Functional Characterization and Structure-Guided Mutational Analysis of the Transsulfuration Enzyme Cystathionine γ-Lyase from Toxoplasma gondii
Abstract
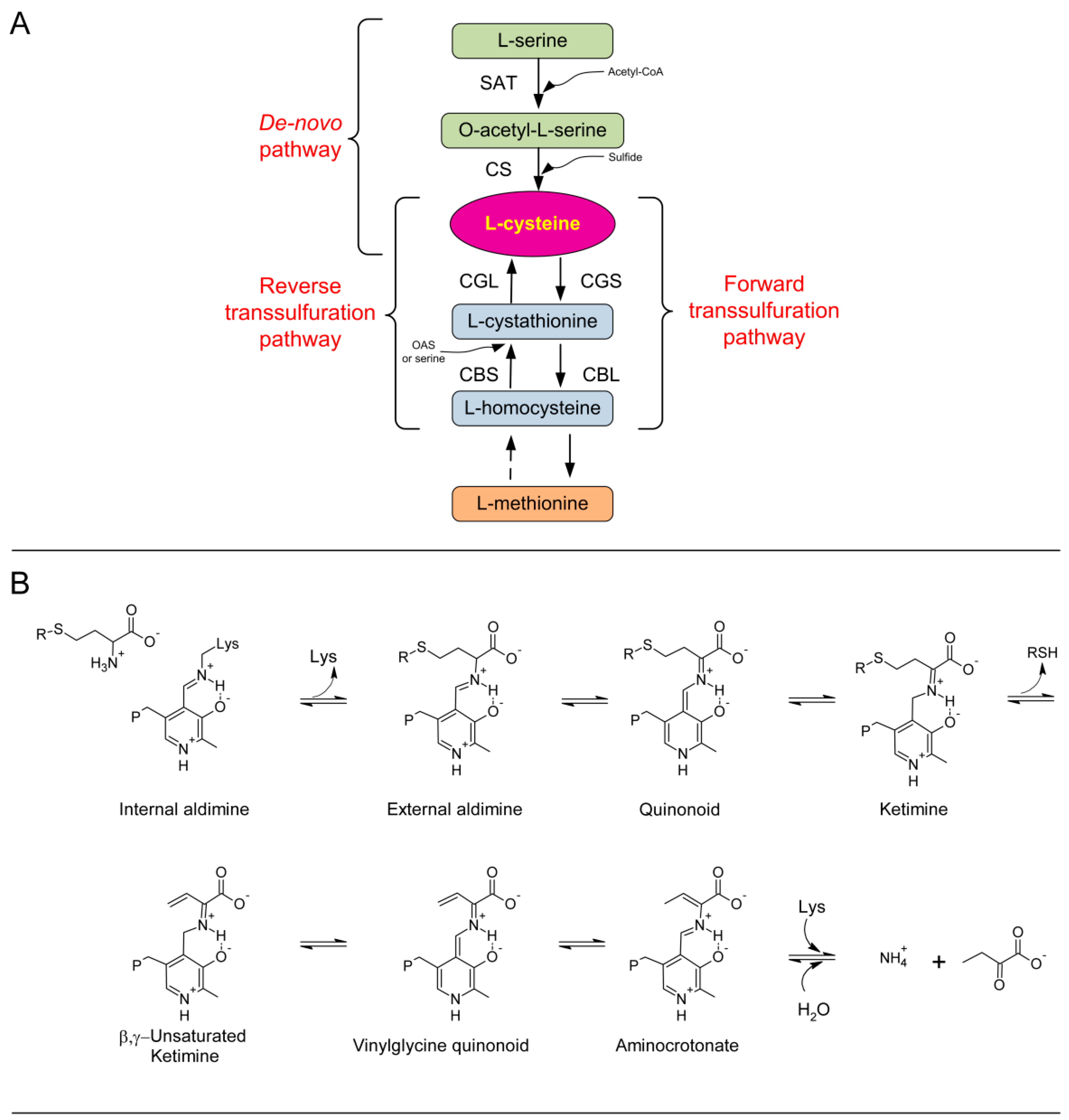
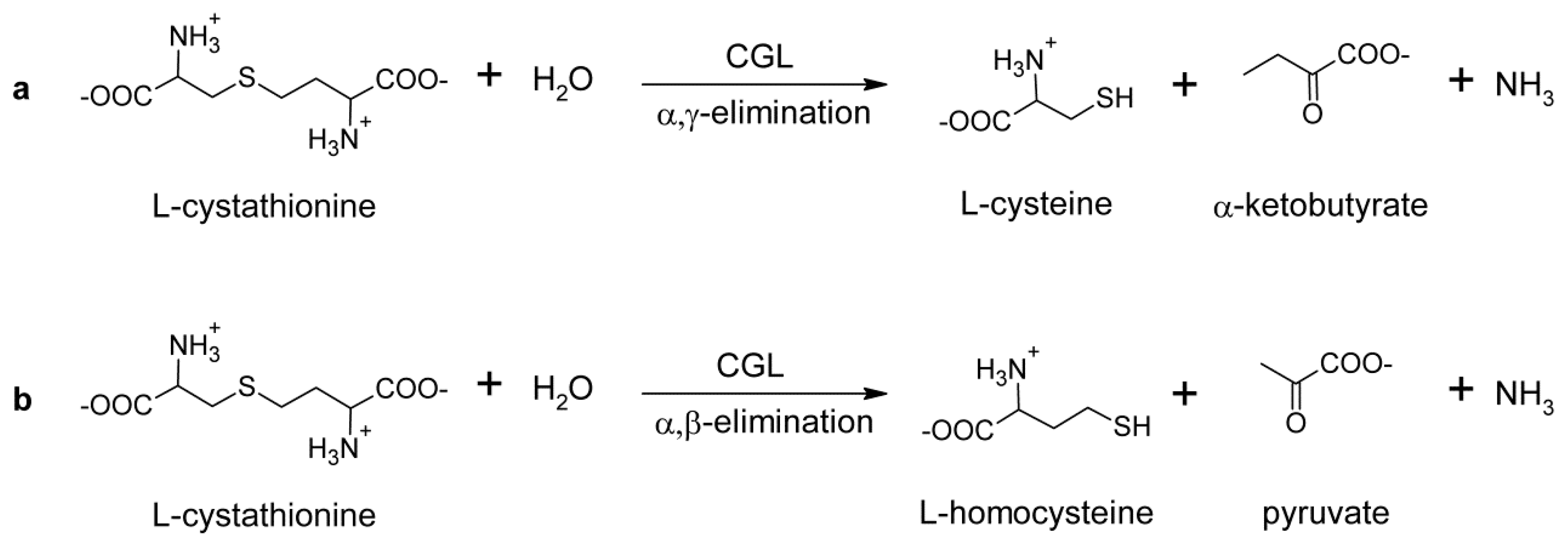
1. Introduction
2. Results
