Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim.
Abstract
1. Introduction
2. Results
2.1. Identification of the WRKY Proteins in Zanthoxylum bungeanum
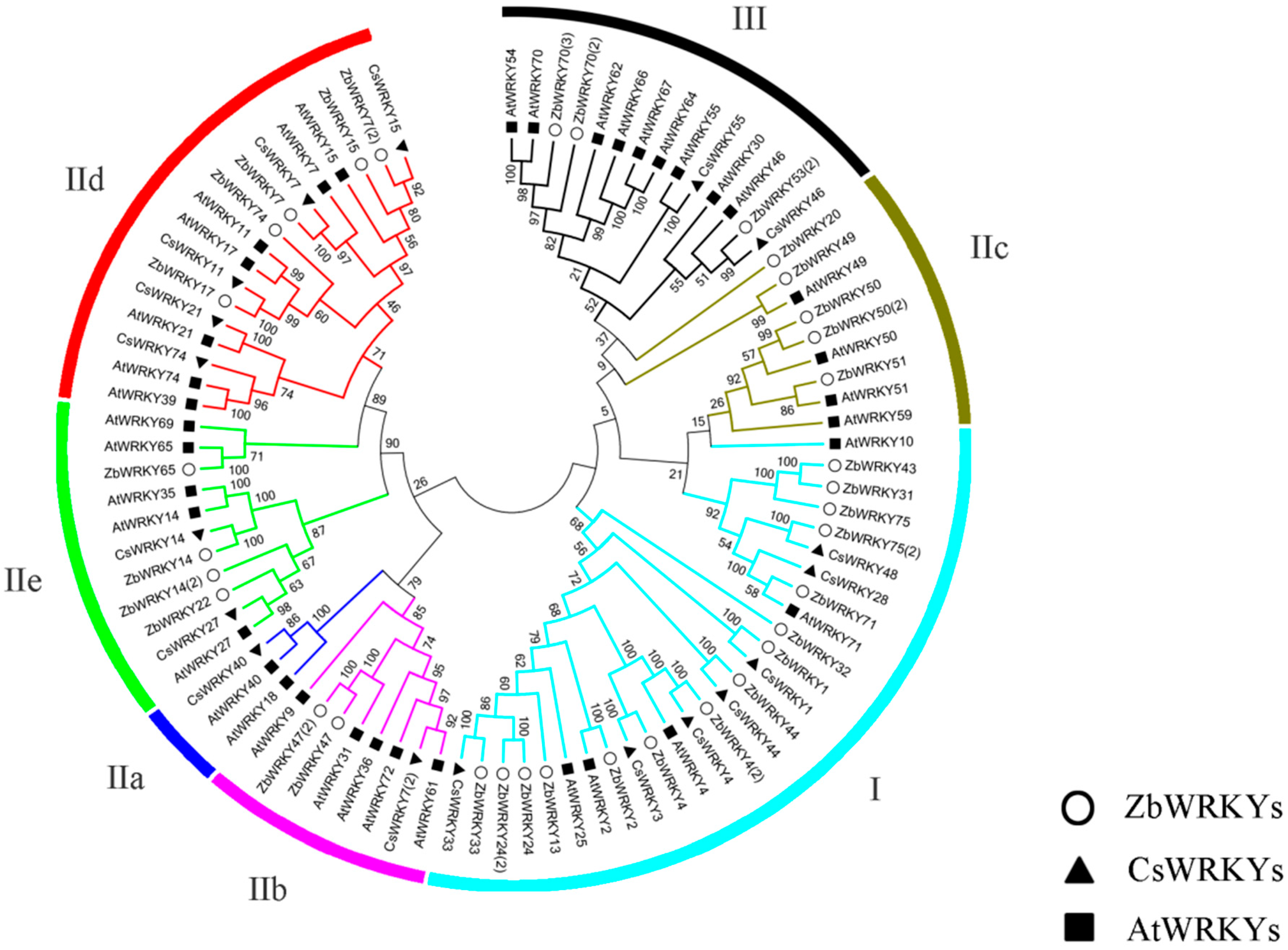
2.2. ZbWRKY Gene Phylogenetic Tree Analysis and Group Identification
2.3. Conserved Domain Analysis of ZbKWRKY Gene
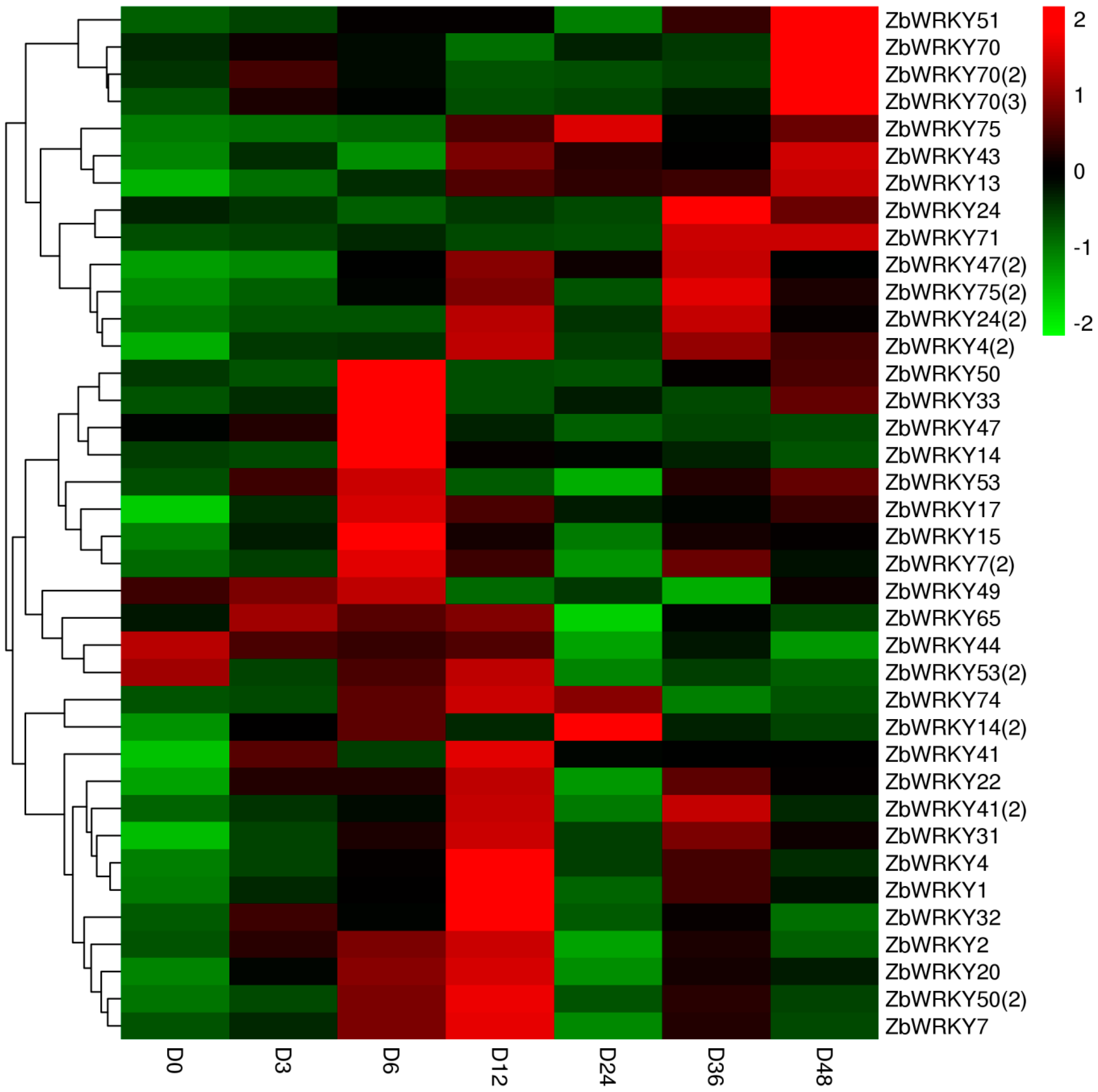
2.4. ZbWRKY Gene Expression Analysis
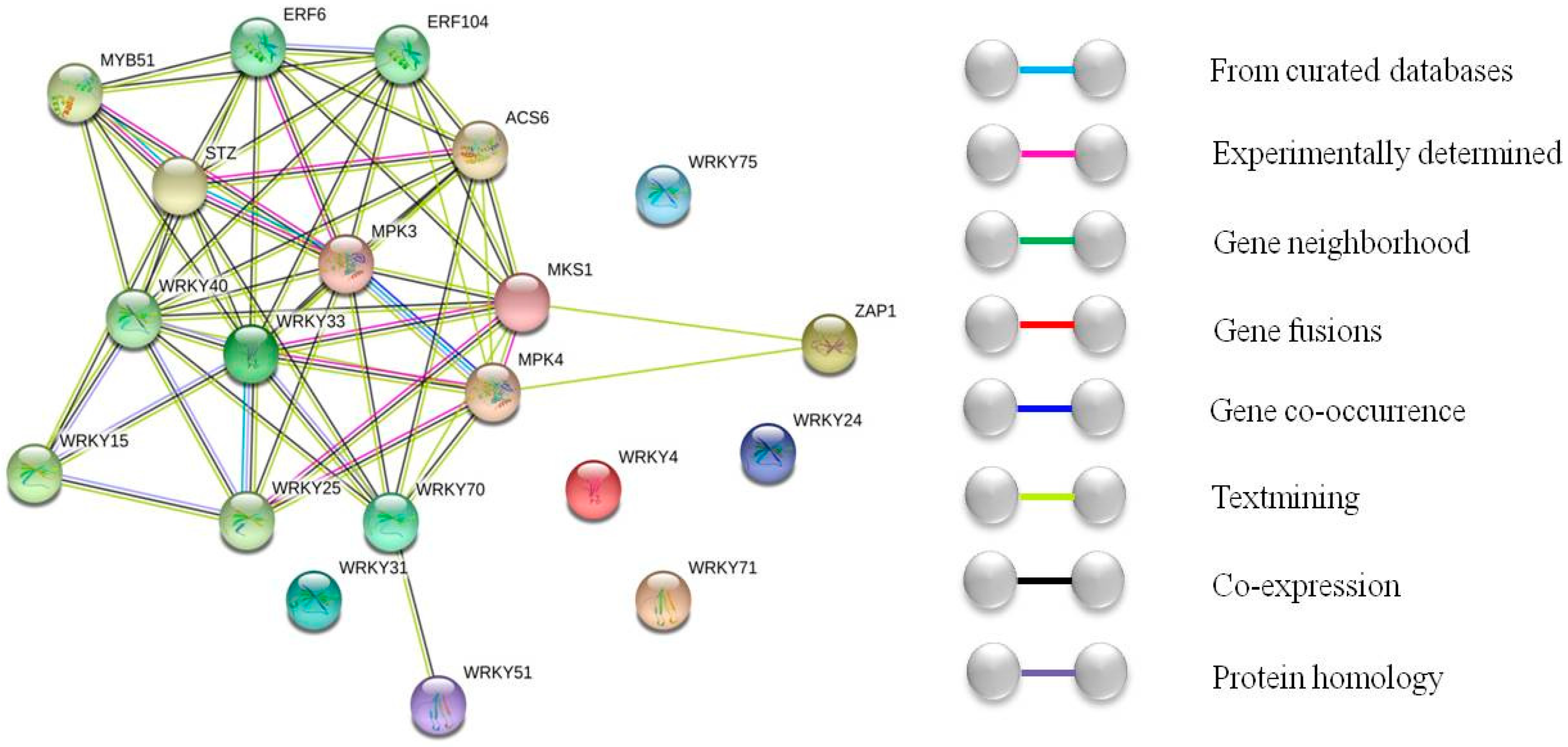
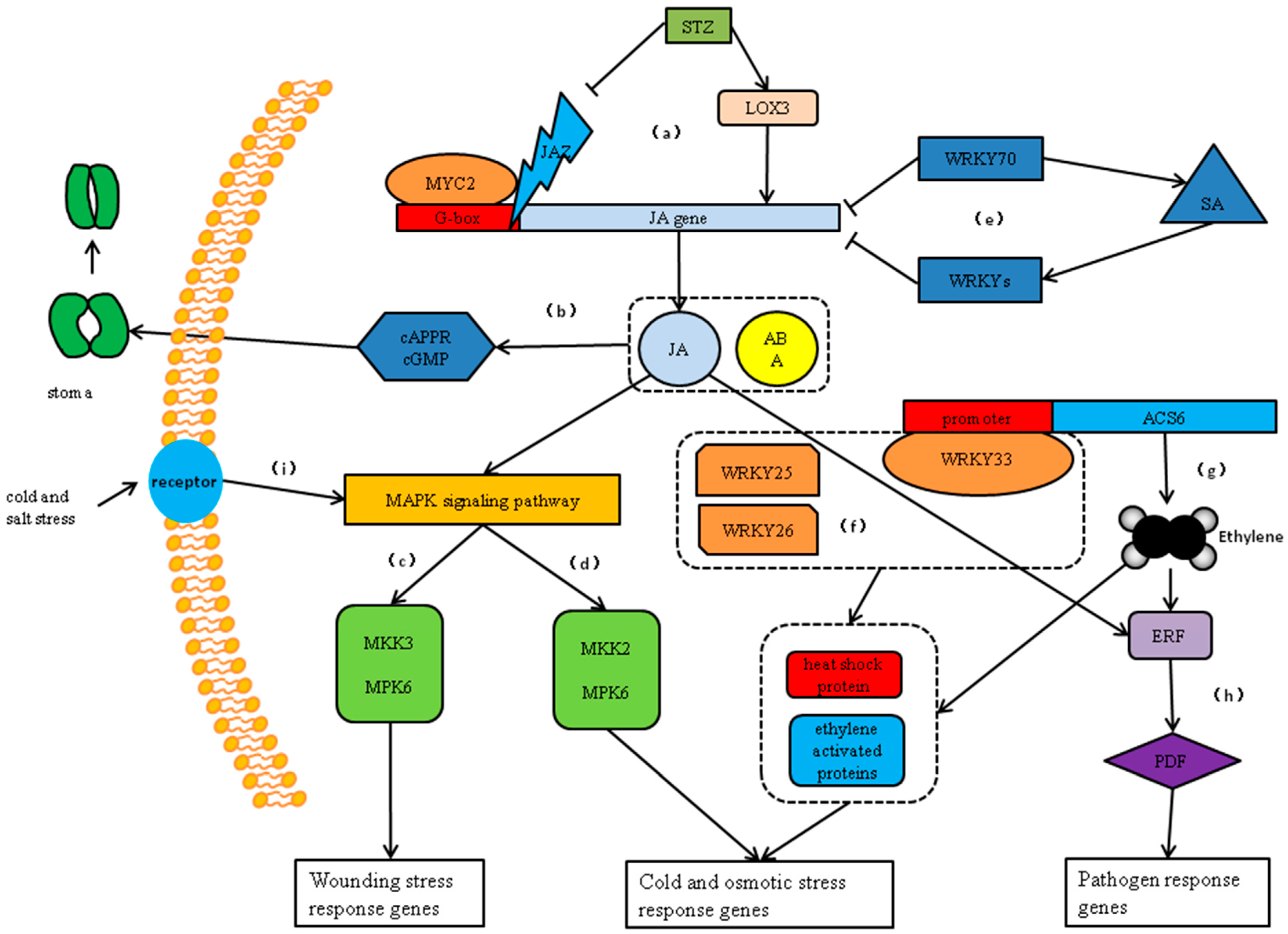
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Total RNA Extraction and Reverse Transcription
4.2.2. Bioinformatics Analysis
4.2.3. Identification and Screening of Transcription Factor Family Genes
4.2.4. Real-Time qPCR
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schwechheimer, C.; Zourelidou, M.; Bevan, M.W. Plant transcription factor studies. Annu. Rev. Plant Biol. 1998, 49, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 224, 563–571. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the ArabidopsisWRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.H.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.; Cocivera, M. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2010, 31, 86–96. [Google Scholar]
- Zhang, C.-Q.; Xu, Y.; Lu, Y.; Yu, H.-X.; Gu, M.-H.; Liu, Q.-Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Şahin-Çevik, M. A WRKY transcription factor gene isolated from Poncirus trifoliata shows differential responses to cold and drought stresses. Plant Omics 2012, 5, 438–445. [Google Scholar]
