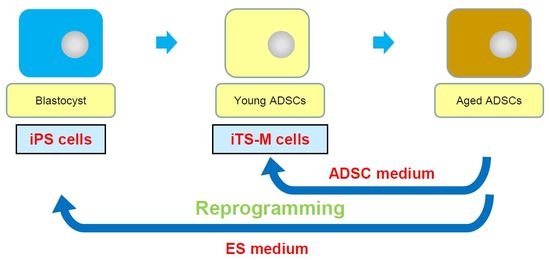
Induction of Expandable Adipose-Derived Mesenchymal Stem Cells from Aged Mesenchymal Stem Cells by a Synthetic Self-Replicating RNA
Abstract
1. Introduction
2. Results
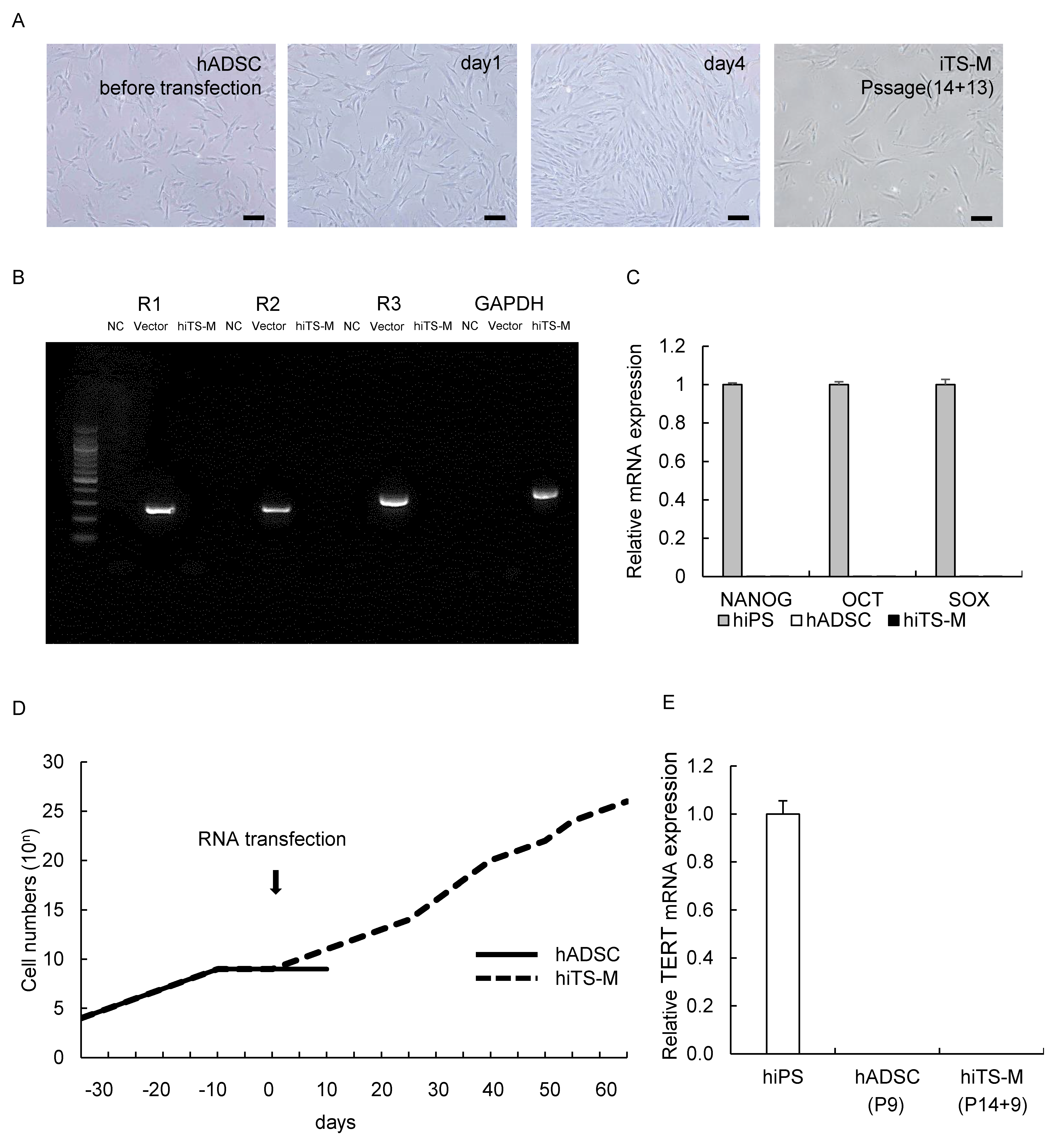
2.1. Generation of hiTS-M Cells from Human ADSCs by an RNA Expression Vector
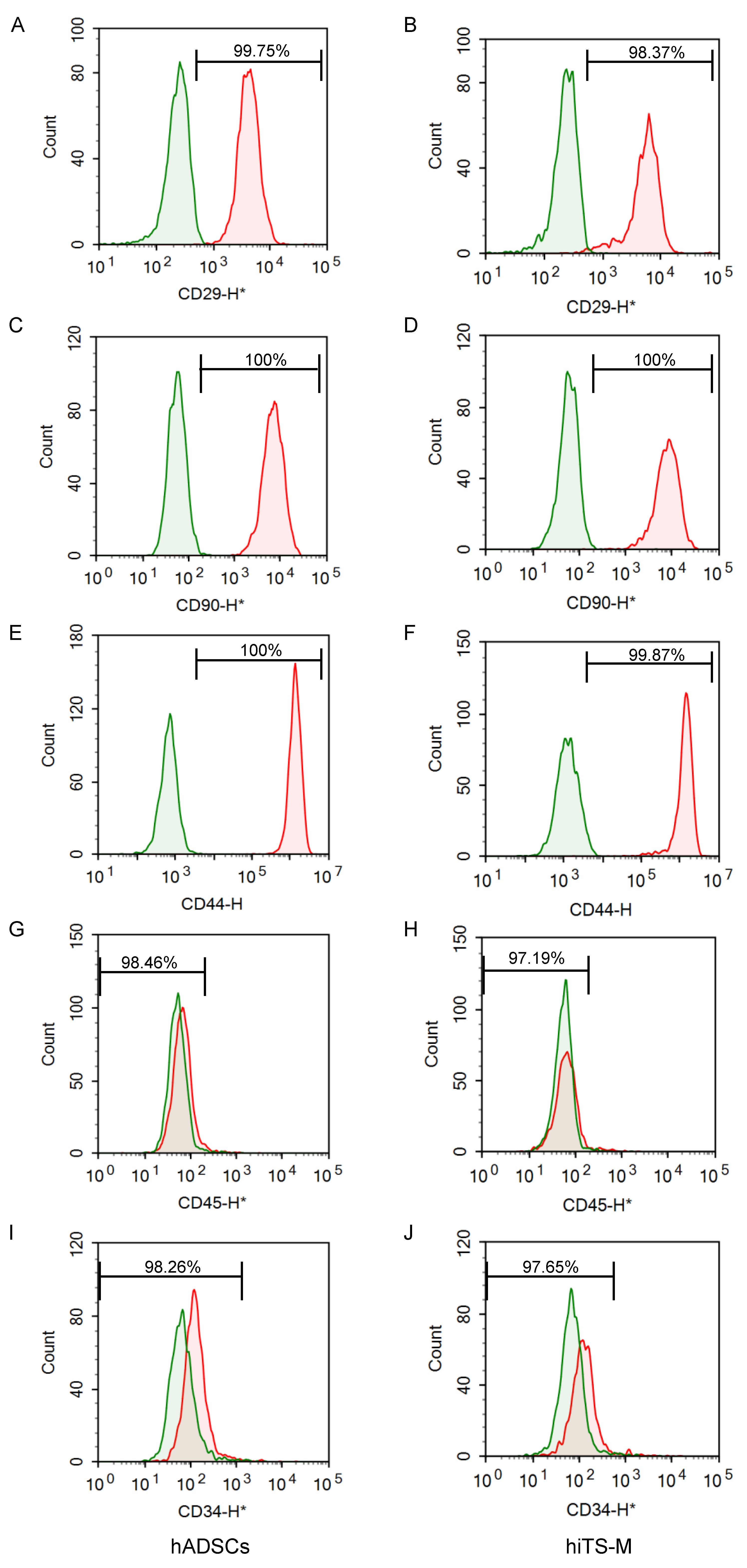
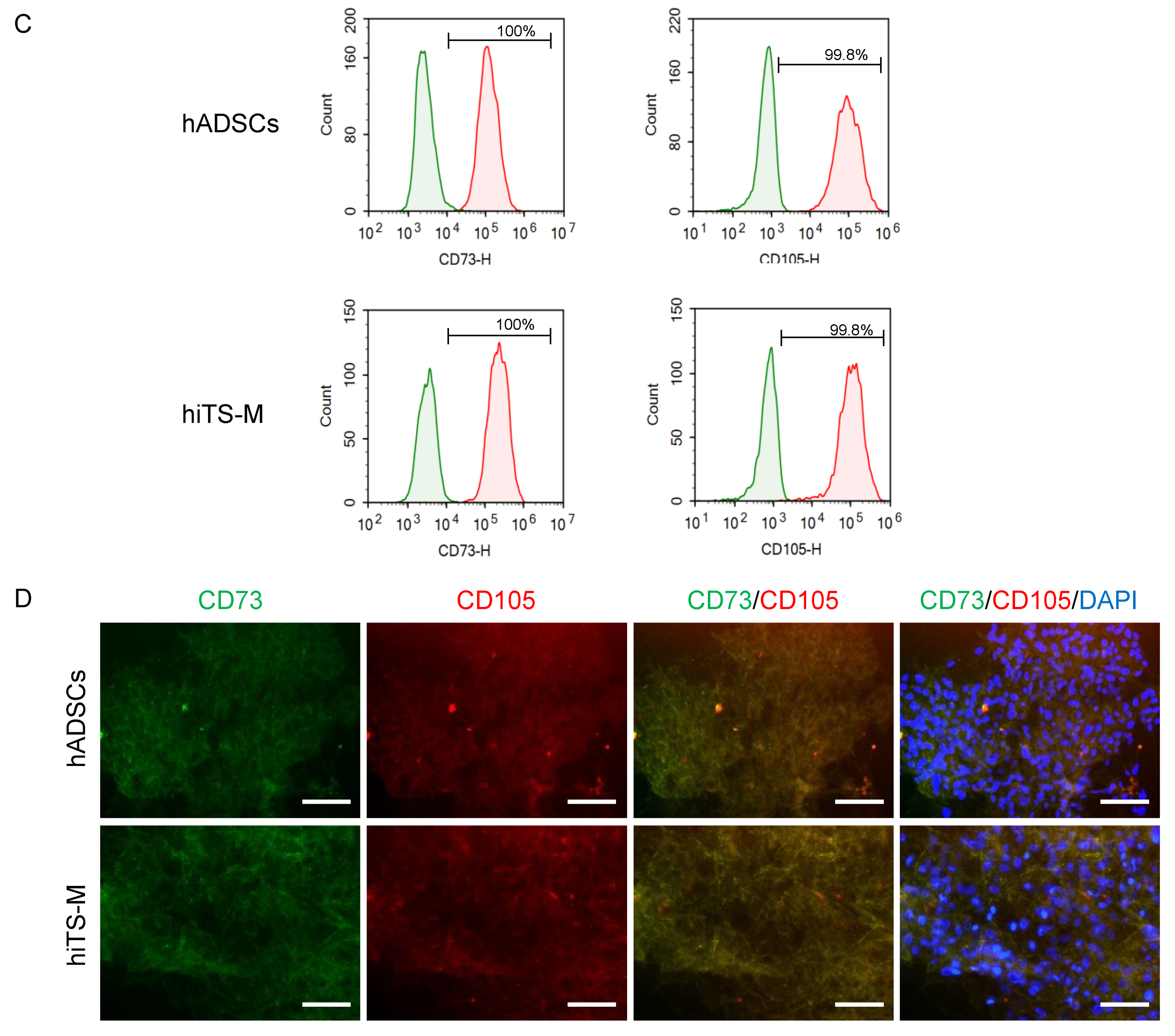
2.2. Characterization of hiTS-M Cells Transfected with the RNA Vector
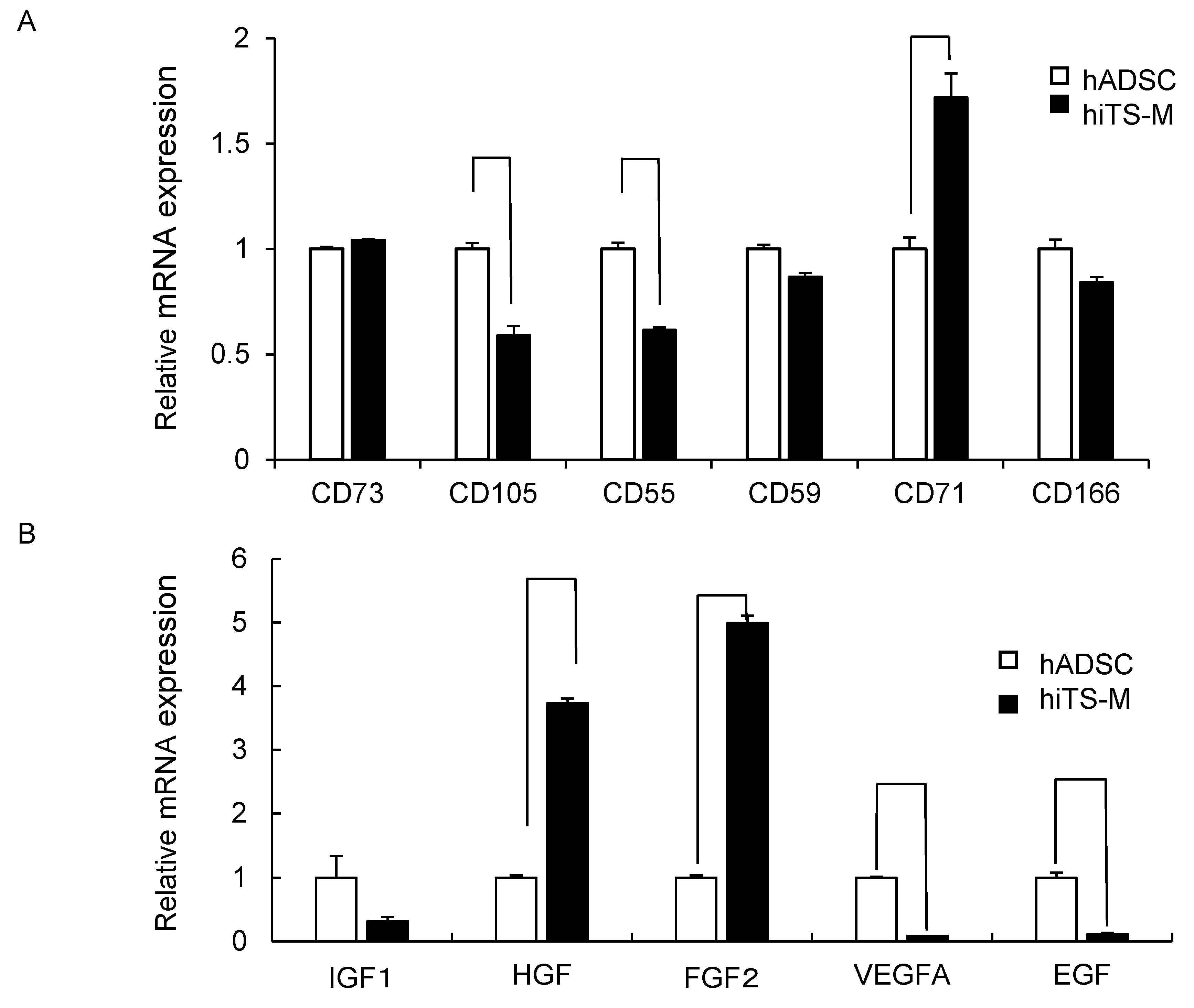
2.3. Genes and Proteins Expressed in hiTS-M Cells
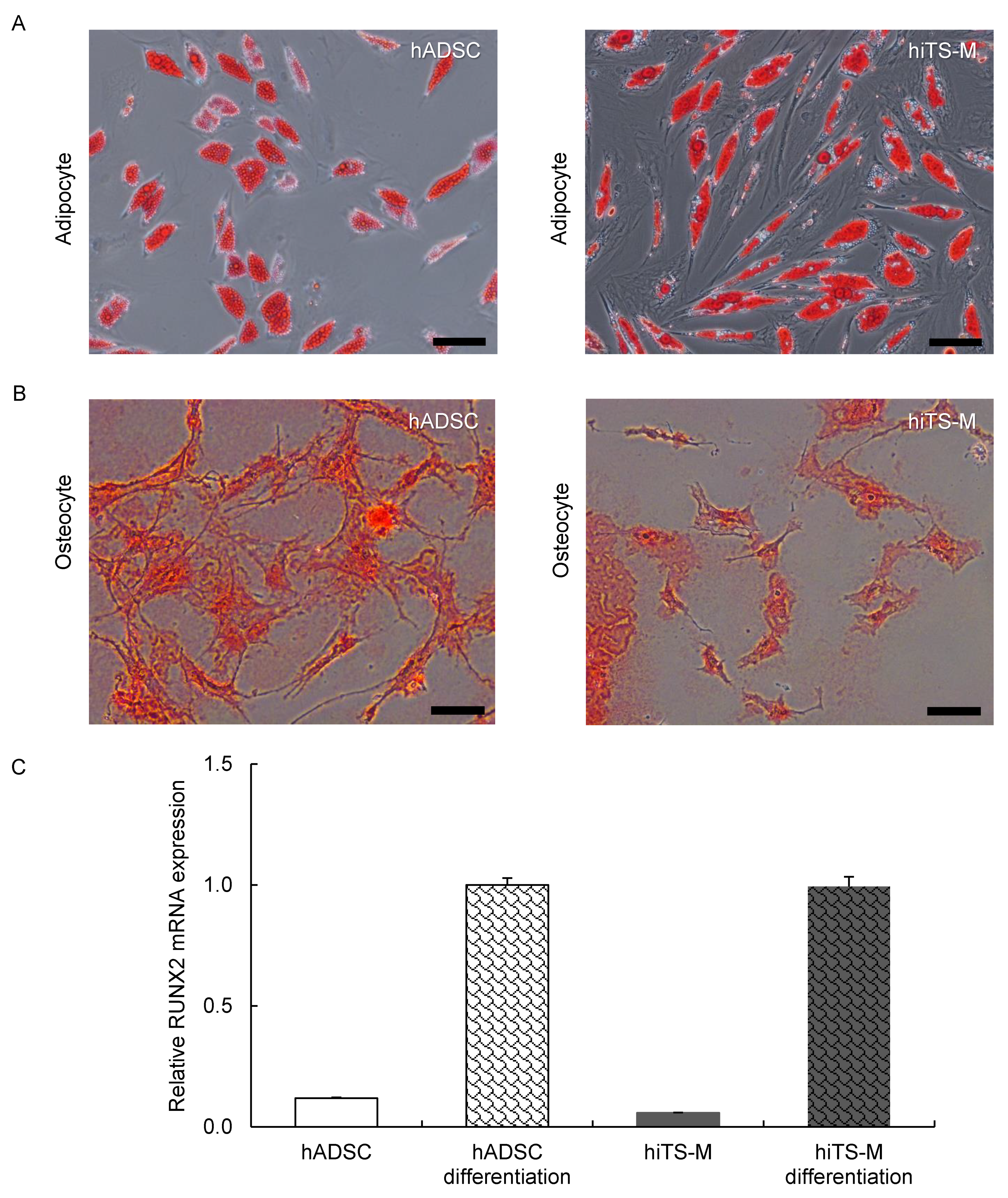
2.4. Analysis of the Differentiation Potential of hiTS-M Cells
2.5. Bisulfite Sequencing of the Genomic Promoter Regions of OCT4 and NANOG in hiPS Cells, hADSCs, and hiTS-M Cells
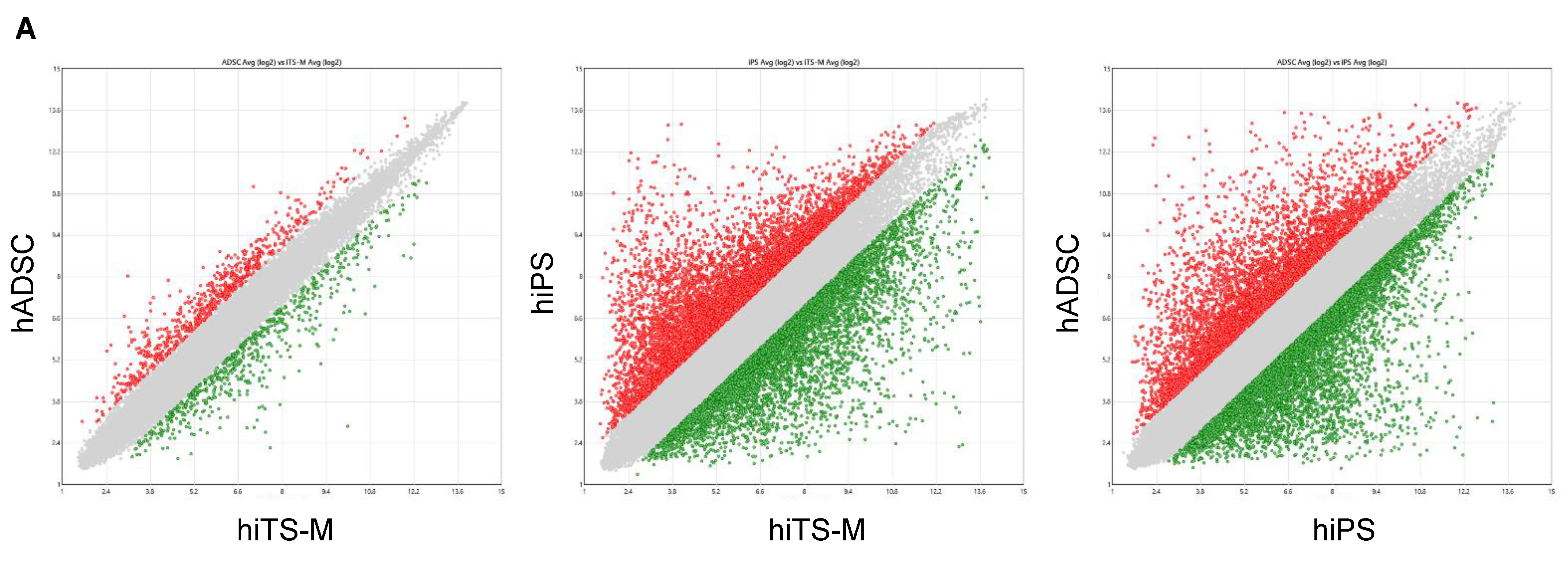
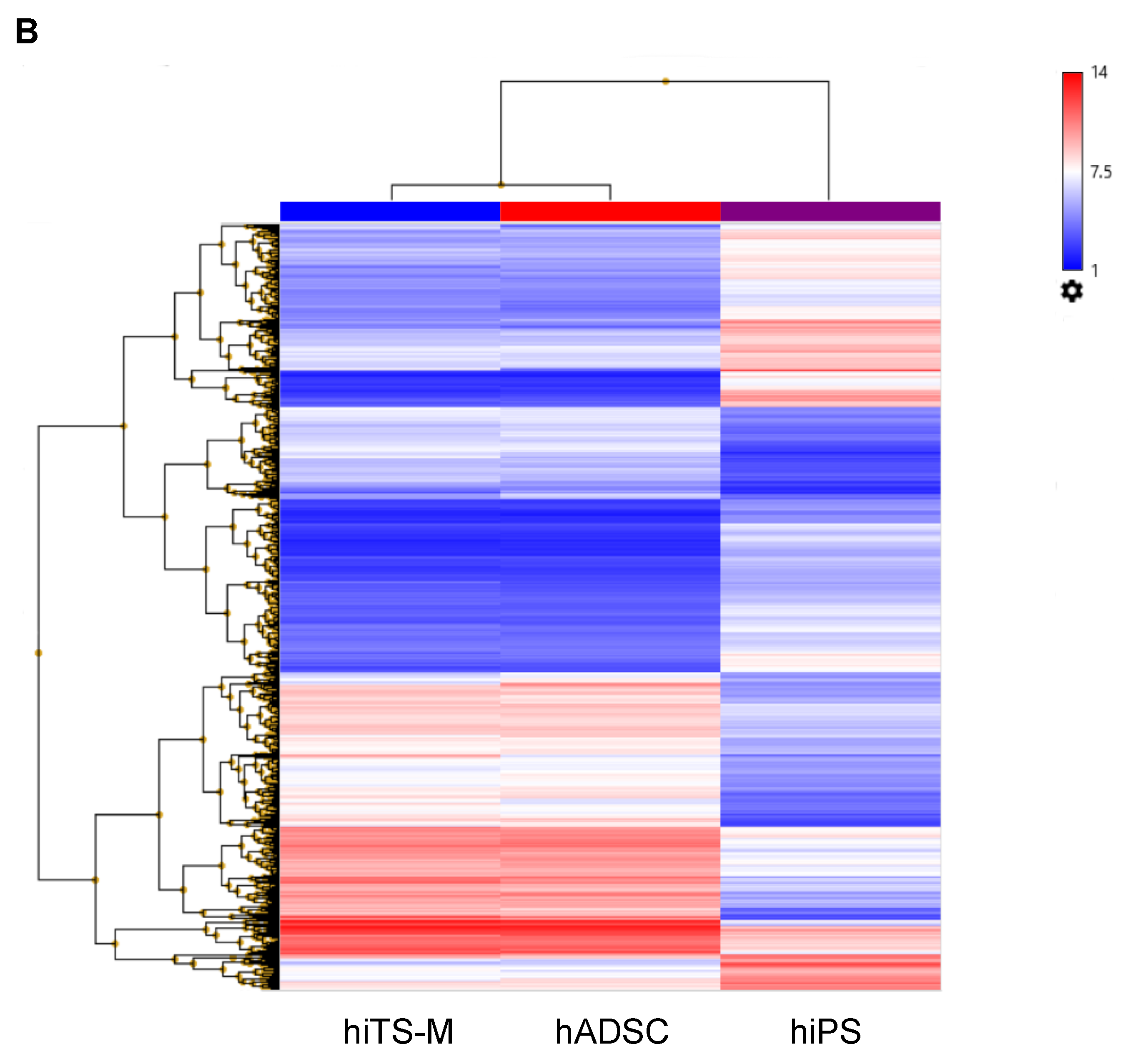
2.6. Microarray Analysis
3. Discussion
4. Materials and Methods
4.1. Culture of hADSCs
4.2. Generation of hiTS-M Cells Using an RNA Replicon
4.3. RT-PCR and qRT-PCR Analyses
4.4. Flow Cytometry
4.5. Immunostaining
4.6. Cell Differentiation
4.7. Bisulfite Genomic Sequencing
4.8. Microarray Analysis
