Diagnosis of Breast Hyperplasia and Evaluation of RuXian-I Based on Metabolomics Deep Belief Networks
Abstract
:1. Introduction
2. Results
2.1. Dataset
2.1.1. Chemicals and Reagents
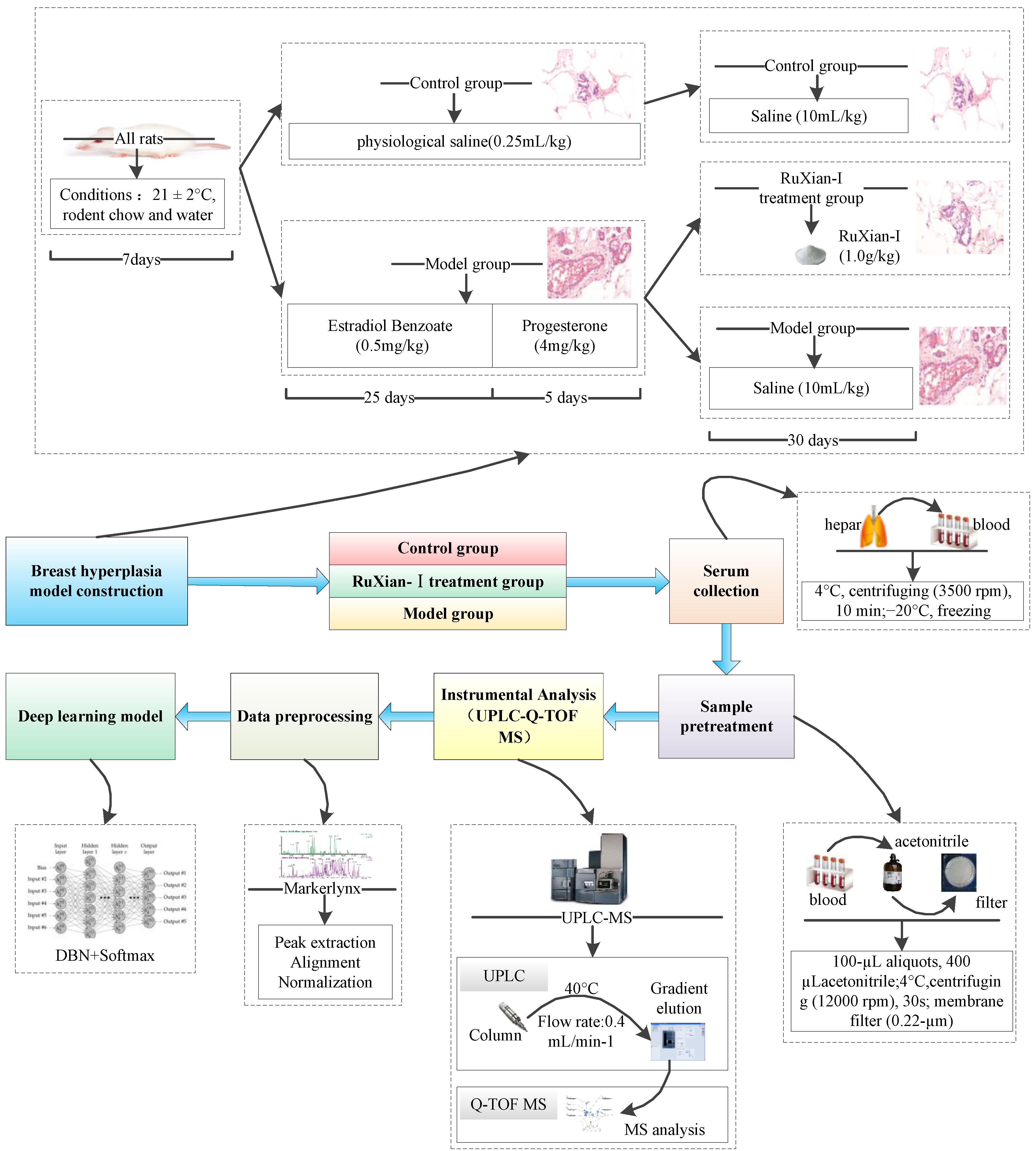
2.1.2. Breast Hyperplasia Model Construction and Treatment
2.1.3. UPLC-MS Conditions
2.1.4. Data Analysis
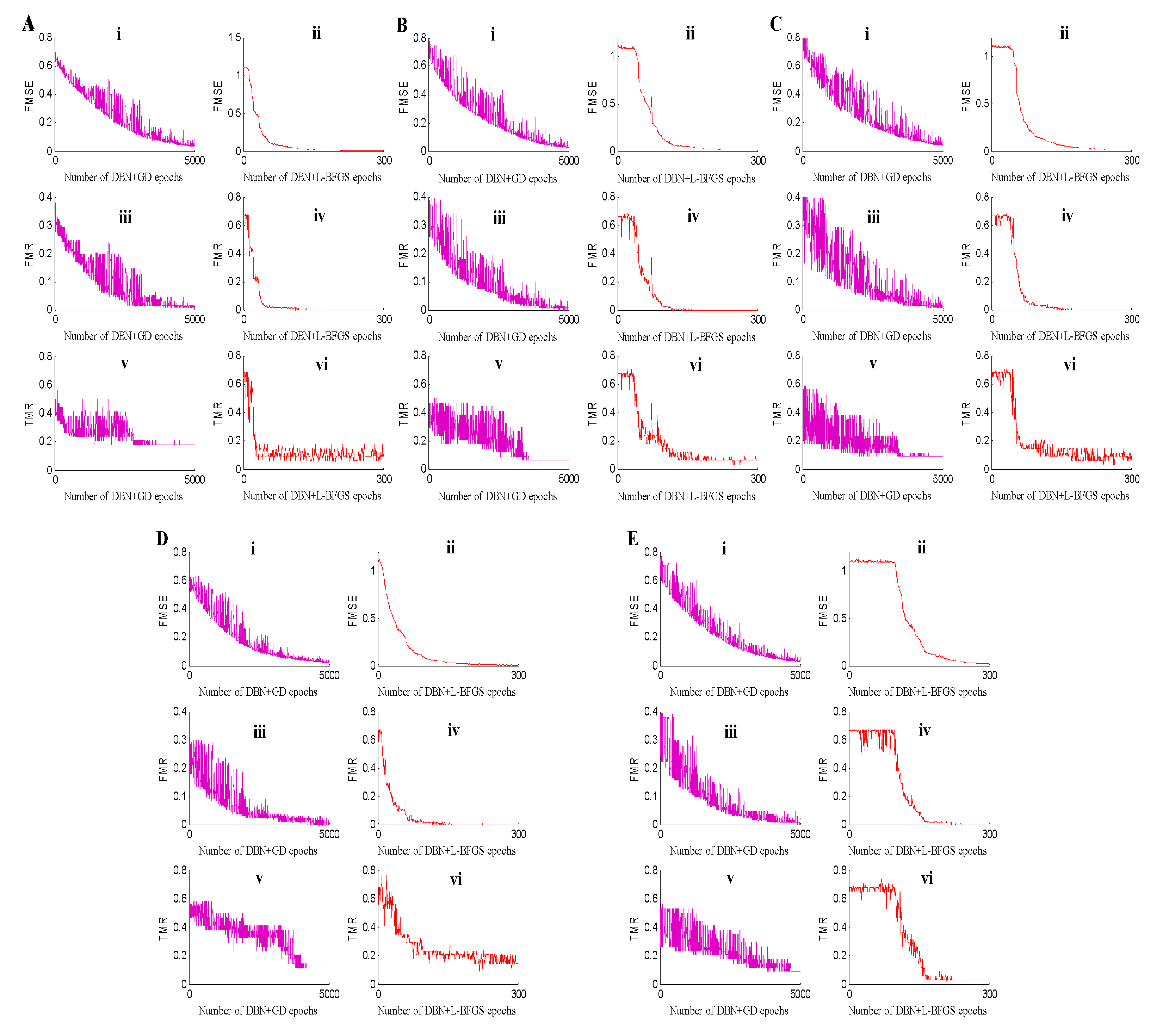
2.2. Classification Experiment
2.2.1. Classification Experiment of the Positive Spectrum Data
2.2.2. Classification Experiment of the Negative Spectrum Data
2.2.3. Classification Experiments across Different Training and Test Datasets for the Positive Spectrum Data
3. Discussion
4. Methods
4.1. Metabolomics Deep Belief Network
4.2. Dropout
4.3. DBN + Softmax Regression
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Ethics Statement
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Polat, A.K.; Kanbour-Shakir, A.; Menekse, E.; Levent, B.F.; Johnson, R. The Role of Mol. Biomarkers for Predicting Adjacent Breast Cancer of Atypical Ductal Hyperplasia Diagnosed on Core Biopsy. Cancer Biomark. 2016, 17, 293–300. [Google Scholar] [CrossRef]
- Amin, A.L.; Purdy, A.C.; Mattingly, J.D.; Kong, A.L.; Termuhlen, P.M. Benign Breast Disease. Surg. Clin. N. Am. 2013, 93, 299–308. [Google Scholar] [CrossRef]
- Cao, S.; Zhu, X.W.; Zhang, C.; Qian, H.; Schuttler, H.B.; Gong, J.P.; Xu, Y. Competition between DNA Methylation, Nucleotide Synthesis, and Antioxidation in Cancer Versus Normal Tissues. Cancer Res. 2017, 77, 4185–4195. [Google Scholar] [CrossRef] [PubMed]
- Pitschmann, A.S.; Purevsuren, A.; Obmann, D.; Natsagdorj, D.; Gunbilig, S.; Narantuya, C.K.; Glasl, S. Traditional Mongolian Medicine: History and Status Quo. Phytochem. Rev. 2013, 12, 943–959. [Google Scholar] [CrossRef]
- Zhang, J.F.; Liu, J.; Gong, G.H.; Zhang, B.; Wei, C.X. Mongolian Medicine RuXian-I Treatment of Estrogen-Induced Mammary Gland Hyperplasia in Rats Related to TCTP Regulating Apoptosis. Evid.-Based Complement. Altern. Med. 2019, 2019, 1907263. [Google Scholar] [CrossRef]
- Hancock, J.T. The Role of Redox Mechanisms in Cell Signalling. Mol. Biotechnol. 2009, 43, 162–166. [Google Scholar] [CrossRef]