- Ullah, A.; Sun, H.; Hakim; Yang, X.; Zhang, X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant 2018, 162, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Choi, L.M.; Lee, G.J.; Kim, Y.J.; Paek, K.H. Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon Tobacco mosaic virus infection. Plant Sci. 2012, 197, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Z.; Dong, J.; Wang, T. Overexpression of MtWRKY76 increases both salt and drought tolerance in Medicago truncatula. Environ. Exp. Bot. 2016, 123, 50–58. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Guo, R.; Wang, Y.; Guo, C.; Li, Z.; Chen, Z.; Gao, H.; Wang, X. Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tissue Organ Cult. 2017, 132, 359–370. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Huang, D.M.; Zhao, G.H.; Chen, Z.D.; Kan, J.Q. The food culture of Chinese prickly ash. China Condiment 2006, 75–81. [Google Scholar]
- Shuming, L.; Jiaqian, S.; Zhenyi, D.; Diandian, W.; Gang, Z.; Bingyin, S. Effects of Drought Stress on the Root Morphology and Water Use Efficiency of Zanthoxylum bungeanum. Sci. Silvae Sin. 2013, 49, 30–35. [Google Scholar]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Li, Y.; Wang, F.; Cheng, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011, 23, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.H.; Eschen-Lippold, L.; Pecher, P.; Hoehenwarter, W.; Sinha, A.K.; Scheel, D.; Lee, J. Regulation of WRKY46 Transcription Factor Function by Mitogen-Activated Protein Kinases in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 61. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.; Zhu, S.; Qiu, J.-L.; Micheelsen, P.; Rocher, A.; Petersen, M.; et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005, 24, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.