TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism
Abstract
1. Introduction
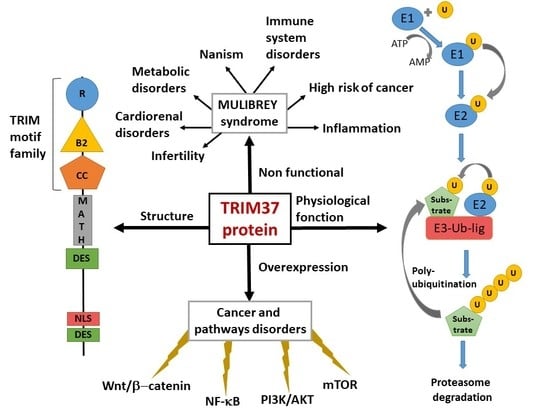
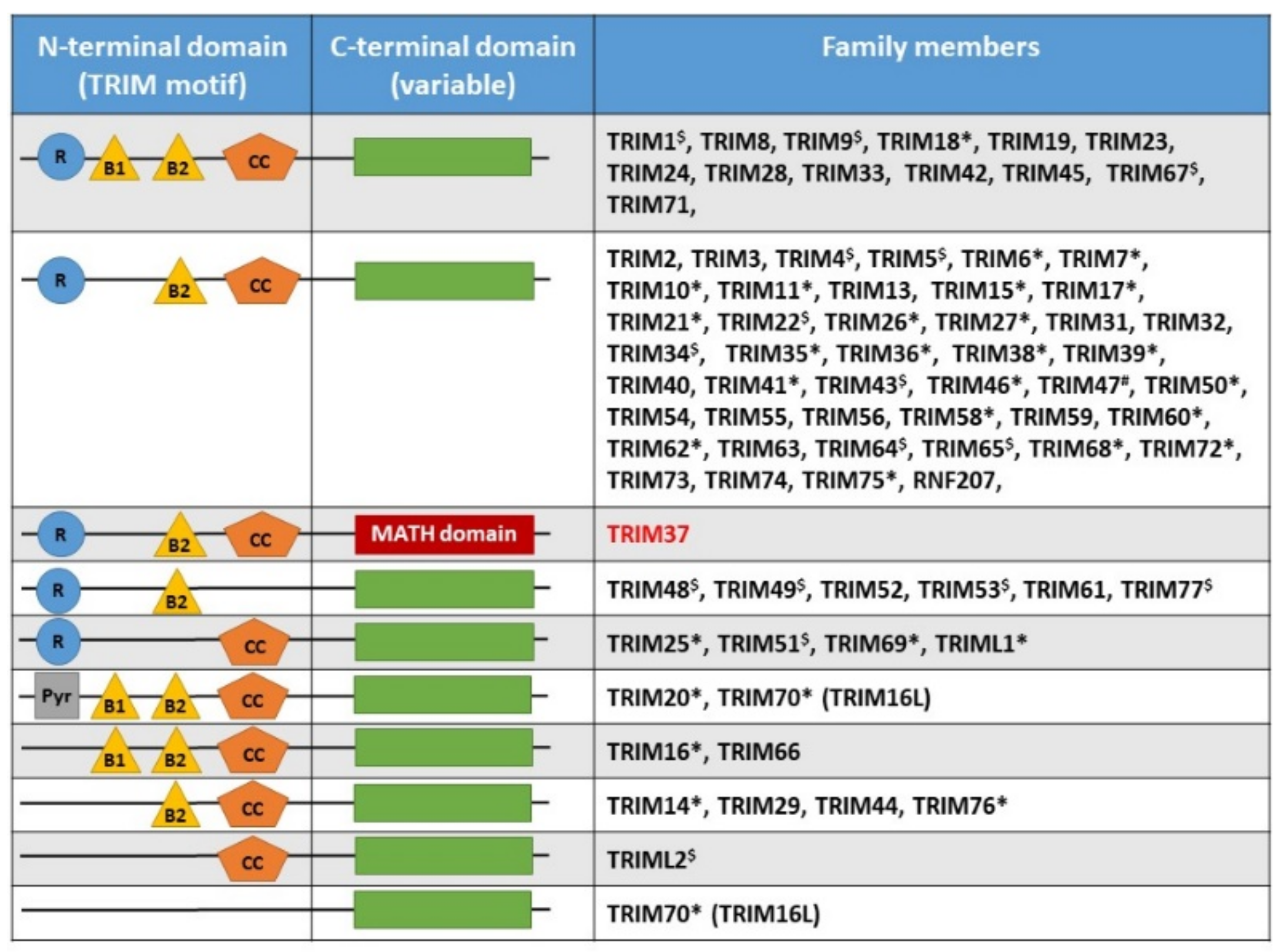
2. TRIM37, Organization of the Gene, Gene Expression, Protein Structure
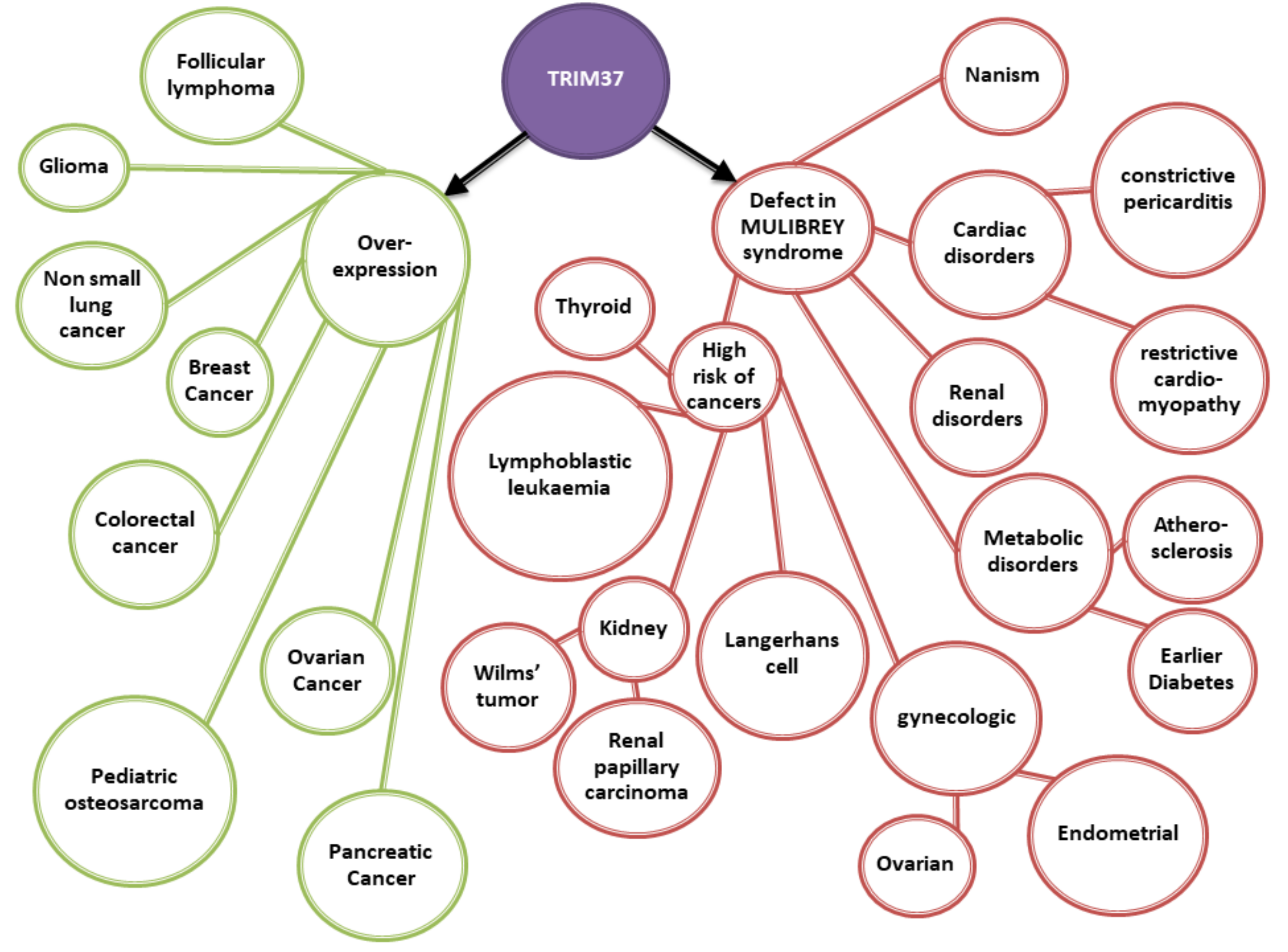
3. MULIBREY Nanism is the Consequence of TRIM37 Mutations
4. MULIBREY Nanism and Cardiorenal Manifestations
5. TRIM37 Variants and the MULIBREY Syndrome, an Inflammatory Disease? The Role of TRIM37 in Innate Immunity and Antiviral Defense
6. Infertility Problems in Both Sexes Are Found in MULIBREY Syndrome
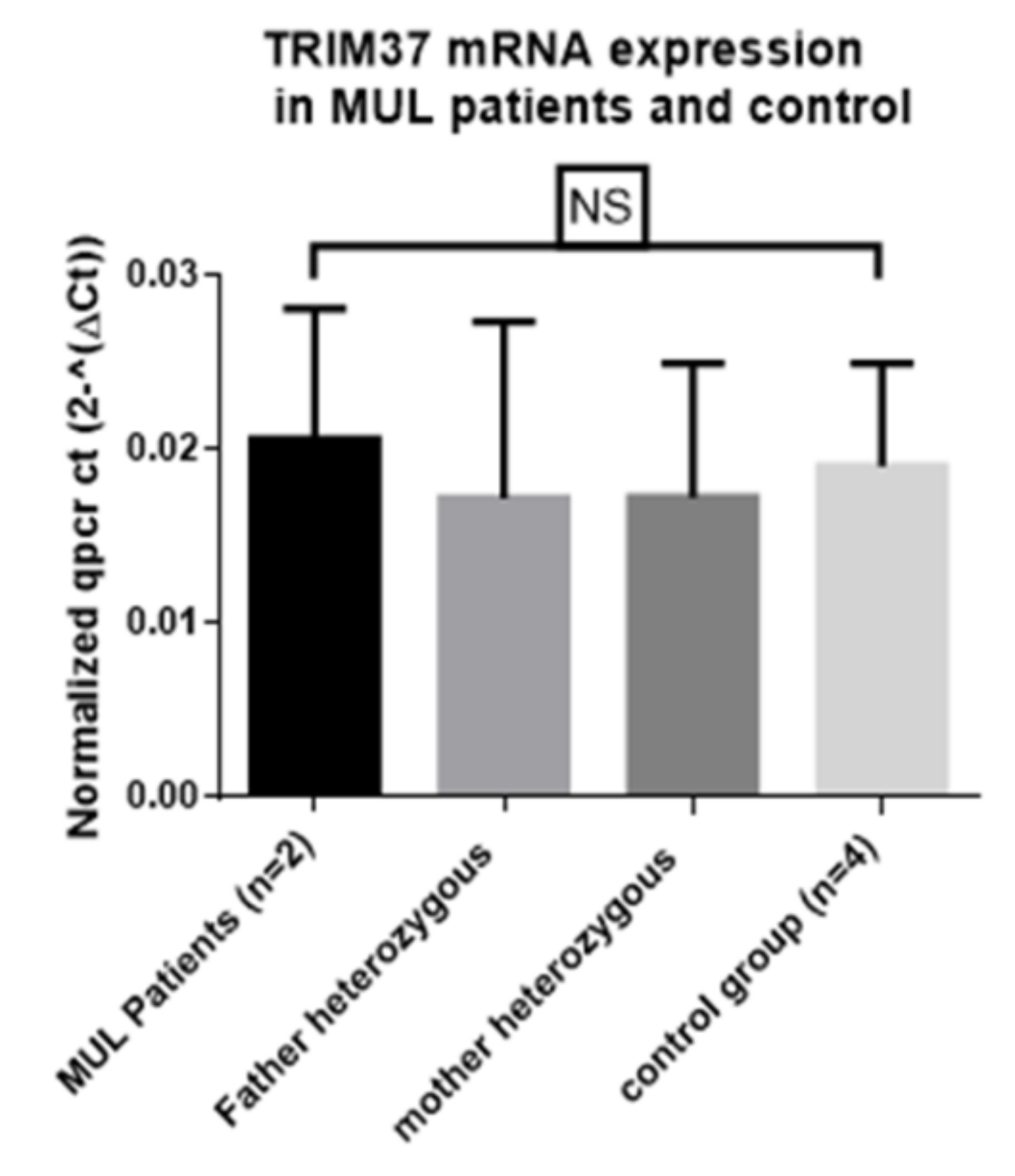
7. Nonsense-Mediated Decay is not Implicated in TRIM37 mRNA Expression in MULIBREY Syndrome
8. TRIM37 Expression is Strongly Associated with Cell Proliferation and Cancer
8.1. Cancerous Process is Associated with an Overexpression of TRIM37 in Various Tissues
8.2. Accumulating In Vitro Evidences Point Toward Several Roles for TRIM37 in the Cell Cycle, Including in Peroxysomes and Centrioles
9. Concluding Remarks and Prospective
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Grutter, C.; Briand, C.; Capitani, G.; Mittl, P.R.; Papin, S.; Tschopp, J.; Grutter, M.G. Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases. FEBS Lett. 2006, 580, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Singh, R. TRIM family proteins: Emerging class of RING E3 ligases as regulator of NF-κB pathway. Biol. Cell 2015, 107, 22–40. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Bonekamp, N.A.; Islinger, M. Fission and proliferation of peroxisomes. Biochim. Biophys. Acta 2012, 1822, 1343–1357. [Google Scholar] [CrossRef] [PubMed]
- Hamalainen, R.H.; Joensuu, T.; Kallijarvi, J.; Lehesjoki, A.E. Characterisation of the Mulibrey nanism-associated TRIM37 gene: Transcription initiation, promoter region and alternative splicing. Gene 2006, 366, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Perheentupa, J.; Autio, S.; Leisti, S.; Raitta, C. Mulibrey-nanism: Dwarfism with muscle, liver, brain and eye involvement. Acta Paediatr. Scand. Suppl. 1970, 206, 274. [Google Scholar] [CrossRef]
- Fokkema, I.F.; Taschner, P.E.; Schaafsma, G.C.; Celli, J.; Laros, J.F.; den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]
- Zapata, J.M.; Martinez-Garcia, V.; Lefebvre, S. Phylogeny of the TRAF/MATH domain. Adv. Exp. Med. Biol. 2007, 597, 1–24. [Google Scholar]
