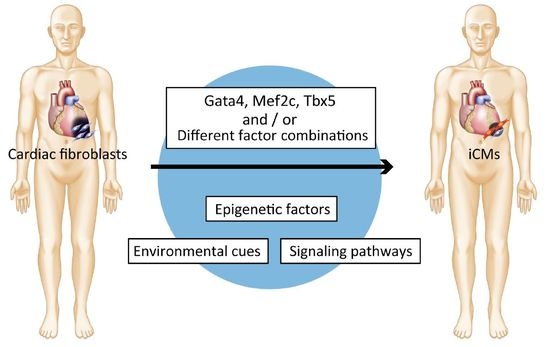
Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration
Abstract
1. Introduction
2. Discovery of Direct Cardiac Reprogramming by Gata4, Mef2c, and Tbx5
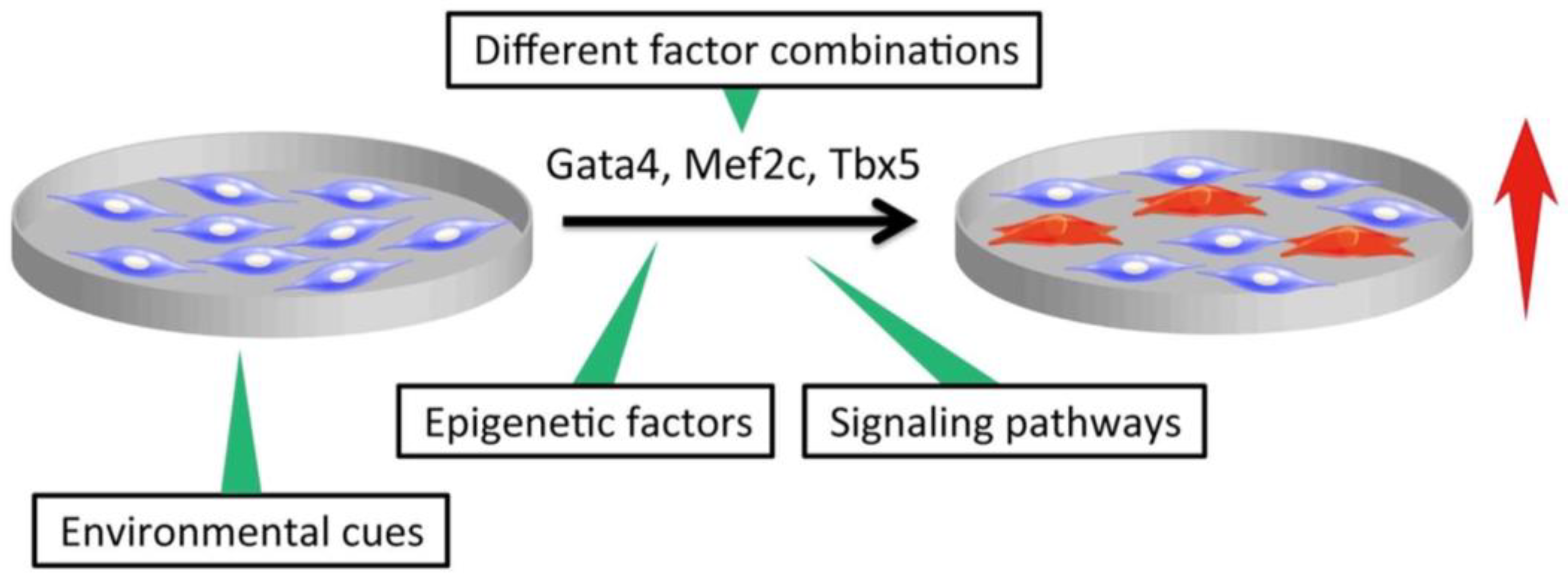
3. Improvement of Direct Cardiac Reprogramming and Cardiac Cell Maturation Efficiencies
3.1. Different Factor Combinations
3.2. Modification of Signaling Pathways and Environmental Cues
3.3. Epigenetic Factors
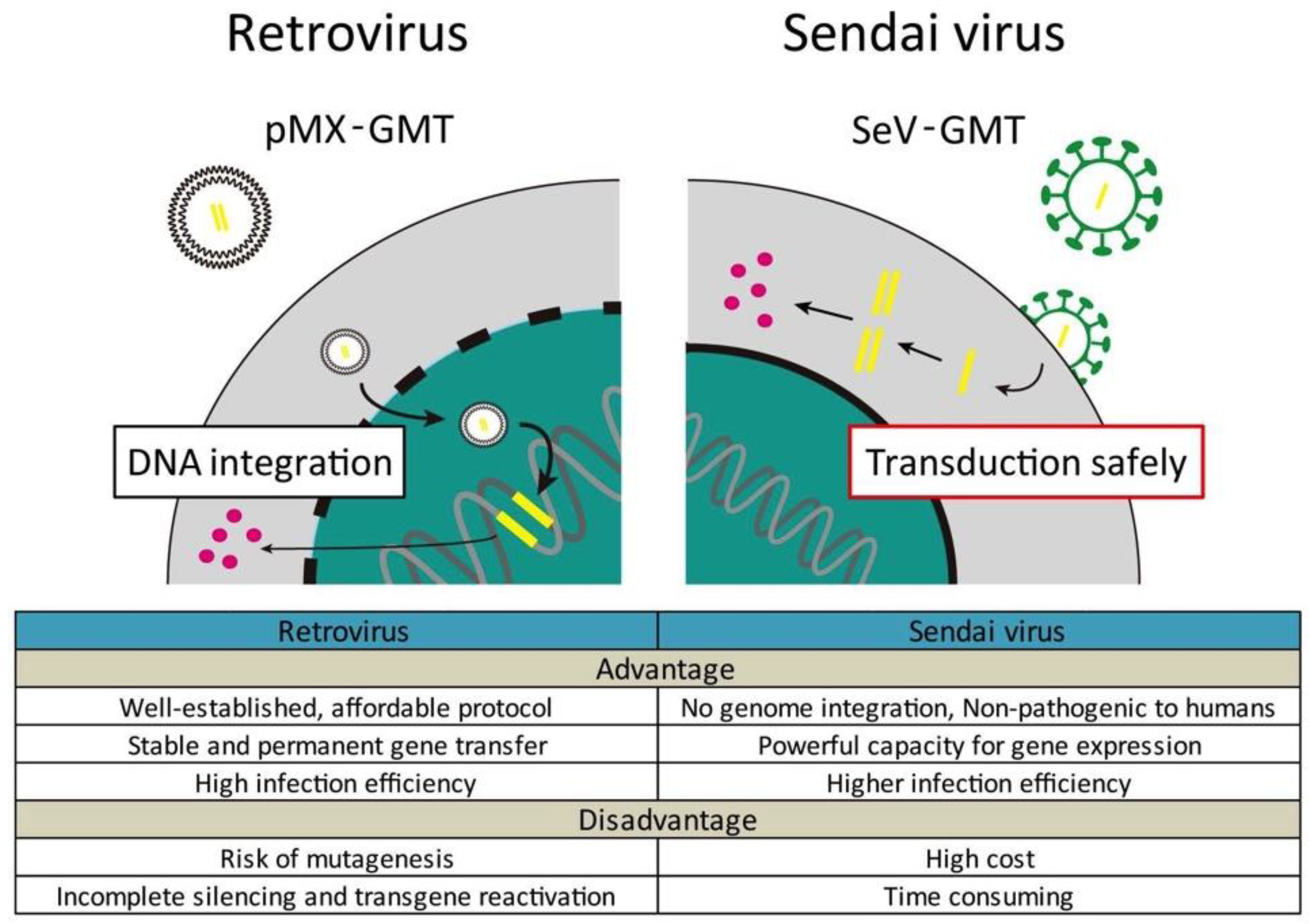
4. Cardiac Reprogramming In Vivo
5. Future Directions and Challenges
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berry, M.F.; Engler, A.J.; Woo, Y.J.; Pirolli, T.J.; Bish, L.T.; Jayasankar, V.; Morine, K.J.; Gardner, T.J.; Discher, D.E.; Sweeney, H.L. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H2196–H2203. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Bose, S.; Jain, K.G.; Bhargava, B.; Airan, B. TGFbeta1 contributes to cardiomyogenic-like differentiation of human bone marrow mesenchymal stem cells. Int. J. Cardiol. 2013, 163, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, T.; Zheng, H.; Cabral-da-Silva, M.; Sanada, F.; Ide-Iwata, N.; Ogorek, B.; Ferreira-Martins, J.; Arranto, C.; D’Amario, D.; del Monte, F.; et al. Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation 2011, 123, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, L. Exogenous Nkx2.5- or GATA-4-transfected rabbit bone marrow mesenchymal stem cells and myocardial cell co-culture on the treatment of myocardial infarction in rabbits. Mol. Med. Rep. 2015, 12, 2607–2621. [Google Scholar] [CrossRef] [PubMed]
- Nagata, H.; Ii, M.; Kohbayashi, E.; Hoshiga, M.; Hanafusa, T.; Asahi, M. Cardiac Adipose-Derived Stem Cells Exhibit High Differentiation Potential to Cardiovascular Cells in C57BL/6 Mice. Stem. Cells Transl. Med. 2016, 5, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Tanosaki, S.; Someya, S.; Fujita, J.; Fukuda, K. Manipulation of Pluripotent Stem Cell Metabolism for Clinical Application. Curr. Stem. Cell Rep. 2017, 3, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.W.H.; Lassar, A.B. Expression of a Single Transfected cDNA Converts Fibroblasts to Myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Sadahiro, T.; Yamanaka, S.; Ieda, M. Direct cardiac reprogramming: Progress and challenges in basic biology and clinical applications. Circ. Res. 2015, 116, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Krane, M.; Deutsch, M.A.; Wang, L.; Rav-Acha, M.; Gregoire, S.; Engels, M.C.; Rajarajan, K.; Karra, R.; Abel, E.D.; et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Furtado, M.B.; Costa, M.W.; Pranoto, E.A.; Salimova, E.; Pinto, A.R.; Lam, N.T.; Park, A.; Snider, P.; Chandran, A.; Harvey, R.P.; et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ. Res. 2014, 114, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Ieda, M. Critical factors for cardiac reprogramming. Circ. Res. 2012, 111, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J.; et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012, 151, 1617–1632. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Markoulaki, S.; Hanna, J.H.; Faddah, D.A.; Buganim, Y.; Kim, J.; Ganz, K.; Steine, E.J.; Cassady, J.P.; Creyghton, M.P.; et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 2011, 9, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007, 2, 3081–3089. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.; Khattak, S.; Poulet, C.; Lindemann, D.; Tanaka, E.M.; Ravens, U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 2012, 53, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Umei, T.C.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Kurotsu, S.; Tamura, F.; Osakabe, R.; et al. Single-construct polycistronic doxycycline-inducible vectors improve direct cardiac reprogramming and can be used to identify the critical timing of transgene expression. Int. J. Mol. Sci. 2017, 18, 1805. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.J.; Lubczyk, C.; Bhakta, M.; Zang, T.; Fernandez-Perez, A.; McAnally, J.; Bassel-Duby, R.; Olson, E.N.; Munshi, N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 2014, 141, 4267–4478. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Katoku-Kikyo, N.; Keirstead, S.A.; Kikyo, N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc. Res. 2013, 100, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Tani, T.; Kikyo, N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 2010, 54, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Masserdotti, G.; Gascon, S.; Gotz, M. Direct neuronal reprogramming: Learning from and for development. Development 2016, 143, 2494–24510. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, F.; Wu, M.; Wang, Z.Z. Development of hematopoietic stem and progenitor cells from human pluripotent stem cells. J. Cell. Biochem. 2015, 116, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Zakikhan, K.; Pournasr, B.; Nassiri-Asl, M.; Baharvand, H. Enhanced direct conversion of fibroblasts into hepatocyte-like cells by Kdm2b. Biochem. Biophys. Res. Commun. 2016, 474, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation 2017, 135, 978–995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.; Hashimoto, H.; Zhou, H.; Morales, M.G.; Chen, B.; Bassel-Duby, R.; Olson, E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017, 8, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Carag-Krieger, C.; Johnson, C.P.; Raab, M.; Tang, H.Y.; Speicher, D.W.; Sanger, J.W.; Sanger, J.M.; Discher, D.E. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: Scar-like rigidity inhibits beating. J. Cell Sci. 2008, 121, 3794–3802. [Google Scholar] [CrossRef] [PubMed]