- Krallinger, M.; Morgan, A.; Smith, L.; Leitner, F.; Tanabe, L.; Wilbur, J.; Lynette, H.; Alfonso, V. Evaluation of Text-Mining Systems for Biology: Overview of the Second Biocreative Community Challenge. Genome Biol. 2008, 9, S1. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Mohanram, K.; Cara, A.D.; Micheli, G.D.; Xenarios, I. Modeling Stochasticity and Robustness in Gene Regulatory Networks. Bioinformatics 2009, 25, i101–i109. [Google Scholar] [CrossRef]
- Sridhar, S.F.; Lam, G.E.; Blelloch, R.R.; Schwartz, R. Mixed Integer Linear Programming for Maximum-Parsimony Phylogeny Inference. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008, 5, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Fiehn, O.; Kopka, J.; Peter, D.; Altmann, T.; Trethewey, R.N.; Willmitzer, L.W. Metabolite Profiling for Plant Functional Genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.B.; Yuan, N.Y.; Lu, W.N.; Wen, W. Three Computational Tools for Predicting Bacterial Essential Genes. In Gene Essentiality: Methods and Protocols; Lu, L.J., Ed.; Springer: New York, NY, USA, 2015; pp. 205–217. [Google Scholar]
- Gromski, P.S.; Xu, Y.; Correa, E.; Ellis, D.I.; Turner, M.L.; Goodacre, R. A Comparative Investigation of Modern Feature Selection and Classification Approaches for the Analysis of Mass Spectrometry Data. Anal. Chim. Acta 2014, 829, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J. High-Throughput DNA Sequencing--Concepts and Limitations. BioEssays 2010, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, H.; Khan, H.; Birol, I. Ntcard: A Streaming Algorithm for Cardinality Estimation in Genomics Data. Bioinformatics 2017, 33, 1324–1330. [Google Scholar] [CrossRef]
- Lin, X.; Wang, Q.; Yin, P.; Tang, L.; Tan, Y. A Method for Handling Metabonomics Data from Liquid Chromatography/Mass Spectrometry: Combinational Use of Support Vector Machine Recursive Feature Elimination, Genetic Algorithm and Random Forest for Feature Selection. Metabolomics 2011, 4, 549–558. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Chang, C.; Jiang, M.; Wang, X.; Xu, L. Study the Therapeutic Mechanism of Amomum Compactum in Gentamicin-Induced Acute Kidney Injury Rat Based on a Back Propagation Neural Network Algorithm. J. Chromatogr. B 2017, 1040, 81–88. [Google Scholar] [CrossRef]
