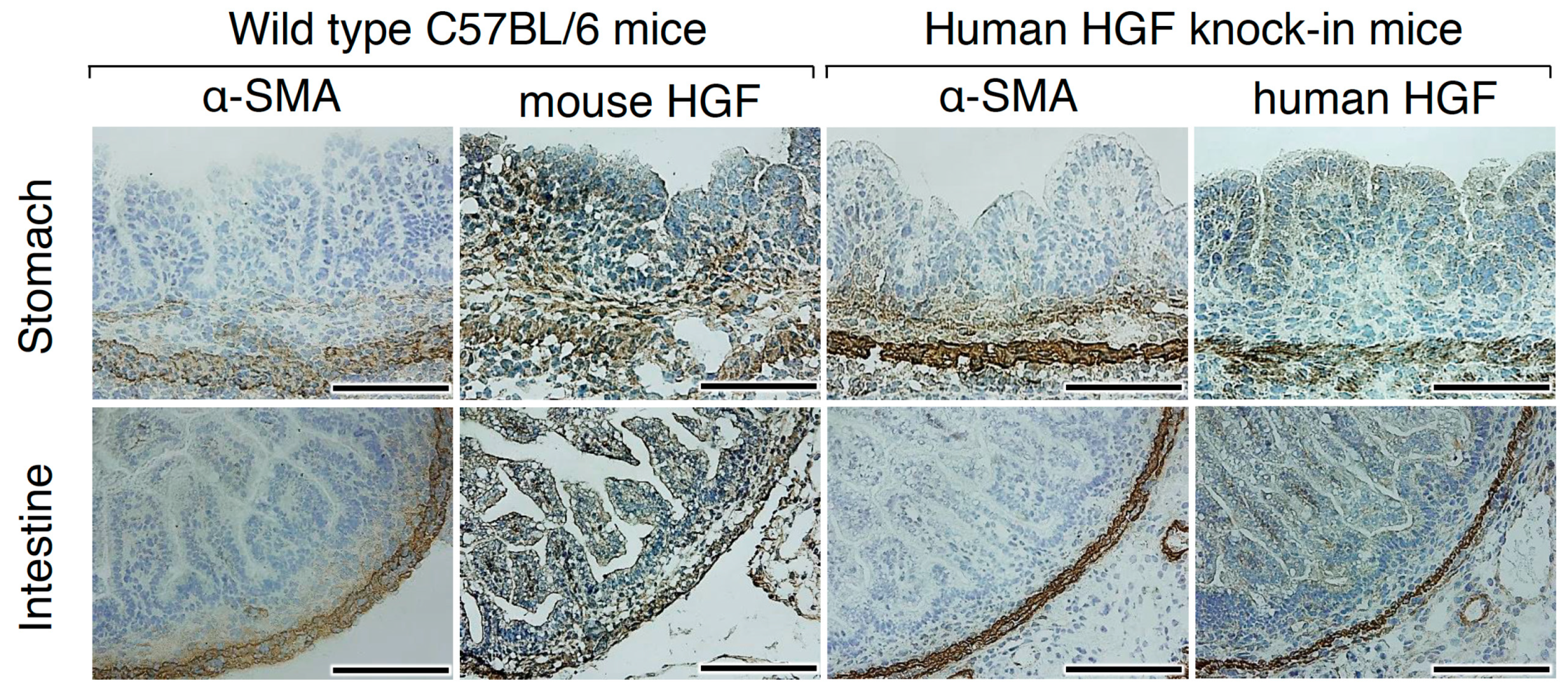
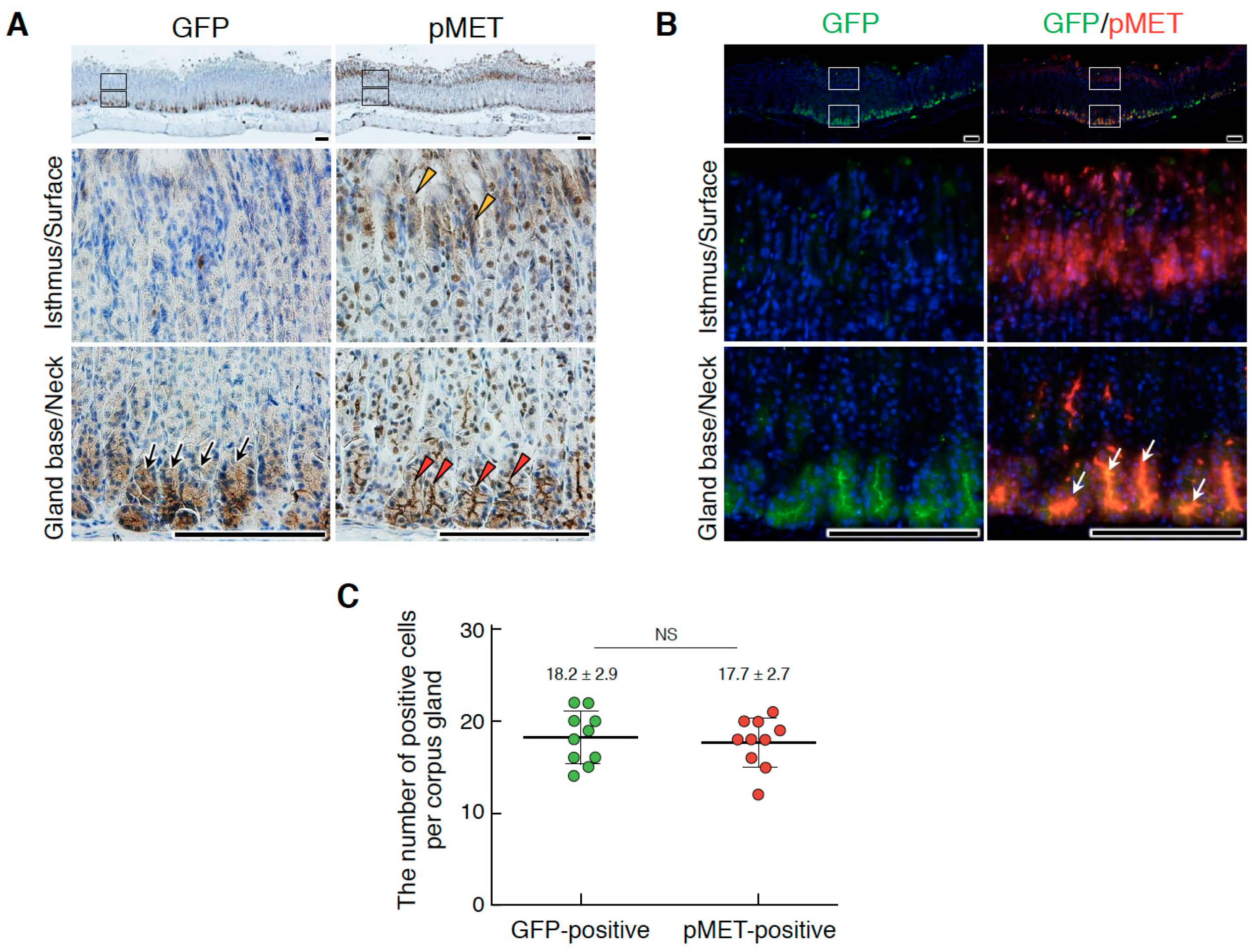
2.2. HGF and MET Receptor Localizations in Human Knock-in Mice
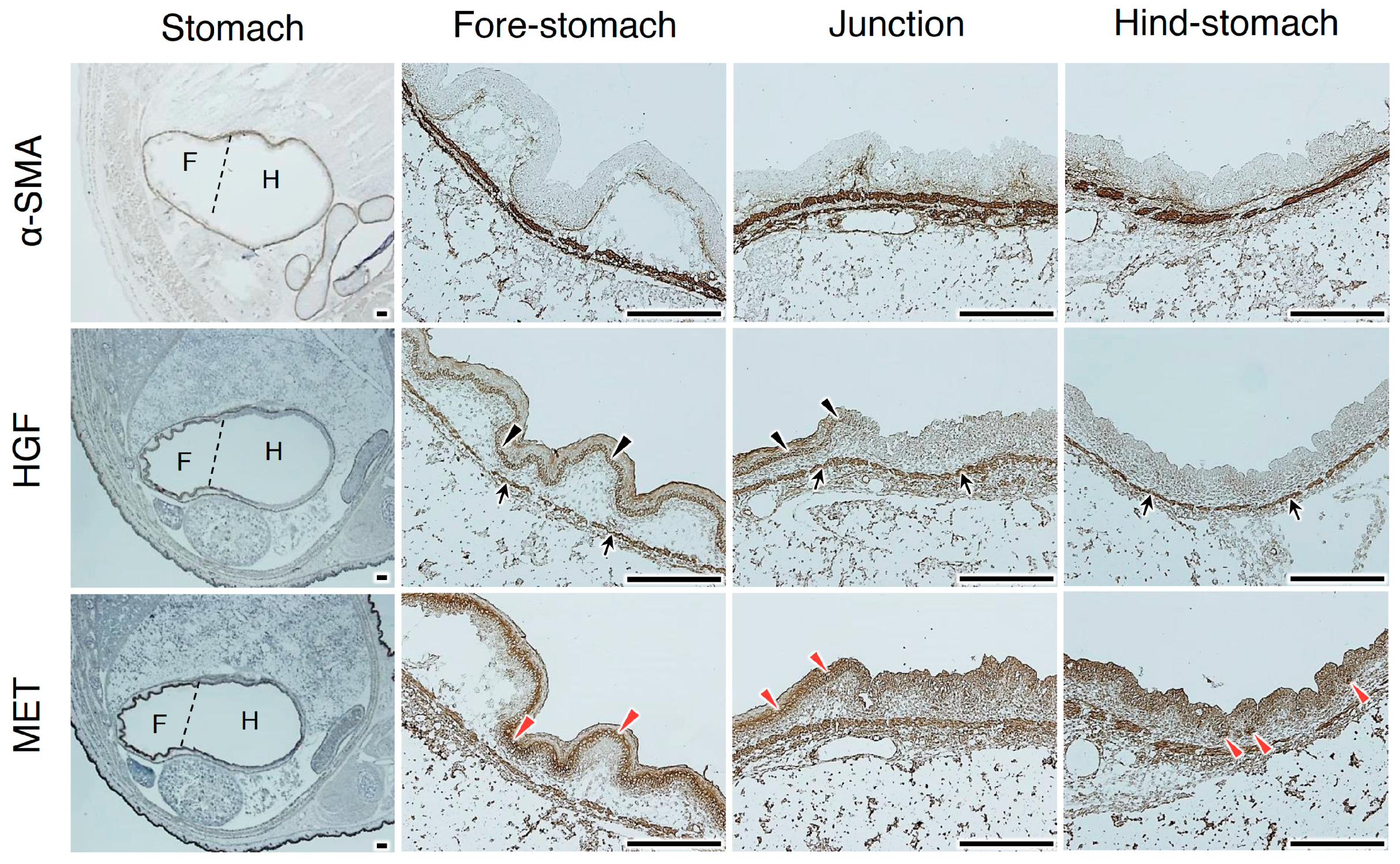
2.3. tcHGF and Phosphorylated MET Receptor in the Developing Stomach
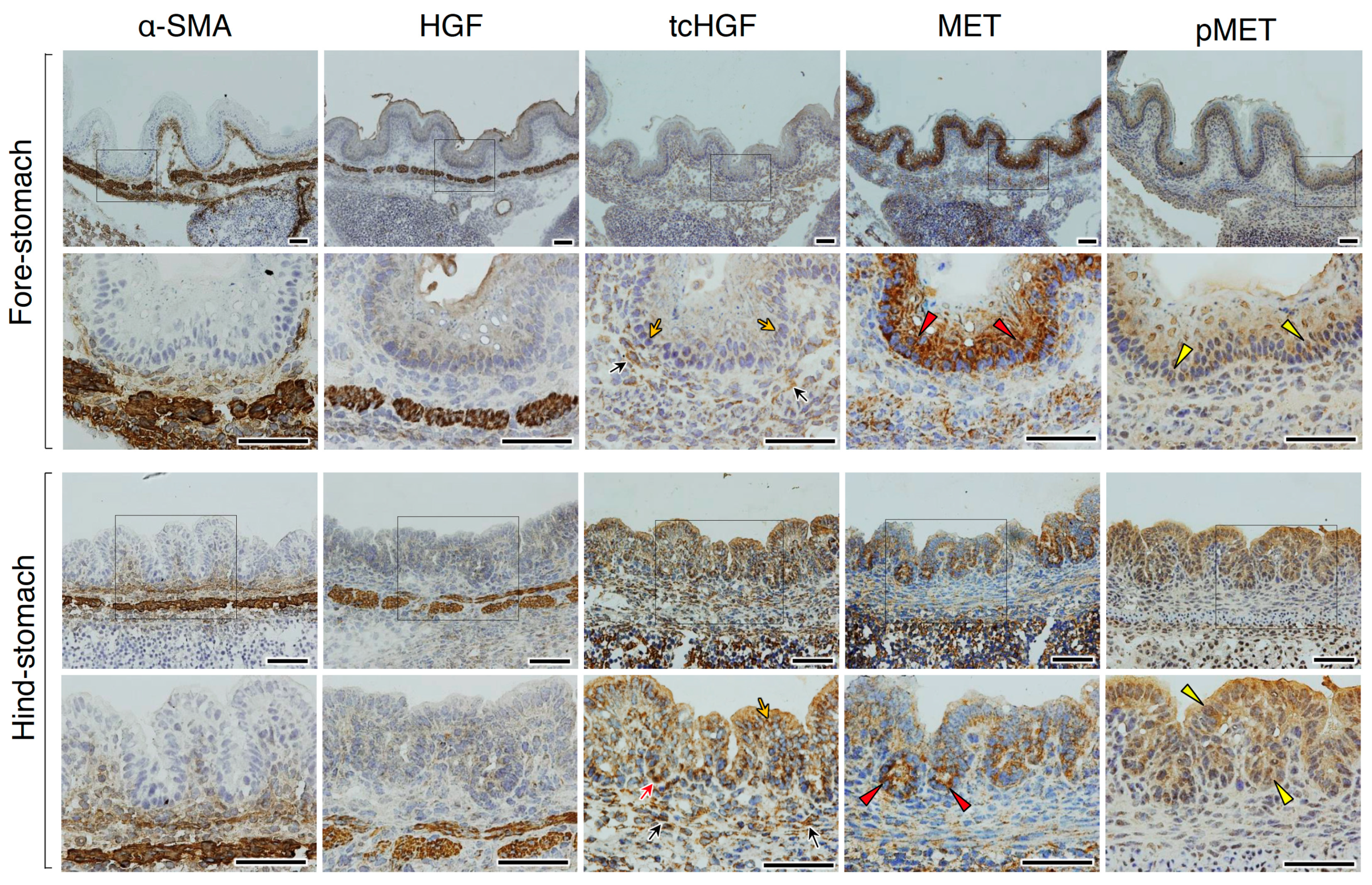
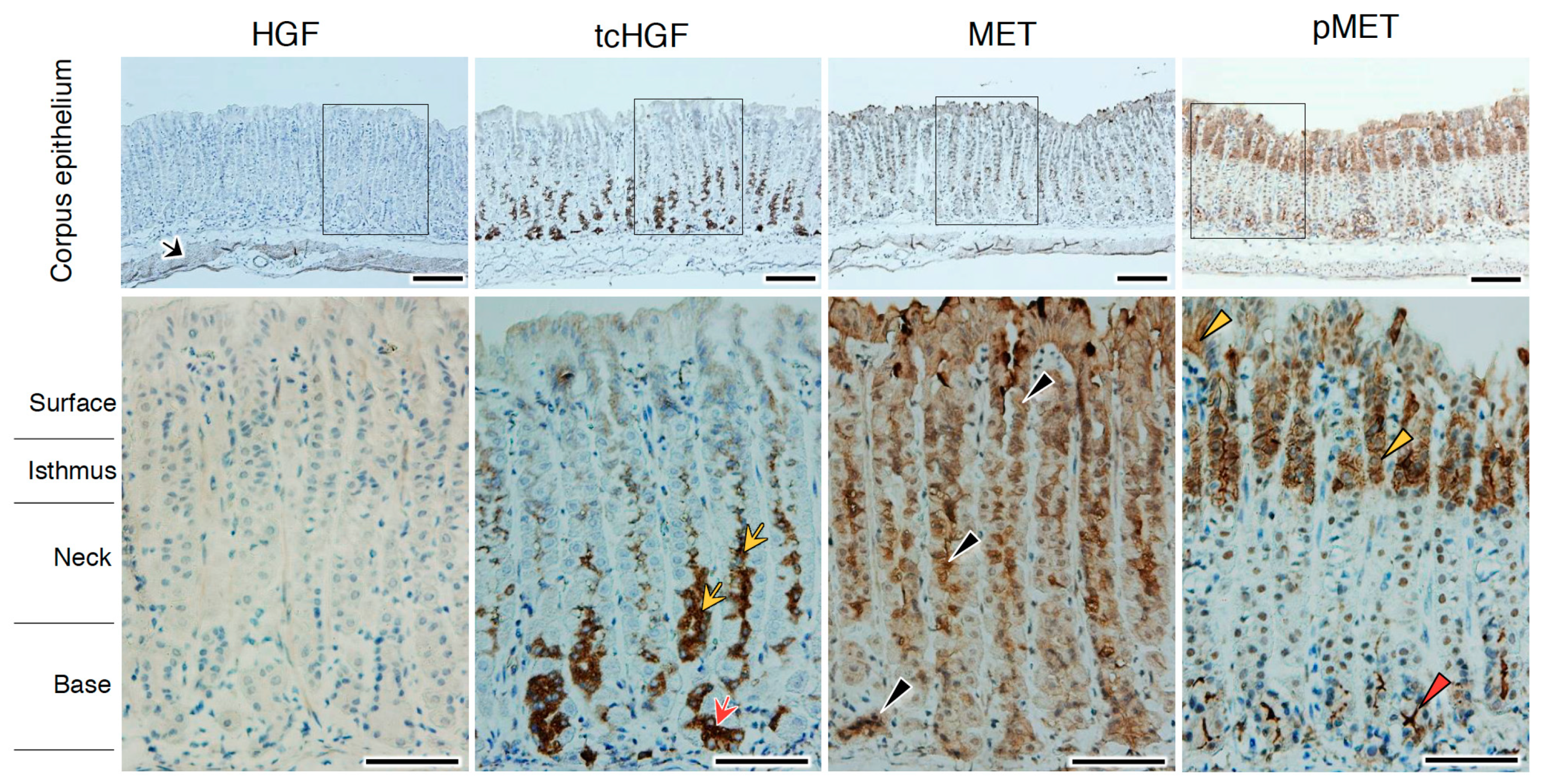
2.4. tcHGF and Phosphorylated MET Receptor in the Adult Stomach
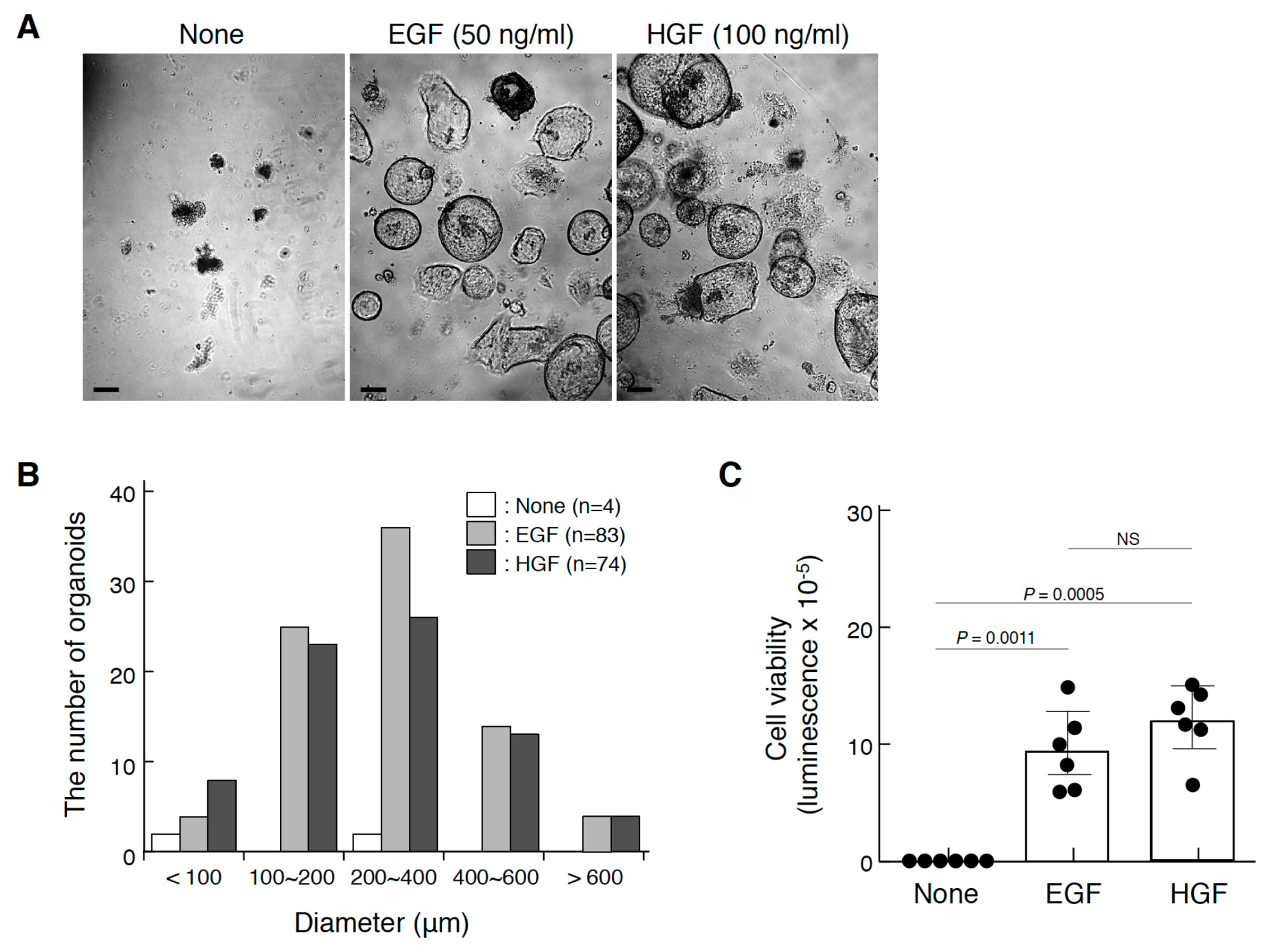
2.5. Promotion of Gastric Organoids
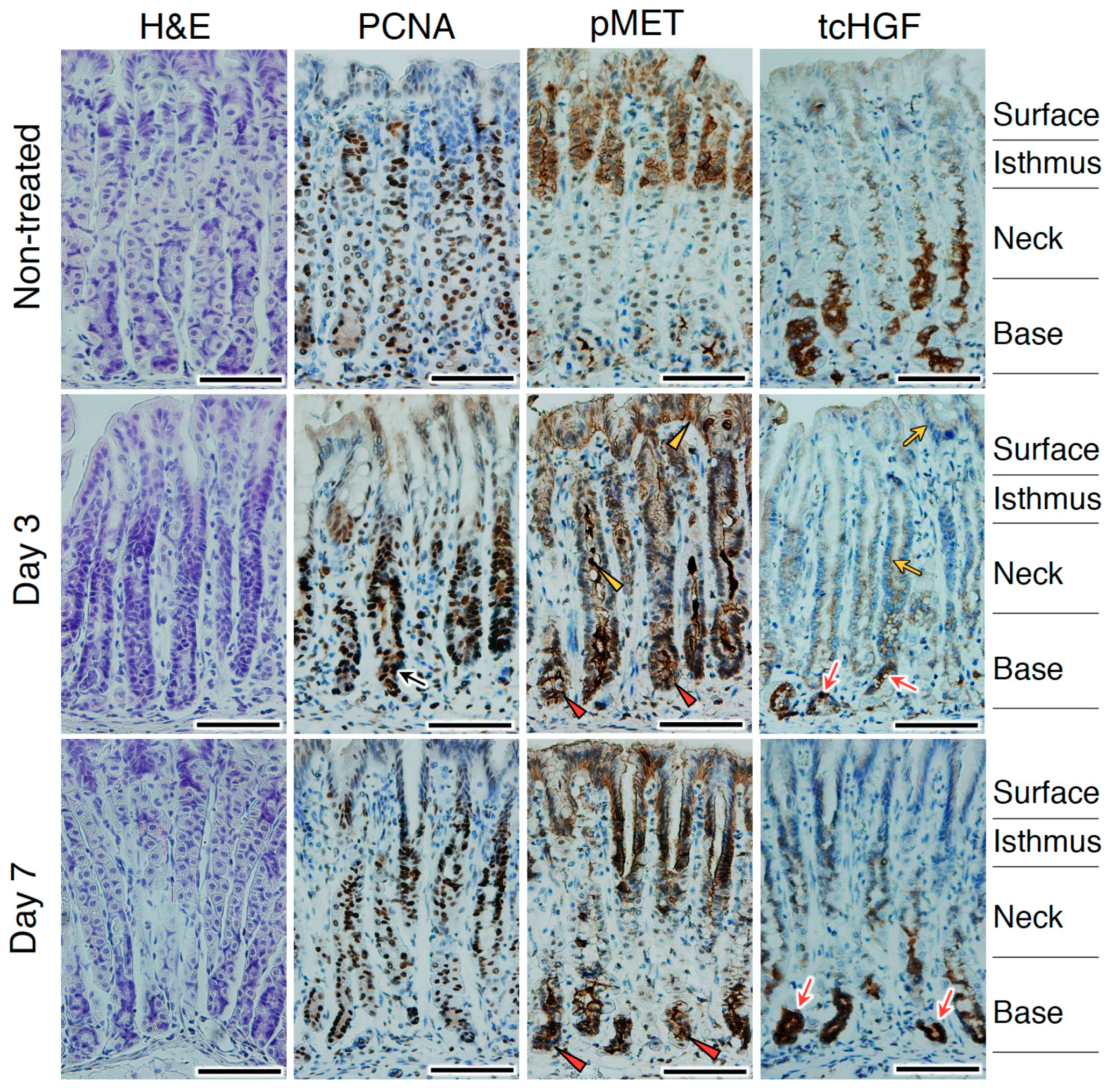
2.6. Changes in tcHGF and pMET Distributions Post-Injury
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Recombinant HGF
4.3. Western Blotting
4.4. Immunohistochemical Staining
4.5. Organoid Culture and Viable Cell Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Birchmeier, C.; Gherardi, E. Developmental roles of HGF/SF and its receptor, the c-met tyrosine kinase. Trends Cell Biol. 1998, 8, 404–410. [Google Scholar] [CrossRef]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Aoki, S.; Matsumoto, K. Hepatocyte growth factor and met in drug discovery. J. Biochem. 2015, 157, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Montesano, R.; Matsumoto, K.; Nakamura, T.; Orci, L. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 1991, 67, 901–908. [Google Scholar] [CrossRef]