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedenstein, A.J.; Petrakova, K.V.; Kurolesova, A.I.; Frolova, G.P. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 1968, 6, 230. [Google Scholar] [CrossRef] [PubMed]
- Zuk, P.A. Human Adipose Tissue Is a Souce of Multipotent Stem Cells. Mol. Biol. Cell 2002, 13, 4279–4295. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Ohgushi, H.; Caplan, A.I. Stem cell technology and bioceramics: From cell to gene engineering. J. Biomed. Mater. Res. 1999, 48, 913–927. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal stem cells. J. Orthop. Res. 1991, 9, 641–650. [Google Scholar] [CrossRef] [PubMed]
- King, S.N.; Hanson, S.E.; Hematti, P.; Thibeault, S.L. Current applications of mesenchymal stem cells for tissue replacement in otolaryngology-head and neck surgery. Am. J. Stem Cells 2012, 1, 225–238. [Google Scholar] [PubMed]
- Mathe, G.; Bernard, J.; Schwarzenberg, L.; Larriue, M.J.; Lalanne, C.M.; Dutrieux, A.; Denoix, P.F.; Sur-mont, J.; Schwarzmann, V.; Ceoara, B. Trial treatment of patients afflicted with acute leukemia in remission with total irradiation followed by homologous bone marrow transfusion. Rev. Fr. Etud. Clin. Biol. 1959, 4, 675–704. [Google Scholar] [PubMed]