- Blanzieri, E.; Melgani, F. Nearest Neighbor Classification of Remote Sensing Images with the Maximal Margin Principle. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1804–1811. [Google Scholar] [CrossRef]
- Aliakbarzadeh, G.; Sereshti, H.; Parastar, H. Pattern Recognition Analysis of Chromatographic Fingerprints of Crocus Sativus, L. Secondary Metabolites Towards Source Identification and Quality Control. Anal. Bioanal. Chem. 2016, 408, 3295–3307. [Google Scholar]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A Tutorial Review: Metabolomics and Partial Least Squares-Discriminant Analysis--a Marriage of Convenience or a Shotgun Wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Hrydziuszko, O.; Mark, R.V. Missing Values in Mass Spectrometry Based Metabolomics: An Undervalued Step in the Data Processing Pipeline. Metabolomics 2012, 8, 161–174. [Google Scholar] [CrossRef]
- Bewick, V.; Liz, C.; Jonathan, B. Statistics Review 14: Logistic Regression. Crit. Care 2005, 9, 112–118. [Google Scholar] [CrossRef]
- Berk, R.; Lawrence, B.; Andreas, B.; Zhang, K.; Zhao, L. Valid Post-Selection Inference. Ann. Stat. 2013, 41, 802–837. [Google Scholar] [CrossRef]
- Ge, H.; Sun, L.; Tan, G.; Chen, Z.; Chen, C.L.P. Cooperative Hierarchical Pso with Two Stage Variable Interaction Reconstruction for Large Scale Optimization. IEEE Trans. Cybern. 2017, 47, 2809–2823. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. Erratum To: Deepcpg: Accurate Prediction of Single-Cell DNA Methylation States Using Deep Learning. Genome Biol. 2017, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G.E. Deep Learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Cireşan, D.; Meier, U.; Schmidhuber, J. Multi-Column Deep Neural Networks for Image Classification. Proc. Conf. Comput. Robot Vis. 2012, 157, 3642–3649. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R. Discovering Binary Codes for Documents by Learning Deep Generative Models. Top. Cogn. Sci. 2011, 3, 74–91. [Google Scholar] [CrossRef]
- Huang, C.; Gong, W.; Fu, W.; Feng, D. A Research of Speech Emotion Recognition Based on Deep Belief Network and Svm. Math. Probl. Eng. 2014, 2014, 749604. [Google Scholar] [CrossRef]