- Gupta, I.; Varshney, N.K.; Khan, S. Emergence of Members of TRAF and DUB of Ubiquitin Proteasome System in the Regulation of Hypertrophic Cardiomyopathy. Front. Genet. 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Trevino, S.R.; Scholtz, J.M.; Pace, C.N. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J. Mol. Biol. 2007, 366, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Avela, K.; Lipsanen-Nyman, M.; Idanheimo, N.; Seemanova, E.; Rosengren, S.; Makela, T.P.; Perheentupa, J.; Chapelle, A.D.; Lehesjoki, A.E. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in Mulibrey nanism. Nat. Genet. 2000, 25, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Jobic, F.; Morin, G.; Vincent-Delorme, C.; Cadet, E.; Cabry, R.; Mathieu-Dramard, M.; Copin, H.; Rochette, J.; Jedraszak, G. New intragenic rearrangements in non-Finnish Mulibrey nanism. Am. J. Med. Genet. A 2017, 173, 2782–2788. [Google Scholar] [CrossRef] [PubMed]
- Mozzillo, E.; Cozzolino, C.; Genesio, R.; Melis, D.; Frisso, G.; Orrico, A.; Lombardo, B.; Fattorusso, V.; Discepolo, V.; della Casa, R.; et al. Mulibrey nanism: Two novel mutations in a child identified by Array CGH and DNA sequencing. Am. J. Med. Genet. A 2016, 170, 2196–2199. [Google Scholar] [CrossRef] [PubMed]
- Kallijarvi, J.; Lahtinen, U.; Hamalainen, R.; Lipsanen-Nyman, M.; Palvimo, J.J.; Lehesjoki, A.E. TRIM37 defective in Mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp. Cell Res. 2005, 308, 146–155. [Google Scholar] [CrossRef]
- Hamalainen, R.H.; Avela, K.; Lambert, J.A.; Kallijarvi, J.; Eyaid, W.; Gronau, J.; Ignaszewski, A.P.; McFadden, D.; Sorge, G.; Lipsanen-Nyman, M.; et al. Novel mutations in the TRIM37 gene in Mulibrey nanism. Hum. Mutat. 2004, 23, 522. [Google Scholar] [CrossRef]
- Jagiello, P.; Hammans, C.; Wieczorek, S.; Arning, L.; Stefanski, A.; Strehl, H.; Epplen, J.T.; Gencik, M. A novel splice site mutation in the TRIM37 gene causes Mulibrey nanism in a Turkish family with phenotypic heterogeneity. Hum. Mutat. 2003, 21, 630–635. [Google Scholar] [CrossRef]
- Yasuhara, J.; Omori, S.; Maeda, J.; Nakagawa, N.; Kamada, M.; Kosaki, K.; Aeba, R.; Yamagishi, H. Successful Total Pericardiectomy for Constrictive Pericarditis in the First Series of Japanese Patients with Mulibrey nanism. Can. J. Cardiol. 2018, 34, 690.e5–690.e8. [Google Scholar] [CrossRef]