-L.; Fiil, B.K.; Petersen, K.; Nielsen, H.B.; Botanga, C.J.; Thorgrimsen, S.; Palma, K.; Suarez-Rodriguez, M.C.; Sandbech-Clausen, S.; Lichota, J.; et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008, 27, 2214–2221. [Google Scholar] [CrossRef]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants a new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Balbi, V.; Devoto, A. Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008, 177, 301–318. [Google Scholar] [CrossRef]
- Farmer, E.E.; Alméras, E.; Krishnamurthy, V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 2003, 6, 372–378. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Dabi, M.; Sapara, K.K.; Joshi, P.S.; Agarwal, P.K. Ectopic Expression of JcWRKY Transcription Factor Confers Salinity Tolerance via Salicylic Acid Signaling. Front Plant Sci. 2016, 7, 1541. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, X.; Zhang, K.; An, H.; Hu, K.; Wen, J.; Shen, J.; Ma, C.; Yi, B.; Tu, J.; et al. Comparative Analysis of the Brassica napus Root and Leaf Transcript Profiling in Response to Drought Stress. Int. J. Mol. Sci. 2015, 16, 18752–18777. [Google Scholar] [CrossRef]
- Takahashi, F.; Yoshida, R.; Ichimura, K.; Mizoguchi, T.; Seo, S.; Yonezawa, M.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 2007, 19, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Gunapati, S.; Naresh, R.; Ranjan, S.; Nigam, D.; Hans, A.; Verma, P.C.; Gadre, R.; Pathre, U.V.; Sane, A.P.; Sane, V.A. Expression of GhNAC2 from G. herbaceum, improves root growth and imparts tolerance to drought in transgenic cotton and Arabidopsis. Sci. Rep. 2016, 6, 24978. [Google Scholar] [CrossRef]
- Hossain, M.A.; Ye, W.; Munemasa, S.; Nakamura, Y.; Mori, I.C.; Murata, Y. Cyclic adenosine 5′-diphosphoribose (cADPR) cyclic guanosine 3′,5′-monophosphate positively function in Ca2+ elevation in methyl jasmonate-induced stomatal closure, cADPR is required for methyl jasmonate-induced ROS accumulation NO production in guard cells. Plant Biol. 2014, 16, 1140–1144. [Google Scholar]