- Putin, E.; Mamoshina, P.; Aliper, A.; Korzinkin, M.; Moskalev, A.; Kolosov, A.; Ostrovskiy, A.; Cantor, C.; Vijg, J.; Zhavoronkov, A. Deep Biomarkers of Human Aging: Application of Deep Neural Networks to Biomarker Development. Aging 2016, 8, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, G.; Wei, Z. A Limited-Memory Bfgs Algorithm Based on a Trust-Region Quadratic Model for Large-Scale Nonlinear Equations. PLoS ONE 2015, 10, e0120993. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sheridan, R.P.; Liaw, A.; Dahl, G.E.; Svetnik, V. Deep Neural Nets as a Method for Quantitative Structure-Activity Relationships. J. Chem. Inf. Model. 2015, 55, 263–274. [Google Scholar] [CrossRef]
- Smolensky, P. Information Processing in Dynamical Systems: Foundations of Harmony Theory. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition; Rumelhart, D.E., McClelland, J.L., PDP Research Group, Eds.; MIT Press: Cambridge, MA, USA, 1986; Volume 1, pp. 194–281. [Google Scholar]
- Hinton, G.E. A Practical Guide to Training Restricted Boltzmann Machines. In Neural Networks: Tricks of the Trade, 2nd ed.; Montavon, G., Orr, G.B., Müller, K.-R., Eds.; Springer: Berlin, Germeny, 2012; pp. 599–619. [Google Scholar]
- Hinton, G.E. Training Products of Experts by Minimizing Contrastive Divergence. Neural Comput. 2002, 14, 1771–1800. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.E.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Hinton, G.E.; Sejnowski, T.J. Learning and Relearning in Boltzmann Machines. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition; Rumelhart, D.E., McClelland, J.L., PDP Research Group, Eds.; MIT Press: Cambridge, MA, USA, 1986; Volume 1, pp. 282–317. [Google Scholar]
- Wager, S.; Wang, S.; Liang, P. Dropout Training as Adaptive Regularization. In Proceedings of the 26th International Conference on Neural Information Processing Systems; Curran Associates Inc.: Lake Tahoe, NV, USA, 2013; Volume 1, pp. 351–359. [Google Scholar]
- Andrew, N.G.; Ngiam, J.; Chuan, Y.F.; Mai, Y.; Sue, C. UFLDL Tutorial, Stanford University. Available online: http://deeplearning.stanford.edu/wiki/index.php/UFLDL_Tutorial (accessed on 20 April 2019).