- Soriano, J.V.; Pepper, M.S.; Nakamura, T.; Orci, L.; Montesano, R. Hepatocyte growth factor stimulates extensive development of branching duct-like structures by cloned mammary gland epithelial cells. J. Cell Sci. 1995, 108, 413–430. [Google Scholar] [PubMed]
- Weidner, K.M.; Behrens, J.; Vandekerckhove, J.; Birchmeier, W. Scatter factor: Molecular characteristics and effect on the invasiveness of epithelial cells. J. Cell Biol. 1990, 111, 2097–2108. [Google Scholar] [CrossRef]
- Nakamura, S.; Gohda, E.; Matsunaga, T.; Yamamoto, I.; Minowada, J. Production of hepatocyte growth factor by human haematopoietic cell lines. Cytokine 1994, 6, 285–294. [Google Scholar] [CrossRef]
- Matsumoto, K.; Okazaki, H.; Nakamura, T. Novel function of prostaglandins as inducers of gene expression of HGF and putative mediators of tissue regeneration. J. Biochem. 1995, 117, 458–464. [Google Scholar] [CrossRef]
- Yant, J.; Buluwela, L.; Niranjan, B.; Gusterson, B.; Kamalati, T. In vivo effects of hepatocyte growth factor/scatter factor on mouse mammary gland development. Exp. Cell Res. 1998, 241, 476–481. [Google Scholar] [CrossRef]
- Ohmichi, H.; Koshimizu, U.; Matsumoto, K.; Nakamura, T. Hepatocyte growth factor (HGF) acts as a mesenchyme-derived morphogenic factor during fetal lung development. Development 1998, 125, 1315–1324. [Google Scholar]
- Ikari, T.; Hiraki, A.; Seki, K.; Sugiura, T.; Matsumoto, K.; Shirasuna, K. Involvement of hepatocyte growth factor in branching morphogenesis of murine salivary gland. Dev. Dyn. 2003, 228, 173–184. [Google Scholar] [CrossRef]