- Hopper, D.W.; Ghan, R.; Schlauch, K.A.; Cramer, G.R. Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol. 2016, 16, 118. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.; Van Wees, S.C. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Liu, X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression Stabilities of Ten Candidate Reference Genes for RT-qPCR in Zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar] [CrossRef] [PubMed]


| Number | Name | Group | Structural Domain | Homologous Gene of Arabidopsis thaliana |
|---|---|---|---|---|
| 1 | ZbWRKY43 | I | WRKYGQK + WRKYGQK + C2H2 | KDO64417.1 |
| 2 | ZbWRKY13 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006464513.1 |
| 3 | ZbWRKY47 | II-b | WRKYGQK + C2H2 | XP_006492428.1 |
| 4 | ZbWRKY75 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006436871.1 |
| 5 | ZbWRKY41 | III | WRKYGQK + C2HC | XP_011022522.1 |
| 6 | ZbWRKY74 | II-d | WRKYGQK + C2H2 | KDO54401.1 |
| 7 | ZbWRKY24 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006431962.1 |
| 8 | ZbWRKY24(2) | I | WRKYGQK + WRKYGQK + C2H2 | XP_006431962.1 |
| 9 | ZbWRKY22 | II-e | WRKYGQK + C2H2 | XP_006444950.1 |
| 10 | ZbWRKY70 | III | WRKYGKK mutation | XP_006435943.1 |
| 11 | ZbWRKY49 | II-c | WRKYGQK + C2H2 | XP_006480891.1 |
| 12 | ZbWRKY50 | II-c | WRKYGQK + C2H2 | XP_006474455.1 |
| 13 | ZbWRKY50(2) | II-c | WRKYGQK + C2H2 | XP_006474454.1 |
| 14 | ZbWRKY71 | I | WRKYGQK + WRKYGQK + C2H2 | KDO78100.1 |
| 15 | ZbWRKY70(2) | III | WRKYQGQK + C2HC | XP_006481203.1 |
| 16 | ZbWRKY65 | II-e | WRKYGQK + C2H2 | XP_006467614.1 |