- Wang, Z.; Batu, D.L.; Saixi, Y.L.; Zhang, B.; Ren, L.Q. 2d-Dige Proteomic Analysis of Changes in Estrogen/Progesterone-Induced Rat Breast Hyperplasia Upon Treatment with the Mongolian Remedy Ruxian-I. Molecules 2011, 16, 3048–3065. [Google Scholar] [CrossRef] [PubMed]
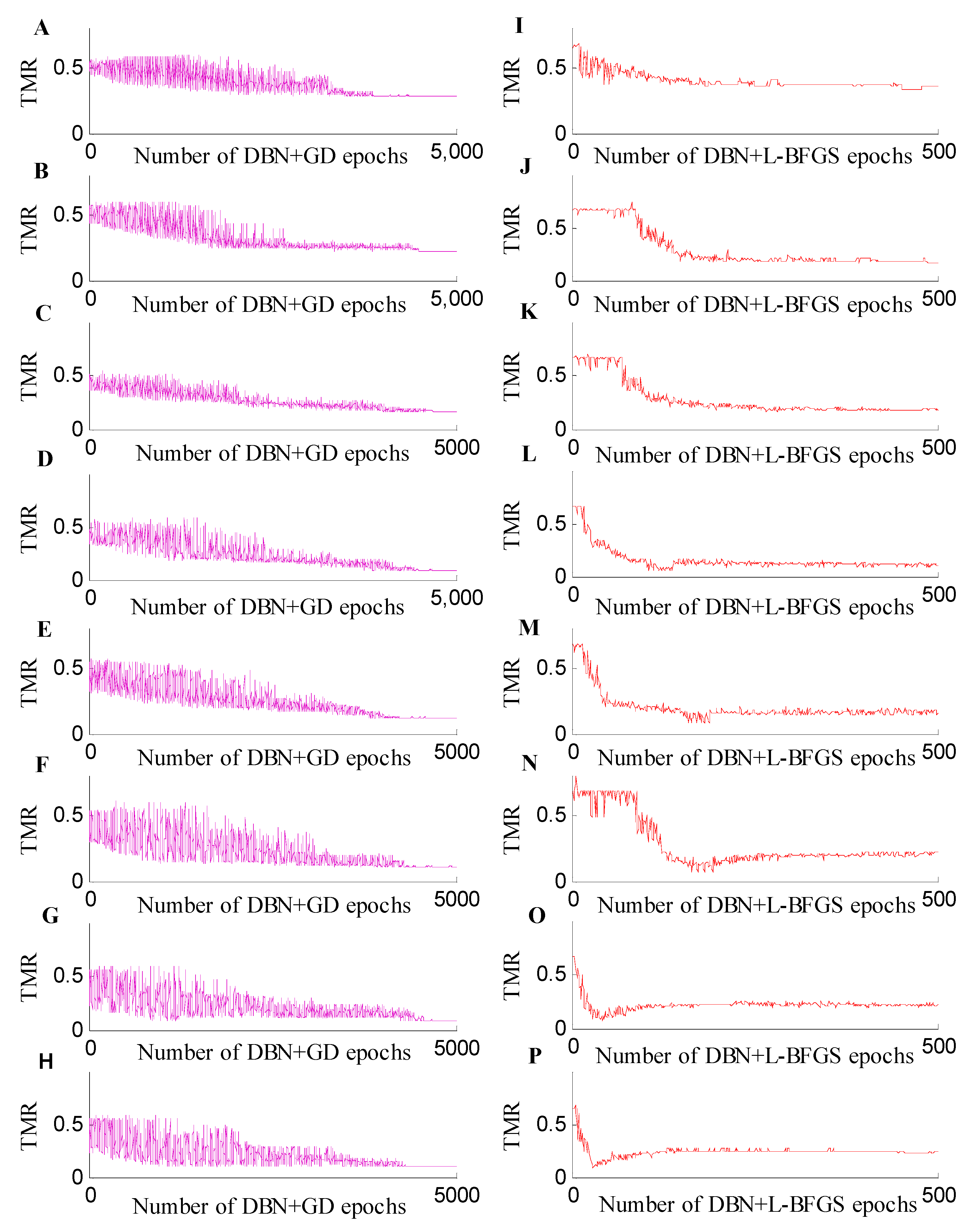

| Group | BPNN | KNN | SVM | DBN+GD+Softmax | DBN+L-BFGS+Softmax |
|---|---|---|---|---|---|
| 1 | 79.41 | 58.82 | 64.71 | 88.24 | 94.12 |
| 2 | 73.53 | 85.29 | 61.76 | 94.12 | 97.06 |
| 3 | 76.47 | 82.35 | 85.29 | 91.18 | 97.06 |
| 4 | 76.47 | 79.41 | 67.65 | 88.24 | 91.18 |
| 5 | 79.41 | 82.35 | 61.76 | 91.18 | 97.06 |
| Mean | 77.06 | 77.64 | 68.23 | 90.59 | 95.30 |
| Group | BPNN | KNN | SVM | DBN+GD+Softmax | DBN+L-BFGS+Softmax |
|---|---|---|---|---|---|
| 1 | 94.12 | 97.06 | 100.00 | 100.00 | 100.00 |
| 2 | 88.24 | 79.41 | 94.12 | 100.00 | 94.12 |
| 3 | 91.18 | 91.18 | 82.35 | 94.12 | 94.12 |
| 4 | 91.18 | 94.12 | 61.76 | 97.06 | 82.35 |
| 5 | 73.53 | 79.41 | 52.94 | 73.53 | 76.47 |
| Mean | 87.65 | 88.24 | 78.23 | 92.94 | 89.41 |
| Training Set | Test Set | BPNN | KNN | SVM | DBN+GD+Softmax | DBN+L-BFGS+Softmax |
|---|---|---|---|---|---|---|
| 50 | 118 | 59.32 | 33.05 | 39.83 | 72.03 | 66.95 |
| 60 | 108 | 68.52 | 70.37 | 40.74 | 77.78 | 83.33 |
| 70 | 98 | 74.49 | 83.67 | 40.82 | 83.67 | 83.67 |
| 80 | 88 | 84.09 | 84.09 | 46.59 | 92.05 | 94.31 |