- Ishibe, S.; Karihaloo, A.; Ma, H.; Zhang, J.; Marlier, A.; Mitobe, M.; Togawa, A.; Schmitt, R.; Czyczk, J.; Kashgarian, M.; et al. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development 2009, 136, 337–345. [Google Scholar] [CrossRef]
- Calvi, C.; Podowski, M.; Lopez-Mercado, A.; Metzger, S.; Misono, K.; Malinina, A.; Dikeman, D.; Poonyagariyon, H.; Ynalvez, L.; Derakhshandeh, R.; et al. Hepatocyte growth factor, a determinant of airspace homeostasis in the murine lung. PLoS Genet. 2013, 9, e1003228. [Google Scholar] [CrossRef]
- Uehara, Y.; Minowa, O.; Mori, C.; Shiota, K.; Kuno, J.; Noda, T.; Kitamura, N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 1995, 373, 702–705. [Google Scholar] [CrossRef]
- Schmidt, C.; Bladt, F.; Goedecke, S.; Brinkmann, V.; Zschiesche, W.; Sharpe, M.; Gherardi, E.; Birchmeier, C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 1995, 373, 699–702. [Google Scholar] [CrossRef]
- Bladt, F.; Riethmacher, D.; Isenmann, S.; Aguzzi, A.; Birchmeier, C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 1995, 376, 768–771. [Google Scholar] [CrossRef]
- Borowiak, M.; Garratt, A.N.; Wüstefeld, T.; Strehle, M.; Trautwein, C.; Birchmeier, C. Met provides essential signals for liver regeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 10608–10613. [Google Scholar] [CrossRef]