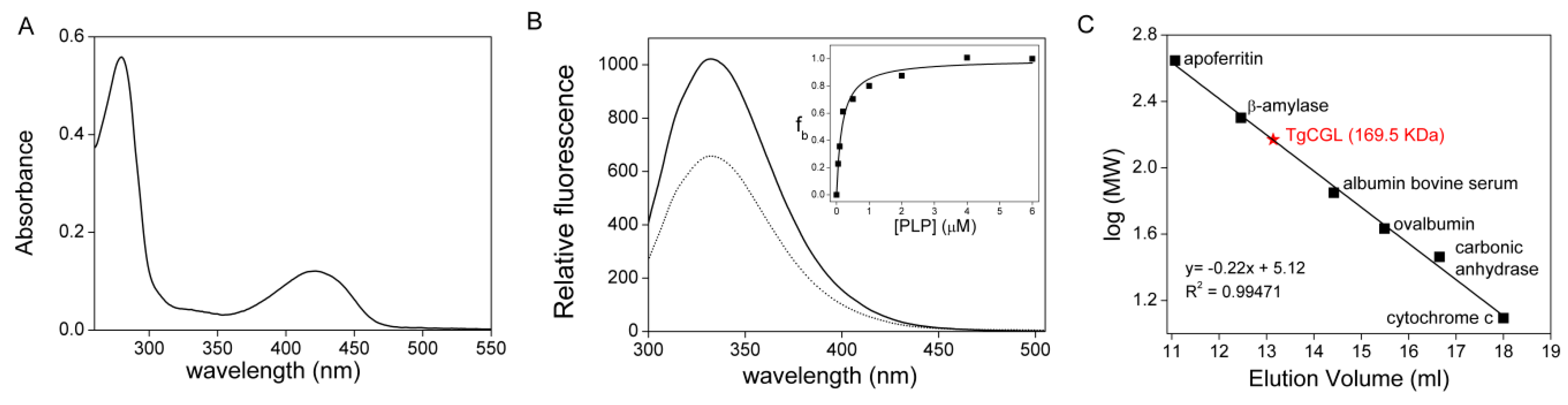
2.1. Properties of Recombinant TgCGL
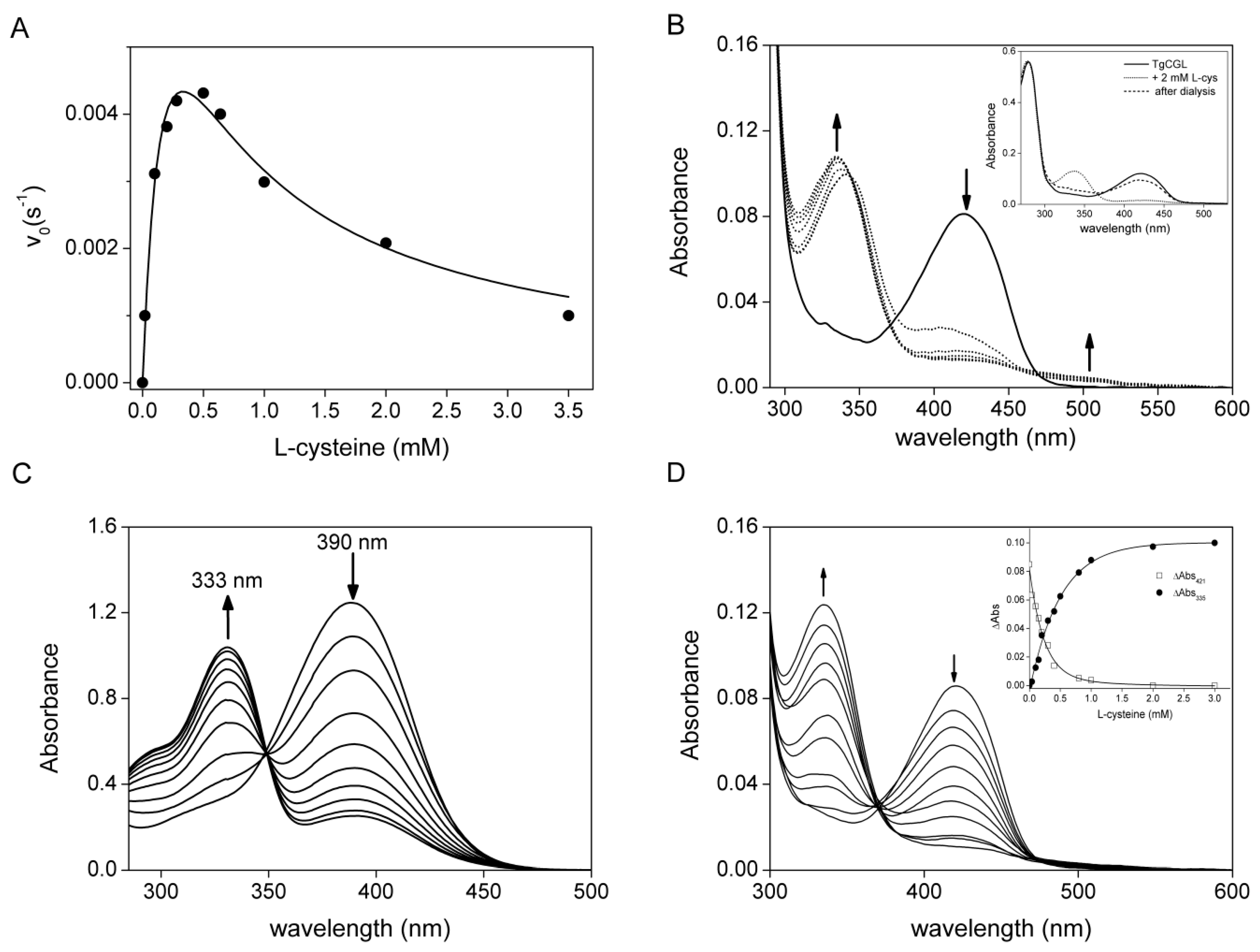
2.2. Kinetic Properties of TgCGL