- Chopra, A.; Lin, V.; McCollough, A.; Atzet, S.; Prestwich, G.D.; Wechsler, A.S.; Murray, M.E.; Oake, S.A.; Kresh, J.Y.; Janmey, P.A. Reprogramming cardiomyocyte mechanosensing by crosstalk between integrins and hyaluronic acid receptors. J. Biomech. 2012, 45, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Sia, J.; Yu, P.; Srivastava, D.; Li, S. Effect of biophysical cues on reprogramming to cardiomyocytes. Biomaterials 2016, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dal-Pra, S.; Mirotsou, M.; Jayawardena, T.M.; Hodgkinson, C.P.; Bursac, N.; Dzau, V.J. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 2016, 6, 38815. [Google Scholar] [CrossRef] [PubMed]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef] [PubMed]
- Chronis, C.; Fiziev, P.; Papp, B.; Butz, S.; Bonora, G.; Sabri, S.; Ernst, J.; Plath, K. Cooperative binding of transcription factors orchestrates reprogramming. Cell 2017, 168, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.R.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 2013, 155, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Ang, C.E.; Davila, J.; Pak, C.; Mall, M.; Lee, Q.Y.; Ahlenius, H.; Jung, S.W.; Sudhof, T.C.; Wernig, M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014, 3, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Van Oevelen, C.; Collombet, S.; Vicent, G.; Hoogenkamp, M.; Lepoivre, C.; Badeaux, A.; Bussmann, L.; Sardina, J.L.; Thieffry, D.; Beato, M.; et al. C/EBPα activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Rep. 2015, 5, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Sardina, J.L.; van Oevelen, C.; Collombet, S.; Kallin, E.M.; Vicent, G.P.; Lu, J.; Thieffry, D.; Beato, M.; Graf, T. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014, 506, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Bossard, P.; Zaret, K.S. GATA transcription factors as potentiators of gut endoderm differentiation. Development 1998, 125, 4909–4917. [Google Scholar] [PubMed]
- Donaghey, J.; Thakurela, S.; Charlton, J.; Chen, J.S.; Smith, Z.D.; Gu, H.; Pop, R.; Clement, K.; Stamenova, E.K.; Karnik, R.; et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018, 50, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Zheng, M.; Wang, L.; Zhou, Y.; Yin, C.; Liu, J.; Qian, L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016, 16, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Dal-Pra, S.; Hodgkinson, C.P.; Mirotsou, M.; Kirste, I.; Dzau, V.J. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by miR combo. Circ. Res. 2017, 120, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Liu, J.; et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Sauls, K.; Greco, T.M.; Wang, L.; Zou, M.; Villasmil, M.; Qian, L.; Cristea, I.M.; Conlon, F.L. Initiating events in direct cardiomyocyte reprogramming. Cell Rep. 2018, 22, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Kikyo, N. Inhibitors of suppressive histone modification promote direct reprogramming of fibroblasts to cardiomyocyte-like cells. Cardiovasc. Res. 2014, 102, 188–190. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Inagawa, K.; Miyamoto, K.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Umei, T.; Wada, R.; Katsumata, Y.; Kaneda, R.; Nakade, K.; et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Finch, E.A.; Zhang, L.; Zhang, H.; Hodgkinson, C.P.; Pratt, R.E.; Rosenberg, P.B.; Mirotsou, M.; Dzau, V.J. MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015, 116, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct in vivo reprogramming with Sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell 2018, 22, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.O.M. Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine. Curr. Gene Ther. 2012, 5, 410–416. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Chen, K.Y.; Naumova, A.V.; Muskheli, V.; Fugate, J.A.; Dupras, S.K.; Reinecke, H.; Xu, C.; Hassanipour, M.; Police, S.; et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007, 25, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Methot, D.; Poppa, V.; Fujio, Y.; Walsh, K.; Murry, C.E. Cardiomyocyte grafting for cardiac repair: Graft cell death and anti-death strategies. J. Mol. Cell. Cardiol. 2001, 33, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Caspi, O.; Huber, I.; Kehat, I.; Habib, M.; Arbel, G.; Gepstein, A.; Yankelson, L.; Aronson, D.; Beyar, R.; Gepstein, L. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J. Am. Coll. Cardiol. 2007, 50, 1884–1893. [Google Scholar] [CrossRef] [PubMed]
- Blum, B.; Benvenisty, N. The tumorigenicity of human embryonic stem cells. Adv. Cancer Res. 2008, 100, 133–158. [Google Scholar] [PubMed]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Welch, J.D.; Ma, H.; Zhou, Y.; Vaseghi, H.R.; Yu, S.; Wall, J.B.; Alimohamadi, S.; Zheng, M.; et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 2017, 551, 100–104. [Google Scholar] [CrossRef] [PubMed]
| Different Factor Combinations | Original Cell | Markers and Efficiency |
|---|---|---|
| Gata4, Mef2c, Tbx5 (GMT) [11] | CF, TTF | α-MHC+ 20%; cTnT+ 30% of α-MHC; α-Actinin+ most of cTnT+ |
| Gata4, Mef2c, Tbx5 (GMT) [15] | adult TTF, adult CF | α-MHC+ 0% |