- Kumpf, M.; Hamalainen, R.H.; Hofbeck, M.; Baden, W. Refractory congestive heart failure following delayed pericardectomy in a 12-year-old child with Mulibrey nanism due to a novel mutation in TRIM37. Eur. J. Pediatr. 2013, 172, 1415–1418. [Google Scholar] [CrossRef]
- Doganc, T.; Konuk, B.E.Y.; Alpan, N.; Konuk, O.; Hamalainen, R.H.; Lehesjoki, A.E.; Tekin, M. A novel mutation in TRIM37 is associated with Mulibrey nanism in a Turkish boy. Clin. Dysmorphol. 2007, 16, 173–176. [Google Scholar] [PubMed]
- Lipsanen-Nyman, M.; Perheentupa, J.; Rapola, J.; Sovijarvi, A.; Kupari, M. Mulibrey heart disease: Clinical manifestations, long-term course, and results of pericardiectomy in a series of 49 patients born before 1985. Circulation 2003, 107, 2810–2815. [Google Scholar] [CrossRef]
- Kivisto, S.; Lipsanen-Nyman, M.; Kupari, M.; Hekali, P.; Lauerma, K. Cardiac involvement in Mulibrey nanism: Characterization with magnetic resonance imaging. J. Cardiovasc. Magn. Reson. 2004, 6, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Baczewski, K.; Czajkowski, M.; Olszewski, K.; Stadnik, A.; Stazka, J. A stone was lifted from her heart: Pericardial constriction in 28-year-old patient with Mulibrey nanism. Kardiol. Pol. 2016, 74, 192. [Google Scholar] [CrossRef] [PubMed]
- Eerola, A.; Jokinen, E.; Pihkala, J.I. Serum levels of natriuretic peptides in children with various types of loading conditions. Scand. Cardiovasc. J. 2009, 43, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Karlberg, N.; Jalanko, H.; Kallijarvi, J.; Lehesjoki, A.E.; Lipsanen-Nyman, M. Insulin resistance syndrome in subjects with mutated RING finger protein TRIM37. Diabetes 2005, 54, 3577–3581. [Google Scholar] [CrossRef] [PubMed]
- Balaji, V.; Pokrzywa, W.; Hoppe, T. Ubiquitylation Pathways In Insulin Signaling and Organismal Homeostasis. Bioessays 2018, 40, e1700223. [Google Scholar] [CrossRef]
- Sivunen, J.; Karlberg, S.; Lohi, J.; Karlberg, N.; Lipsanen-Nyman, M.; Jalanko, H. Renal findings in patients with Mulibrey nanism. Pediatr. Nephrol. 2017, 32, 1531–1536. [Google Scholar] [CrossRef]
- Trang, A.; Aguilar, D. Treating Disease Mechanisms in Patients With Heart Failure and Diabetes Mellitus. Curr. Heart Fail. Rep. 2017, 14, 445–453. [Google Scholar] [CrossRef]