2.3. Spectral Characterization of the Interaction between TgCGL and l-cys
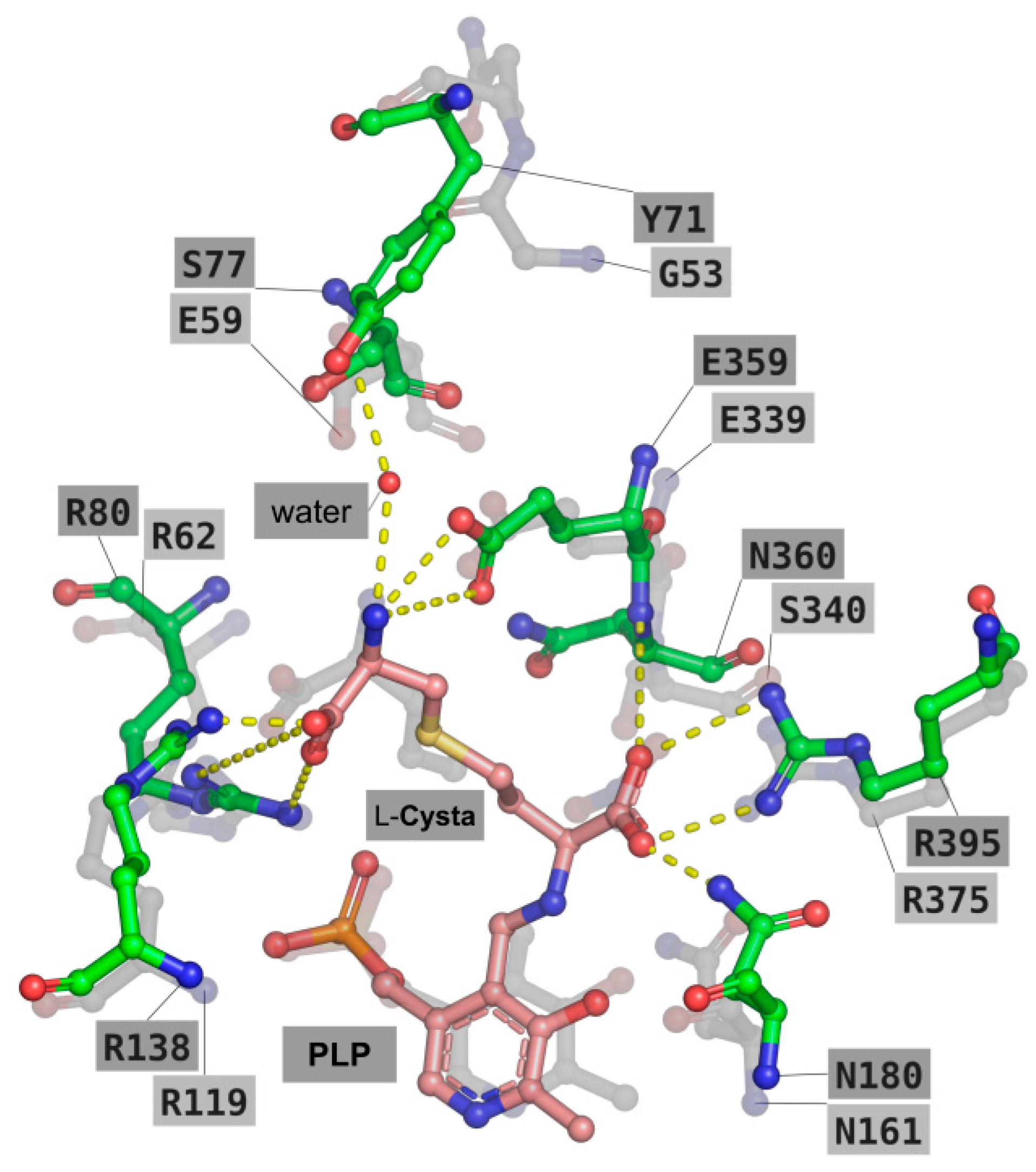
2.4. Molecular Modelling