- Mathe, G.; Jammet, H.; Pendic, B.; Schwarzenberg, L.; Duplan, J.F.; Maupin, B.; Lataret, R.; Larriue, M.J.; Kalic, D.; Djukic, Z. Transfusion and grafts of homologous bonemarrow in humans after accidental high dose irradiation. Rev. Fr. Etud. Clin. Biol. 1959, 4, 226–238. [Google Scholar] [PubMed]
- Kumar, A.; D’Souza, S.S.; Moskvin, O.V.; Toh, H.; Wang, B.; Zhang, J.; Swanson, S.; Guo, L.W.; Thomson, J.A.; Slukvin, I.I. Specification and Diversification of Pericytes and Smooth Muscle Cells from Mesenchymoangioblasts. Cell Rep. 2017, 19, 1902–1916. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Salimath, B.P.; Schieker, M.; Stark, G.B.; Finkenzeller, G. Inhibition of metastasis-associated gene 1 expression affects proliferation and osteogenic differentiation of immortalized human mesenchymal stem cells. Cell Prolif. 2011, 44, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Gaur, M.; Dobke, M.; Lunyak, V.V. Mesenchymal Stem Cells from Adipose Tissue in Clinical Applications for Dermatological Indications and Skin Aging. Int. J. Mol. Sci. 2017, 18, 208. [Google Scholar] [CrossRef] [PubMed]