| 17 | ZbWRKY7 | II-d | WRKYGQK + C2H2 | KDO84087.1 |
| 18 | ZbWRKY31 | I | WRKYGQK + WRKYGQK + C2H2 | KDO54715.1 |
| 19 | ZbWRKY15 | II-d | WRKYGQK + C2H2 | XP_006425967.1 |
| 20 | ZbWRKY7(2) | II-d | WRKYGQK + C2H2 | XP_006494483.1 |
| 21 | ZbWRKY17 | II-d | WRKYGQK + C2H2 | XP_006483548.1 |
| 22 | ZbWRKY33 | I | WRKYGQK + WRKYGQK + C2H2 | XP_012089749.1 |
| 23 | ZbWRKY4 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006482724.1 |
| 24 | ZbWRKY53 | III | WRKYQGQK + C2HC | XP_006491244.1 |
| 25 | ZbWRKY75(2) | I | WRKYGQK + WRKYGQK + C2H2 | AEO31515.1 |
| 26 | ZbWRKY70(3) | III | WRKYQGQK + C2HC | KDO67551.1 |
| 27 | ZbWRKY14 | II-e | WRKYGQK + C2H2 | XP_006448817.1 |
| 28 | ZbWRKY20 | II-c | WRKYGQK + C2H2 | XP_006452514.1 |
| 29 | ZbWRKY47(2) | II-b | WRKYGQK + C2H2 | XP_006444621.1 |
| 30 | ZbWRKY32 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006453624.1 |
| 31 | ZbWRKY44 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006428919.1 |
| 32 | ZbWRKY53(2) | III | WRKYQGQK + C2HC | XP_006439376.1 |
| 33 | ZbWRKY2 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006474751.1 |
| 34 | ZbWRKY4(2) | I | WRKYGQK + WRKYGQK + C2H2 | KDO83046.1 |
| 35 | ZbWRKY1 | I | WRKYGQK + WRKYGQK + C2H2 | XP_006447745.1 |
| 36 | ZbWRKY41(2) | III | WRKYQGQK + C2HC | XP_006490540.1 |
| 37 | ZbWRKY51 | II-c | WRKYGQK + C2H2 | XP_006464492.1 |
| 38 | ZbWRKY14(2) | II-e | WRKYGQK + C2H2 | KDO76342.1 |
| Number | Name | F/R | Primer Sequences | Tm | Product Length | Homologous Sequence of Arabidopsis thaliana |
|---|---|---|---|---|---|---|
| 1 | ZbWRKY43 | F | TTCAGCTTCCCAGTTCTCATT | 56.6 | 144 | KDO64417.1 |
| R | TGTTTCACCTTATCATTACTCCC | 55.8 | ||||
| 2 | ZbWRKY13 | F | TTTTCAGCTTCCCAGTTCTCA | 57.6 | 146 | XP_006464513.1 |