- Huh, C.G.; Factor, V.M.; Sánchez, A.; Uchida, K.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA 2004, 101, 4477–4482. [Google Scholar] [CrossRef]
- Chmielowiec, J.; Borowiak, M.; Morkel, M.; Stradal, T.; Munz, B.; Werner, S.; Wehland, J.; Birchmeier, C.; Birchmeier, W. c-Met is essential for wound healing in the skin. J. Cell Biol. 2007, 177, 151–162. [Google Scholar] [CrossRef]
- Joosten, S.P.J.; Zeilstra, J.; van Andel, H.; Mijnals, R.C.; Zaunbrecher, J.; Duivenvoorden, A.A.M.; van de Wetering, M.; Clevers, H.; Spaargaren, M.; Pals, S.T. MET signaling mediates intestinal crypt-villus development, regeneration, and adenoma formation and is promoted by stem cell CD44 isoforms. Gastroenterology 2017, 153, 1040–1053. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Kataoka, H. Mechanisms of hepatocyte growth factor activation in cancer tissues. Cancers 2014, 6, 1890–1904. [Google Scholar] [CrossRef]
- Matsumoto, K.; Umitsu, M.; De Silva, D.M.; Roy, A.; Bottaro, D.P. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017, 108, 296–307. [Google Scholar] [CrossRef]
- Miyazawa, K.; Shimomura, T.; Naka, D.; Kitamura, N. Proteolytic activation of hepatocyte growth factor in response to tissue injury. J. Biol. Chem. 1994, 269, 8966–8970. [Google Scholar]
- Kataoka, H.; Hamasuna, R.; Itoh, H.; Kitamura, N.; Koono, M. Activation of hepatocyte growth factor/scatter factor in colorectal carcinoma. Cancer Res. 2000, 60, 6148–6159. [Google Scholar]