- Iwazawa, R.; Kozakai, S.; Kitahashi, T.; Nakamura, K.; Hata, K.I. The Therapeutic Effects of Adipose-Derived Stem Cells and Recombinant Peptide Pieces on Mouse Model of DSS Colitis. Cell Transpl. 2018, 27, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei Qomi, R.; Sheykhhasan, M. Adipose-derived stromal cell in regenerative medicine: A review. World J. Stem Cells 2017, 9, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Owczarczyk-Saczonek, A.; Wociór, A.; Placek, W.; Maksymowicz, W.; Wojtkiewicz, J. The Use of Adipose-Derived Stem Cells in Selected Skin Diseases (Vitiligo, Alopecia, and Nonhealing Wounds). Stem Cells Int. 2017, 2017, 4740709. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, N.; Fujii, T.; Iwase, T.; Ohgushi, H.; Itoh, T.; Uematsu, M.; Yamagishi, M.; Mori, H.; Kanagawa, K.; Kitamura, S. Intravenous administration of mesenchymal stem cells improves cardiac function in rats with acute myocardial infarction through angiogenesis and myogenesis. Am. J. Physiol. Heart Circ. Physiol. 2004, 287, 2670–2676. [Google Scholar] [CrossRef] [PubMed]
- Patrick, C.W., Jr. Adipose tissue engineering: The future of breast and soft tissue reconstruction following tumor resection. Semin. Surg. Oncol. 2000, 19, 302–311. [Google Scholar] [CrossRef]
- Patrick, C.W. Breast tissue engineering. Annu. Rev. Biommed. Eng. 2004, 6, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Beahm, E.K.; Walton, R.L.; Patrick, C.W., Jr. Progress in adipose tissue construct development. Clin. Plast. Surg. 2003, 30, 547–558. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Sridharan, R.; Xie, W.; Utikal, J.; Eminli, S.; Arnold, K.; Stadtfeld, M.; Yachechko, R.; Tchieu, J.; Jaenisch, R.; et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, Y.; Kinjo, T.; Noguchi, H. The Development of Cancer through the Transient Overexpression of Reprogramming Factors. Cell Med. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Noguchi, H.; Saitoh, I.; Tsugata, T.; Kataoka, H.; Watanabe, M.; Noguchi, Y. Induction of tissue-specific stem cells by reprogramming factors, and tissue-specific selection. Cell Death Differ. 2015, 22, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, I.; Sato, M.; Soda, M.; Inada, E.; Iwase, Y.; Murakami, T.; Ohshima, H.; Hayasaki, H.; Noguchi, H. Tissue-Specific Stem Cells Obtained by Reprogramming of Non-Obese Diabetic (NOD) Mouse-Derived Pancreatic Cells Confer Insulin Production in Response to Glucose. PLoS ONE 2016, 11, e0163580. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Gros, E.; Li, H.R.; Kumar, S.; Deacon, D.C.; Maron, C.; Muotri, A.R.; Chi, N.C.; Fu, X.D.; Yu, B.D.; et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell 2013, 13, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, H. Characterization of induced tissue-specific stem cells from pancreas by a synthetic self-replicative RNA. Sci. Rep. 2018, 8, 12341. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Miyagi-Shiohira, C.; Nakashima, Y. Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 930. [Google Scholar] [CrossRef] [PubMed]
- Panasophonkul, S.; Samart, P.; Kongon, K.; Sathanawongs, A. Phenotypic characteristics of feline adipose-derived stem cells affected by cell passage number. Pol. J. Vet. Sci. 2017, 20, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Legzdina, D.; Romanauska, A.; Nikulshin, S.; Kozlovska, T.; Berzins, U. Characterization of Senescence of Culture-expanded Human Adipose-derived Mesenchymal Stem Cells. Int. J. Stem Cells 2016, 9, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Nahar, S.; Miyagi-Shiohira, C.; Kinjo, T.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Fujita, J.; Noguchi, H. A Liquid Chromatography with Tandem Mass Spectrometry-Based Proteomic Analysis of Cells Cultured in DMEM 10% FBS and Chemically Defined Medium Using Human Adipose-Derived Mesenchymal Stem Cells. Int. J. Mol. Sci. 2018, 19, 2042. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, H. Evaluation of Serum-Free, Xeno-Free Cryopreservation Solutions for Human Adipose-Derived Mesenchymal Stem Cells. Cell Med. 2016, 9, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, A.; Miyazaki, A.; Kawarabayashi, K.; Shono, M.; Akazawa, Y.; Hasegawa, T.; Ueda-Yamaguchi, K.; Kitamura, T.; Yoshizaki, K.; Fukumoto, S.; et al. Piezo type mechanosensitive ion channel component 1 functions as a regulator of the cell fate determination of mesenchymal stem cells. Sci. Rep. 2017, 7, 17696. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Kitamura, S.; Saitoh, I.; Watanabe, M.; Noguchi, H. Induction of Expandable Adipose-Derived Mesenchymal Stem Cells from Aged Mesenchymal Stem Cells by a Synthetic Self-Replicating RNA. Int. J. Mol. Sci. 2018, 19, 3489. https://doi.org/10.3390/ijms19113489