- Kettunen, K.M.; Karikoski, R.; Hamalainen, R.H.; Toivonen, T.T.; Antonenkov, V.D.; Kulesskaya, N.; Voikar, V.; Holtta-Vuori, M.; Ikonen, E.; Sainio, K.; et al. Trim37-deficient mice recapitulate several features of the multi-organ disorder Mulibrey nanism. Biol. Open 2016, 5, 584–595. [Google Scholar] [CrossRef]
- Corn, J.E.; Vucic, D. Ubiquitin in inflammation: The right linkage makes all the difference. Nat. Struct. Mol. Biol. 2014, 21, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, L.; Zhao, X.; Li, B.; Ren, D.; Yu, L.; Pan, H.; Gong, Q.; Song, L.; Zhou, X.; et al. Tripartite motif-containing 37 (TRIM37) promotes the aggressiveness of non-small-cell lung cancer cells by activating the NF-κB pathway. J. Pathol. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Goru, S.K.; Pandey, A.; Gaikwad, A.B. E3 ubiquitin ligases as novel targets for inflammatory diseases. Pharmacol. Res. 2016, 106, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Jin, H.S.; Aki, D.; Lee, J.; Liu, Y.C. The ubiquitin system in immune regulation. Adv. Immunol. 2014, 124, 17–66. [Google Scholar] [PubMed]
- Uchida, D.; Hatakeyama, S.; Matsushima, A.; Han, H.; Ishido, S.; Hotta, H.; Kudoh, J.; Shimizu, N.; Doucas, V.; Nakayama, K.I.; et al. AIRE functions as an E3 ubiquitin ligase. J. Exp. Med. 2004, 199, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Rajsbaum, R.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Valdiviezo, J.; Shi, M.; Inn, K.S.; Fernandez-Sesma, A.; Jung, J.; Garcia-Sastre, A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 2013, 38, 384–398. [Google Scholar] [CrossRef]
- Van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Tabah, A.A.; Tardif, K.; Mansky, L.M. Anti-HIV-1 activity of Trim 37. J. Gen. Virol. 2014, 95, 960–967. [Google Scholar] [CrossRef]
- Dai, Y.; Li, Y.; Cheng, R.; Gao, J.; Lou, C. TRIM37 inhibits PDGF-BB-induced proliferation and migration of airway smooth muscle cells. Biomed. Pharmacother. 2018, 101, 24–29. [Google Scholar] [CrossRef]
- Haraldsson, A.; van der Burgt, C.J.; Weemaes, C.M.; Otten, B.; Bakkeren, J.A.; Stoelinga, G.B. Antibody deficiency and isolated growth hormone deficiency in a girl with Mulibrey nanism. Eur. J. Pediatr. 1993, 152, 509–512. [Google Scholar] [CrossRef]