- Leushacke, M.; Tan, S.H.; Wong, A.; Swathi, Y.; Hajamohideen, A.; Tan, L.T.; Goh, J.; Wong, E.; Denil, S.L.I.J.; Murakami, K.; et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 2017, 19, 774–786. [Google Scholar] [CrossRef]
- Umitsu, M.; Sakai, K.; Ogasawara, S.; Kaneko, M.K.; Asaki, R.; Tamura-Kawakami, K.; Kato, Y.; Matsumoto, K.; Takagi, J. Probing conformational and functional states of human hepatocyte growth factor by a panel of monoclonal antibodies. Sci. Rep. 2016, 6, 33149. [Google Scholar] [CrossRef]
- Sakai, K.; Passioura, T.; Sato, H.; Ito, K.; Furuhashi, H.; Umitsu, M.; Takagi, J.; Kato, Y.; Mukai, H.; Warashina, S.; et al. Macrocyclic peptide-based inhibition and imaging of hepatocyte growth factor. Nat. Chem. Biol. 2019, 15, 598–606. [Google Scholar] [CrossRef]
- Saenz, J.B.; Burclaff, J.; Mills, J.C. Modeling murine gastric metaplasia through tamoxifen-induced acute parietal cell loss. Methods Mol. Biol. 2016, 329–339. [Google Scholar] [CrossRef]
- Huh, W.J.; Khurana, S.S.; Geahlen, J.H.; Kohli, K.; Waller, R.A.; Mills, J.C. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 2012, 142, 21–24. [Google Scholar] [CrossRef]
- Lyon, M.; Deakin, J.A.; Mizuno, K.; Nakamura, T.; Gallagher, J.T. Interaction of hepatocyte growth factor with heparan sulfate. Elucidation of the major heparan sulfate structural determinants. J. Biol. Chem. 1994, 269, 11216–11223. [Google Scholar]