| R | TGTTTCACCTTATCATTACTCCC | 55.8 | ||||
| 3 | ZbWRKY47 | F | AGAAGCGAGTAACCTTAAAGTAGA | 54.7 | 203 | XP_006492428.1 |
| R | TGACATTCAATGCAGCAGAT | 54.3 | ||||
| 4 | ZbWRKY75 | F | GATGATGGTTATAGATGGAGGA | 53.5 | 202 | XP_006436871.1 |
| R | GTTCAAAGCTGTCAGTGAGTTT | 54 | ||||
| 5 | ZbWRKY41 | F | GAGCAATCTGGACTTTGAGGGA | 60.8 | 186 | XP_011022522.1 |
| R | CCAAGAATGTCTTTTTGACCGT | 58.7 | ||||
| 6 | ZbWRKY74 | F | CAAGCACTTTGTCATCAACTAA | 53.5 | 158 | KDO54401.1 |
| R | ACTCCCAGAATCTTCTCCTCTG | 56.8 | ||||
| 7 | ZbWRKY24 | F | TCATCCTAAGCCTCAATCTACC | 55.7 | 212 | XP_006431962.1 |
| R | ATTTACTCCTCTGTGATCCCTG | 55.3 | ||||
| 8 | ZbWRKY24(2) | F | CAACAAAGAAGAAAGTGGAGAGG | 57.9 | 135 | XP_006431962.1 |
| R | GGATGGCATTAGAATTAACCGAA | 60.5 | ||||
| 9 | ZbWRKY22 | F | GAAACAAGTGGAGCGAAACAGA | 59.6 | 206 | XP_006444950.1 |
| R | CCGGAGATGAGGAAGAGAAAGT | 59.3 | ||||
| 10 | ZbWRKY70 | F | TGAAGATGGTCATGGATGGAGA | 59.9 | 62 | XP_006435943.1 |
| R | TTGGATATTTAGCATTGCGGAT | 59.5 | ||||
| 11 | ZbWRKY49 | F | GGCTTCATCTCCACTTTGCTTAC | 60.1 | 87 | XP_006480891.1 |
| R | TGGTCTTTTTTGGTTTCTTTGTT | 58.1 | ||||
| 12 | ZbWRKY50 | F | CATTTTCGGGGCTGTTCATA | 58.6 | 122 | XP_006474455.1 |
| R | CCTCTCCCTCCCACTTTCAT | 57.9 | ||||
| 13 | ZbWRKY50(2) | F | TATGGGAAGAAGACGGTGAA | 55.4 | 156 | XP_006474454.1 |
| R | ATTGCTTTGGTGAGTGTGGA | 55.9 | ||||
| 14 | ZbWRKY71 | F | GAAGATGGGTATCGGTGGA | 55 | 189 | KDO78100.1 |
| R | CAAGGTTGTGGGAAGTGGA | 56 | ||||
| 15 | ZbWRKY70(2) | F | AAATCTCATTCATAAGTCCAC | 56.4 | 262 | XP_006481203.1 |
| R | GCTTCTGATAATCTTCCCAAC | 53.4 | ||||
| 16 | ZbWRKY65 | F | TTCAAAAGCCGCCAAGACC | 61 | 153 | XP_006467614.1 |
| R | TCCATTTCCCCAAACCACG | 61.1 | ||||
| 17 | ZbWRKY7 | F | TGCCTCCTTTGCCTCTCCA | 61.2 | 147 | KDO84087.1 |