- Karlberg, S.; Tiitinen, A.; Alfthan, H.; Lipsanen-Nyman, M. Premature ovarian insufficiency and early depletion of the ovarian reserve in the monogenic Mulibrey nanism disorder. Hum. Reprod. 2018, 33, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Karlberg, S.; Toppari, J.; Karlberg, N.; Nurmio, M.; Karikoski, R.; Jalanko, H.; Lipsanen-Nyman, M. Testicular failure and male infertility in the monogenic Mulibrey nanism disorder. J. Clin. Endocrinol. Metab. 2011, 96, 3399–3407. [Google Scholar] [CrossRef]
- Azenabor, A.; Ekun, A.O.; Akinloye, O. Impact of Inflammation on Male Reproductive Tract. J. Reprod. Infertil. 2015, 16, 123–129. [Google Scholar] [PubMed]
- Sheldon, I.M.; Owens, S.E.; Turner, M.L. Innate immunity and the sensing of infection, damage and danger in the female genital tract. J. Reprod. Immunol. 2017, 119, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, P.J.; Menezes, J.; Romao, L. The role of alternative splicing coupled to nonsense-mediated mRNA decay in human disease. Int. J. Biochem. Cell Biol. 2017, 91, 168–175. [Google Scholar] [CrossRef]
- M’Baya-Moutoula, E.; Louvet, L.; Molinie, R.; Guerrera, I.C.; Cerutti, C.; Fourdinier, O.; Nourry, V.; Gutierrez, L.; Morliere, P.; Mesnard, F.; et al. A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim. Biophys. Acta 2018, 1864, 2664–2678. [Google Scholar] [CrossRef] [PubMed]
- Karlberg, N.; Karlberg, S.; Karikoski, R.; Mikkola, S.; Lipsanen-Nyman, M.; Jalanko, H. High frequency of tumours in Mulibrey nanism. J. Pathol. 2009, 218, 163–171. [Google Scholar] [CrossRef]
- Karlberg, S.; Lipsanen-Nyman, M.; Lassus, H.; Kallijarvi, J.; Lehesjoki, A.E.; Butzow, R. Gynecological tumors in Mulibrey nanism and role for RING finger protein TRIM37 in the pathogenesis of ovarian fibrothecomas. Mod. Pathol. 2009, 22, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Hamalainen, R.H.; Mowat, D.; Gabbett, M.T.; O’brien, T.A.; Kallijarvi, J.; Lehesjoki, A.E. Wilms’ tumor and novel TRIM37 mutations in an Australian patient with Mulibrey nanism. Clin. Genet. 2006, 70, 473–479. [Google Scholar] [CrossRef]
- Wang, W.; Xia, Z.; Farre, J.C.; Subramani, S. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy 2018, 14, 1574–1585. [Google Scholar] [CrossRef]