- Schuppan, D.; Schmid, M.; Somasundaram, R.; Ackermann, R.; Ruehl, M.; Nakamura, T.; Riecken, E.O. Collagens in the liver extracellular matrix bind hepatocyte growth factor. Gastroenterology 1998, 114, 139–152. [Google Scholar] [CrossRef]
- Fukushima, T.; Uchiyama, S.; Tanaka, H.; Kataoka, H. Hepatocyte growth factor activator: A proteinase linking tissue injury with repair. Int. J. Mol. Sci. 2018, 19, 3435. [Google Scholar] [CrossRef]
- Oberst, M.D.; Singh, B.; Ozdemirli, M.; Dickson, R.B.; Johnson, M.D.; Lin, C.Y. Characterization of matriptase expression in normal human tissues. J. Histochem. Cytochem. 2003, 51, 1017–1025. [Google Scholar] [CrossRef]
- Wang, J.K.; Lee, M.S.; Tseng, I.C.; Chou, F.P.; Chen, Y.W.; Fulton, A.; Lee, H.S.; Chen, C.J.; Johnson, M.D.; Lin, C.Y. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am. J. Physiol. 2009, 297, C459–C470. [Google Scholar] [CrossRef]
- List, K.; Kosa, P.; Szabo, R.; Bey, A.L.; Wang, C.B.; Molinolo, A.; Bugge, T.H. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am. J. Pathol. 2009, 175, 1453–1463. [Google Scholar] [CrossRef]
- Kosa, P.; Szabo, R.; Molinolo, A.A.; Bugge, T.H. Suppression of tumorigenicity-14, encoding matriptase, is a critical suppressor of colitis and colitis-associated colon carcinogenesis. Oncogene 2012, 31, 3679–3695. [Google Scholar] [CrossRef]