| R | CGCTGTCAGTATCCCCTGC | 58.6 | ||||
| 18 | ZbWRKY31 | F | AAAGGAAATGGTAAACGAGG | 54 | 222 | KDO54715.1 |
| R | TCTGAAGAAGAAAGCGAAGG | 55 | ||||
| 19 | ZbWRKY15 | F | GATTGTCTGACATCCCACC | 52 | 147 | XP_006425967.1 |
| R | CATCCAAAGCTCTCTCCAC | 52.2 | ||||
| 20 | ZbWRKY7(2) | F | GGAGAAAGTACGGACAGAAACC | 57.4 | 136 | XP_006494483.1 |
| R | GTGACAACAAGCATTGATGGAT | 57.4 | ||||
| 21 | ZbWRKY17 | F | AACGGGAAGCAAGGAGGAT | 58.5 | 91 | XP_006483548.1 |
| R | TGTACGGCTGAGAGGCGAG | 59.4 | ||||
| 22 | ZbWRKY33 | F | CTTCCTCCTGTTCCCCCTTCT | 61.2 | 257 | XP_012089749.1 |
| R | CGTGGTTCCTGTTGCCTCTTA | 60.3 | ||||
| 23 | ZbWRKY4 | F | ACTTGTTTTCCATCCGTCAC | 54.6 | 158 | XP_006482724.1 |
| R | CCATCATCAGCAGGTTTGTC | 55.8 | ||||
| 24 | ZbWRKY53 | F | TCCTTTCCCAACACTACACCAA | 59.6 | 116 | XP_006491244.1 |
| R | AGAGTCCAGCCGACCTTTACAT | 59.7 | ||||
| 25 | ZbWRKY75(2) | F | ATAATAAGTGGGGTTCTTTTGGTT | 58.1 | 132 | AEO31515.1 |
| R | GTTGTTGTTGATGTTGTAGTGGTG | 57.4 | ||||
| 26 | ZbWRKY70(3) | F | TCCCAACAGAAGTGAAACAAG | 55.2 | 95 | KDO67551.1 |
| R | CACAAAATCCTCCCACATAAT | 54.3 | ||||
| 27 | ZbWRKY14 | F | CCATGTGATTCACCAGTGACG | 59.1 | 249 | XP_006448817.1 |
| R | TCCAGTAGACCTGCTGTTTGC | 57.9 | ||||
| 28 | ZbWRKY20 | F | ACTGAGGTTCGAGTGGGTGAC | 58.6 | 229 | XP_006452514.1 |
| R | CTGCCTGAGGCTTAAAAAAGG | 59 | ||||
| 29 | ZbWRKY47(2) | F | TCCATTTCCCACCATTACCC | 59.4 | 122 | XP_006444621.1 |
| R | ATGAAGCATGTCGTTGCCTT | 58 | ||||
| 30 | ZbWRKY32 | F | ACCAAACAACTTCAACCCAGT | 56.2 | 135 | XP_006453624.1 |
| R | AAGGCTTTATTTCAAACCCGA | 58.7 | ||||
| 31 | ZbWRKY44 | F | CACCCAAAACCTCAACCTCCTA | 60.8 | 292 | XP_006428919.1 |
| R | TTCCTTCATCACATTCCCCACT | 60.8 | ||||
| 32 | ZbWRKY53(2) | F | TTTTTAGGGGGTTTCTCCTC | 55.5 | 162 | XP_006439376.1 |