| 90 | 78 | 78.21 | 82.05 | 48.72 | 88.46 | 92.31 |
| 100 | 68 | 77.94 | 79.41 | 44.12 | 89.71 | 92.65 |
| 110 | 58 | 77.59 | 75.86 | 53.45 | 91.38 | 93.10 |
| 120 | 48 | 81.25 | 70.83 | 54.17 | 89.58 | 91.67 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, M.; Liang, Y.; Pei, Z.; Wang, X.; Zhou, F.; Wei, C.; Feng, X. Diagnosis of Breast Hyperplasia and Evaluation of RuXian-I Based on Metabolomics Deep Belief Networks. Int. J. Mol. Sci. 2019, 20, 2620. https://doi.org/10.3390/ijms20112620
Jiang M, Liang Y, Pei Z, Wang X, Zhou F, Wei C, Feng X. Diagnosis of Breast Hyperplasia and Evaluation of RuXian-I Based on Metabolomics Deep Belief Networks. International Journal of Molecular Sciences. 2019; 20(11):2620. https://doi.org/10.3390/ijms20112620
Chicago/Turabian StyleJiang, Mingyang, Yanchun Liang, Zhili Pei, Xiye Wang, Fengfeng Zhou, Chengxi Wei, and Xiaoyue Feng. 2019. "Diagnosis of Breast Hyperplasia and Evaluation of RuXian-I Based on Metabolomics Deep Belief Networks" International Journal of Molecular Sciences 20, no. 11: 2620. https://doi.org/10.3390/ijms20112620
APA StyleJiang, M., Liang, Y., Pei, Z., Wang, X., Zhou, F., Wei, C., & Feng, X. (2019). Diagnosis of Breast Hyperplasia and Evaluation of RuXian-I Based on Metabolomics Deep Belief Networks. International Journal of Molecular Sciences, 20(11), 2620. https://doi.org/10.3390/ijms20112620