- Miyazawa, K.; Shimomura, T.; Kitamura, A.; Kondo, J.; Morimoto, Y.; Kitamura, N. Molecular cloning and sequence analysis of the cDNA for a human serine protease responsible for activation of hepatocyte growth factor. Structural similarity of the protease precursor to blood coagulation factor XII. J. Biol. Chem. 1993, 268, 10024–10028. [Google Scholar]
- Itoh, H.; Hamasuna, R.; Kataoka, H.; Yamauchi, M.; Miyazawa, K.; Kitamura, N.; Koono, M. Mouse hepatocyte growth factor activator gene: Its expression not only in the liver but also in the gastro-intestinal tract. Biochim. Biophys. Acta 2000, 1491, 295–302. [Google Scholar] [CrossRef]
- Itoh, H.; Naganuma, S.; Takeda, N.; Miyata, S.; Uchinokura, S.; Fukushima, T.; Uchiyama, S.; Tanaka, H.; Nagaike, K.; Shimomura, T.; et al. Regeneration of injured intestinal mucosa is impaired in hepatocyte growth factor activator-deficient mice. Gastroenterology 2004, 127, 1423–1435. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Schroeder, M.D.; Ma, C.; Rando, T.A. HGFA is an injury-regulated systemic factor that induces the transition of stem cells into GAlert. Cell Rep. 2017, 19, 479–486. [Google Scholar] [CrossRef]
- Leung, C.; Tan, S.H.; Barker, N. Recent advances in lgr5+ stem cell research. Trends Cell Biol. 2018, 28, 380–391. [Google Scholar] [CrossRef]
- Vermeulen, L.; De Sousa, E.; Melo, F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef]