| Puro polycistronic MTG (Mef2c, Tbx5, Gata4) [20] | CF | α-MHC+ 16%; cTnT+ 24% |
| Mef2c, Tbx5, Myocd [21] | MEF, TTF, CF | cTnT+ 12% (96% of α-MHC) |
| Gata4, Hand 2, Mef2c, Tbx5 (GHMT) [22] | adult TTF, CF | αMHC+ cTnT+ 9.2% (TTF), 7.5% (CF) |
| Chimeric Mef2c + GHT [26] | MEF | cTnT+ 20.9%; beating iCMs 3.5% |
| Hand2, Nxk2.5, Gata4, Mef2c, Tbx5 (HNGMT) [28] | MEF, CF | GCaMP5 activity+ 1.6% |
| MiR-1, miR-133, miR-208, miR-499 [27] | CF | αMHC+ 1.5% to 7.7% |
| GMT + miR-133 [30] | MEF, CF | αMHC+ 19.4%; cTnT+ 5.4% |
| Signaling Pathways | ||
| MicroRNA cocktail + JAK inhibitor I [29] | CF | αMHC+ ~28% |
| HNGMT + TGFβ inhibitor [35] | MEF, adult CF | cTnT-GCaMP5+ activity: 16.95% |
| GHMT + ROCK/TGFβ inhibitor [36] | MEF, adult CF | cTnT+ ~67%; α-actinin+ ~64% |
| GMT + FGF2, FGF10, VEGF [41] | MEF, TTF | αMHC+ 15%; cTnT+ 5% |
| GMT + WNT/TGFβ inhibitor [37] | CF | αMHC+ 38.3%; cTnT+ 23.4%; cTnT-GCaMP5+ activity 12.5% |
| GHMT + Akt1 (AGHMT) + Notch inhibitor [40] | MEF | cTnT+ 70%; Ca flux+ 40%; beating 45% |
| GHMT + Akt1 (AGHMT) + ZNF281 [39] | TTF | αMHC+ ~33%; cTnT+ ~45% |
| Environmental Cues | ||
| GMT in microgrooved substrate [44] | TTF | αMHC+ 32%; beating more than double |
| Epigenetic Factors | ||
| MGT + Bmi1 [58] | CF | αMHC+ 30%; cTnT+ 28% |
| Different Factor Combinations | Original Cell | Markers and Efficiency |
|---|---|---|
| Gata4, Hand2, Tbx5, Myocd, miR-1, miR-133 [70] | nHFF, DF, CF | cTnT ~19% (nHFF); calcium transient+ |
| Gata4, Mef2c, Tbx5 (GMT) + Mesp1, Myocd [71] | DF, CF | cTnT 5% (CF); sarcomeric structure+ |
| GMT + Esrrg, Mesp1, Myocd, ZFPM2 (7F) [72] | hESC-derived fibroblast | cTnT 13%; Ca flux+, action potential |
| GMT + Mesp1, Myocd, miR-133 [30] | CF | cTnT 23–27% |
| GMT + Myocd, WNT/TGFβ inhibitor [37] | CF | cTnT >2-fold compared with 7F; Calcium transients ~80% |
| SeV-GMT [64] | CF | cTnT 15%; Synchronous with surroundings by cocultivation |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tani, H.; Sadahiro, T.; Ieda, M. Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. Int. J. Mol. Sci. 2018, 19, 2629. https://doi.org/10.3390/ijms19092629
Tani H, Sadahiro T, Ieda M. Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. International Journal of Molecular Sciences. 2018; 19(9):2629. https://doi.org/10.3390/ijms19092629
Chicago/Turabian StyleTani, Hidenori, Taketaro Sadahiro, and Masaki Ieda. 2018. "Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration" International Journal of Molecular Sciences 19, no. 9: 2629. https://doi.org/10.3390/ijms19092629
APA StyleTani, H., Sadahiro, T., & Ieda, M. (2018). Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. International Journal of Molecular Sciences, 19(9), 2629. https://doi.org/10.3390/ijms19092629