- Jiang, J.; Tian, S.; Yu, C.; Chen, M.; Sun, C. TRIM37 promoted the growth and migration of the pancreatic cancer cells. Tumour Biol. 2016, 37, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yu, C.; Chen, M.; Tian, S.; Sun, C. Over-expression of TRIM37 promotes cell migration and metastasis in hepatocellular carcinoma by activating Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2015, 464, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.E.; Gan, J. TRIM37 promotes epithelialmesenchymal transition in colorectal cancer. Mol. Med. Rep. 2017, 15, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Xin, M.; Cheng, H.; Huang, Z.; Hu, T.; Zhang, T.; Wang, J. TRIM37 promotes tumor cell proliferation and drug resistance in pediatric osteosarcoma. Oncol. Lett. 2017, 14, 6365–6372. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.L.; Gao, Y.L.; Wen-Zhong, H. Knockdown of TRIM37 suppresses the proliferation, migration and invasion of glioma cells through the inactivation of PI3K/Akt signaling pathway. Biomed. Pharmacother. 2018, 99, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Guan, H.T.; Dai, Z.J.; Ma, Y.G.; Liu, X.X.; Wang, X.J. Knockdown of Tripartite Motif-Containing Protein 37 (TRIM37) Inhibits the Proliferation and Tumorigenesis in Colorectal Cancer Cells. Oncol. Res. 2017, 25, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Green, M.R. TRIMming down tumor suppressors in breast cancer. Cell Cycle 2015, 14, 1345–1346. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Gazin, C.; Chamberlain, L.; Ou, J.; Zhu, X.; Tushir, J.S.; Virbasius, C.M.; Lin, L.; Zhu, L.J.; Wajapeyee, N.; et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 2014, 516, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Tuna, M.; Smid, M.; Martens, J.W.; Foekens, J.A. Prognostic value of acquired uniparental disomy (aUPD) in primary breast cancer. Breast Cancer Res. Treat. 2012, 132, 189–196. [Google Scholar] [CrossRef]
- Brodtkorb, M.; Lingjaerde, O.C.; Huse, K.; Troen, G.; Hystad, M.; Hilden, V.I.; Myklebust, J.H.; Leich, E.; Rosenwald, A.; Delabie, J.; et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood 2014, 123, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Dimova, I.; Orsetti, B.; Negre, V.; Rouge, C.; Ursule, L.; Lasorsa, L.; Dimitrov, R.; Doganov, N.; Toncheva, D.; Theillet, C. Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array CGH analysis. Tumori 2009, 95, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Song, L.; Zhu, J.; Hu, Y.; Cao, L.; Tan, Z.; Zhang, S.; Li, Z.; Li, J. An ATM/TRIM37/NEMO axis counteracts genotoxicity by activating nuclear-to-cytoplasmic NF-κB signaling. Cancer Res. 2018, 78, 6399–6412. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Balestra, F.R.; Strnad, P.; Fluckiger, I.; Gonczy, P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell 2013, 25, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xia, Z.J.; Farre, J.C.; Subramani, S. TRIM37, a novel E3 ligase for PEX5-mediated peroxisomal matrix protein import. J. Cell Biol. 2017, 216, 2843–2858. [Google Scholar] [CrossRef] [PubMed]
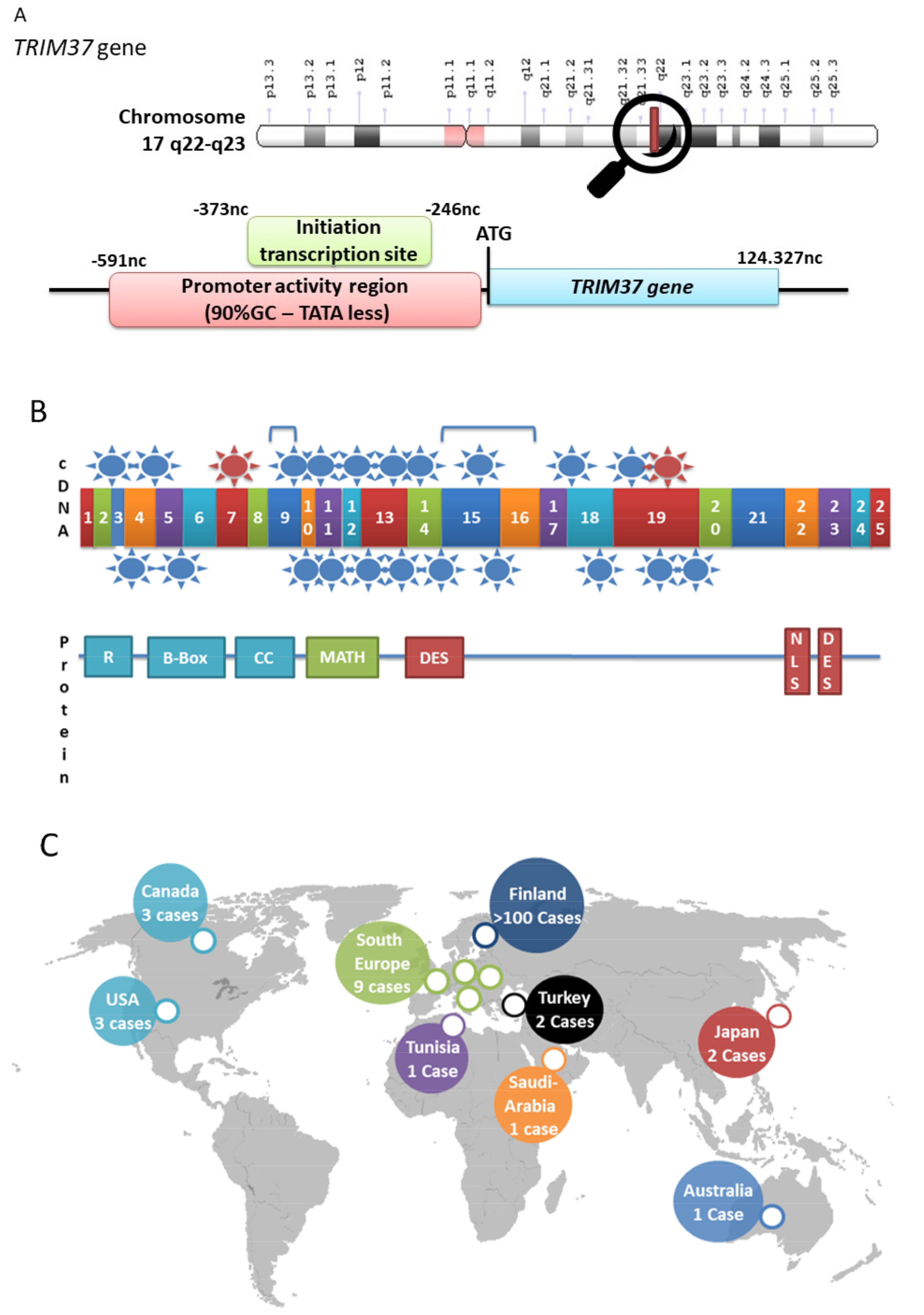
| n° | Mutations | Type | Exon | Country | Publish Cases | References |
|---|---|---|---|---|---|---|
| 1 | c.81delG | Deletion | 2 | French | 1 | [9] |
| 2 | c.181C > T | Nonsense | 4 | French | 1 | [14] |
| 3 | c.227T > C | Missence | 4 | Finnish | 1 | [16] |
| 4 | C.326G > C | Missence | 5 | Australian | 1 | [17] |
| 5 | c.493-2A > G | Splice-site | 7 | Finnish; Fin-major | >100 | [13] |
| 6 | c.685_809del | Large deletion | 9 | French | 1 | [14] |
| 7 | c.745C > T | Nonsense | 9 | Canadian | 1 | [17] |
| 8 | c.810-1G > A | Splice-site | 10 | Turkish | 3 | [18] |
| 9 | c.838-842delACTTT | Deletion | 10 | Czech | 1 | [13] |
| 10 | c.860G > A | Splice-site | 10 | Australian | 1 | [17] |
| 11 | c.874_877delAGAG | Deletion | 11 | Japanese | 1 | [19] |
| 12 | c.965G > T | Missence | 12 | Canadian | 1 | [17] |
| 13 | c.1016C > G | Missence | 13 | Japanese | 1 | [19] |
| 14 | c.1037_1040dupAGAT | Duplication | 13 | Canadian | 1 | [17] |
| 15 | c.1166A > G | Nonsense | 13 | Finnish | 1 | |
| 16 | c.1233delA | Deletion | 14 | German | 1 | [20] |
| 17 | c.1313+507_1668-207del | Large deletion | 15,16 | Italian | 1 | [17] |
| 18 | c.1315_1667del | Large deletion | 15-16 | French | 1 | [14] |
| 19 | c.1346dupA | Duplication | 15 | American | 1 | [13] |
| 20 | c.1411C > T | Nonsense | 15 | Canadian, Tunisian | 1 | [17] |
| 21 | c.1894_1895delGA | Deletion | 18 | Turkish | 1 | [21] |
| 22 | c.1910_1911dupTA | Duplication | 18 | Swiss | 1 | [9] |
| 23 | c.1949-12A > G | Splice-site | 19 | Italian | 2 | [15] |
| 24 | c.2056C > T | Nonsense | 19 | Saudi-Arabian | 1 | [17] |
| 25 | c.2212delG | Deletion | 19 | Finnish; Fin-minor | 1 | [13] |
| 26 | c.1949-12A > G | Splice-site | 19 | Italian | 2 | [15] |