- Fukuta, K.; Matsumoto, K.; Nakamura, T. Multiple biological responses are induced by glycosylation-deficient hepatocyte growth factor. Biochem. J. 2005, 388, 555–562. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jangphattananont, N.; Sato, H.; Imamura, R.; Sakai, K.; Terakado, Y.; Murakami, K.; Barker, N.; Oshima, H.; Oshima, M.; Takagi, J.; et al. Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach. Int. J. Mol. Sci. 2019, 20, 2955. https://doi.org/10.3390/ijms20122955
Jangphattananont N, Sato H, Imamura R, Sakai K, Terakado Y, Murakami K, Barker N, Oshima H, Oshima M, Takagi J, et al. Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach. International Journal of Molecular Sciences. 2019; 20(12):2955. https://doi.org/10.3390/ijms20122955
Chicago/Turabian StyleJangphattananont, Nawaphat, Hiroki Sato, Ryu Imamura, Katsuya Sakai, Yumi Terakado, Kazuhiro Murakami, Nick Barker, Hiroko Oshima, Masanobu Oshima, Junichi Takagi, and et al. 2019. "Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach" International Journal of Molecular Sciences 20, no. 12: 2955. https://doi.org/10.3390/ijms20122955
APA StyleJangphattananont, N., Sato, H., Imamura, R., Sakai, K., Terakado, Y., Murakami, K., Barker, N., Oshima, H., Oshima, M., Takagi, J., Kato, Y., Yano, S., & Matsumoto, K. (2019). Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach. International Journal of Molecular Sciences, 20(12), 2955. https://doi.org/10.3390/ijms20122955