| R | GGCATTTGAATTATTGGCTG | 56 | ||||
| 33 | ZbWRKY2 | F | AGACAGCCTCAGCCTCCAAAC | 60.8 | 183 | XP_006474751.1 |
| R | TCCTCAGATGATGCACCTCCA | 60.9 | ||||
| 34 | ZbWRKY4(2) | F | ATCCGCCACCTCAATCTAA | 55.2 | 201 | KDO83046.1 |
| R | CAGCATCACCCCCTTCTTC | 57.4 | ||||
| 35 | ZbWRKY1 | F | GCTGTTGGAATTGTCGTGTCT | 57.5 | 81 | XP_006447745.1 |
| R | ATCGGATGATTTGTCTTTGGC | 59.1 | ||||
| 36 | ZbWRKY41(2) | F | AACGAAGAAACGAAATACAACA | 55 | 234 | XP_006490540.1 |
| R | CCTATCAGAAGGACAAACAACA | 54.7 | ||||
| 37 | ZbWRKY51 | F | ATCTTGAAGTAATGGATGATGGA | 55.8 | 73 | XP_006464492.1 |
| R | TTATTTGGGTTGTTCTTGACTGA | 57.1 | ||||
| 38 | ZbWRKY14(2) | F | GCAAGGAAACAAGTGGAACG | 58.1 | 214 | KDO76342.1 |
| R | ACTCATCATCAATGGCGGTC | 57.7 | ||||
| UBQ | Ubiquitin extension protein | F | TCGAAGATGGCCGTACATTG | 57.5 | 122 | — |
| R | TCCTCTAAGCCTCAGCACCA | 59.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, X.; Hou, L.; Shi, J.; Yang, T.; Liu, Y.; Wei, A. Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 2019, 20, 68. https://doi.org/10.3390/ijms20010068
Fei X, Hou L, Shi J, Yang T, Liu Y, Wei A. Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim. International Journal of Molecular Sciences. 2019; 20(1):68. https://doi.org/10.3390/ijms20010068
Chicago/Turabian StyleFei, Xitong, Lixiu Hou, Jingwei Shi, Tuxi Yang, Yulin Liu, and Anzhi Wei. 2019. "Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim." International Journal of Molecular Sciences 20, no. 1: 68. https://doi.org/10.3390/ijms20010068
APA StyleFei, X., Hou, L., Shi, J., Yang, T., Liu, Y., & Wei, A. (2019). Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim. International Journal of Molecular Sciences, 20(1), 68. https://doi.org/10.3390/ijms20010068