Miyagi-Shiohira C, Nakashima Y, Kobayashi N, Kitamura S, Saitoh I, Watanabe M, Noguchi H. Induction of Expandable Adipose-Derived Mesenchymal Stem Cells from Aged Mesenchymal Stem Cells by a Synthetic Self-Replicating RNA. International Journal of Molecular Sciences. 2018; 19(11):3489. https://doi.org/10.3390/ijms19113489
Chicago/Turabian StyleMiyagi-Shiohira, Chika, Yoshiki Nakashima, Naoya Kobayashi, Shinji Kitamura, Issei Saitoh, Masami Watanabe, and Hirofumi Noguchi. 2018. "Induction of Expandable Adipose-Derived Mesenchymal Stem Cells from Aged Mesenchymal Stem Cells by a Synthetic Self-Replicating RNA" International Journal of Molecular Sciences 19, no. 11: 3489. https://doi.org/10.3390/ijms19113489
APA StyleMiyagi-Shiohira, C., Nakashima, Y., Kobayashi, N., Kitamura, S., Saitoh, I., Watanabe, M., & Noguchi, H. (2018). Induction of Expandable Adipose-Derived Mesenchymal Stem Cells from Aged Mesenchymal Stem Cells by a Synthetic Self-Replicating RNA. International Journal of Molecular Sciences, 19(11), 3489. https://doi.org/10.3390/ijms19113489