| Cancers | Publications | Signaling Pathways | Known Substrates | Association with: | TRIM37 Expression in Tumor Cells |
|---|---|---|---|---|---|
| Pediatric osteosarcoma | [54] | Wnt/β-catenin | - | Proliferation and drug resistance | Overexpressed |
| Glioma | [55] | PI3K/Akt | - | Proliferation, migration, and invasion | Overexpressed |
| Non-small-cell lung cancer | [32] | NFκB | Ubiquitinates TRAF2 | Apoptosis, angiogenesis, and proliferation | Overexpressed |
| Pancreatic cancer | [51] | Wnt/β-catenin | Binds β-Catenin | Proliferation and migration | Overexpressed |
| Hepatocellular carcinoma | [52] | Wnt/β-catenin | - | Proliferation and migration | Overexpressed |
| Colorectal Cancer | [53] | - | Proliferation, invasion, and migration | Overexpressed | |
| [56] | Wnt/β-catenin | - | Proliferation | Overexpressed | |
| Breast Cancer | [57,58] | PRC1/PRC2 | H2A ubiquitination | Promotes transformation/silencing of tumor suppressor | Overexpressed in ∼40% of breast cancers |
| [59] | - | - | High frequency of chromosomal rearrangement in TRIM37 gene associated with mortality | - | |
| Follicular lymphoma | [60] | NFκB | - | Promotes transformation | Overexpressed |
| Ovarian Cancer | [61] | - | - | High frequency of chromosomal rearrangement in TRIM37 gene | - |
| Esophagus Cancer | [62] | NFkB | Binds TRAF6 and ubiquitinates NEMO | Relapse-free survival and drug resistance | Overexpressed |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brigant, B.; Metzinger-Le Meuth, V.; Rochette, J.; Metzinger, L. TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism. Int. J. Mol. Sci. 2019, 20, 67. https://doi.org/10.3390/ijms20010067
Brigant B, Metzinger-Le Meuth V, Rochette J, Metzinger L. TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism. International Journal of Molecular Sciences. 2019; 20(1):67. https://doi.org/10.3390/ijms20010067
Chicago/Turabian StyleBrigant, Benjamin, Valérie Metzinger-Le Meuth, Jacques Rochette, and Laurent Metzinger. 2019. "TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism" International Journal of Molecular Sciences 20, no. 1: 67. https://doi.org/10.3390/ijms20010067
APA StyleBrigant, B., Metzinger-Le Meuth, V., Rochette, J., & Metzinger, L. (2019). TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism. International Journal of Molecular Sciences, 20(1), 67. https://doi.org/10.3390/ijms20010067