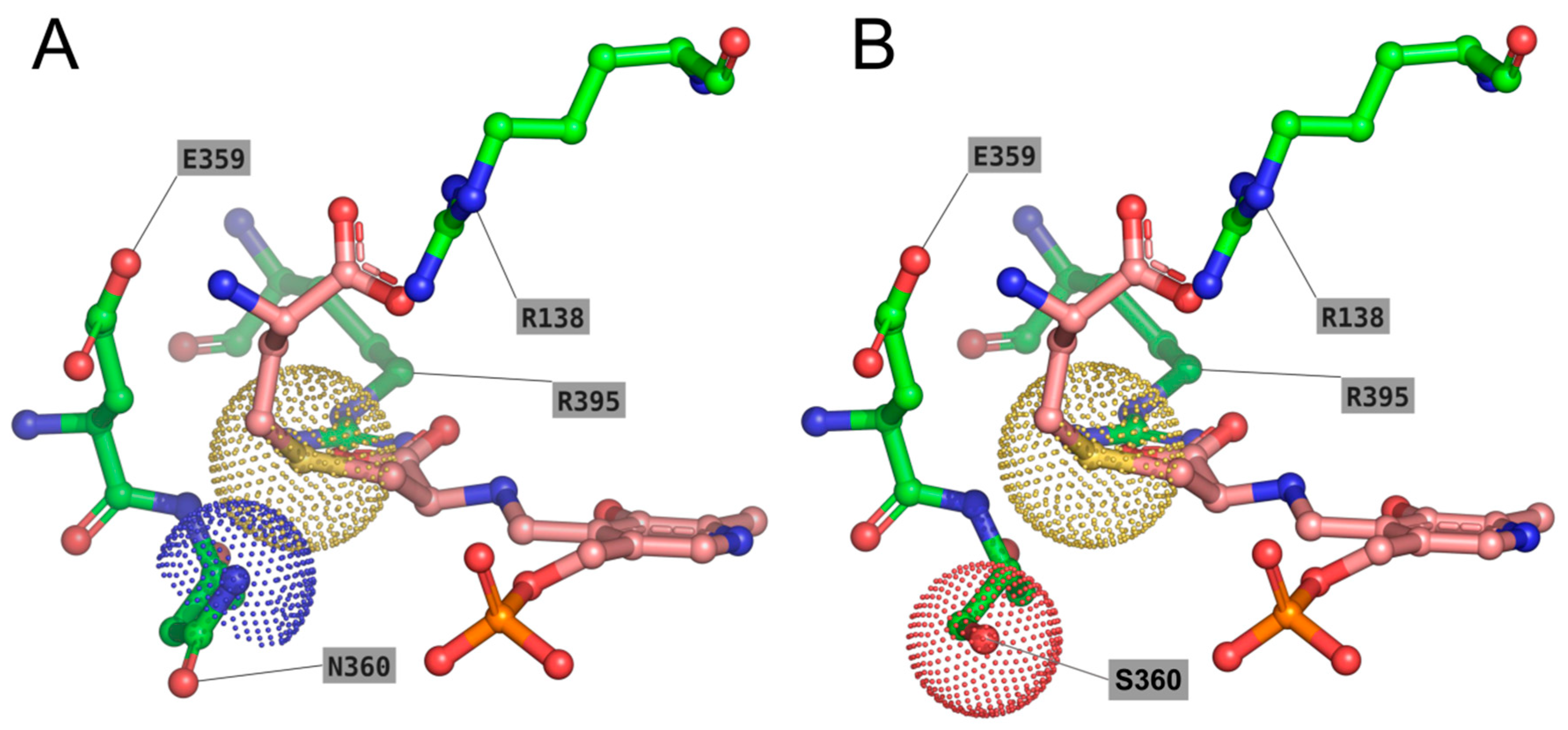
2.5. Deciphering the Importance of Asn360 and Ser77 of TgCGL on Activity and Stability
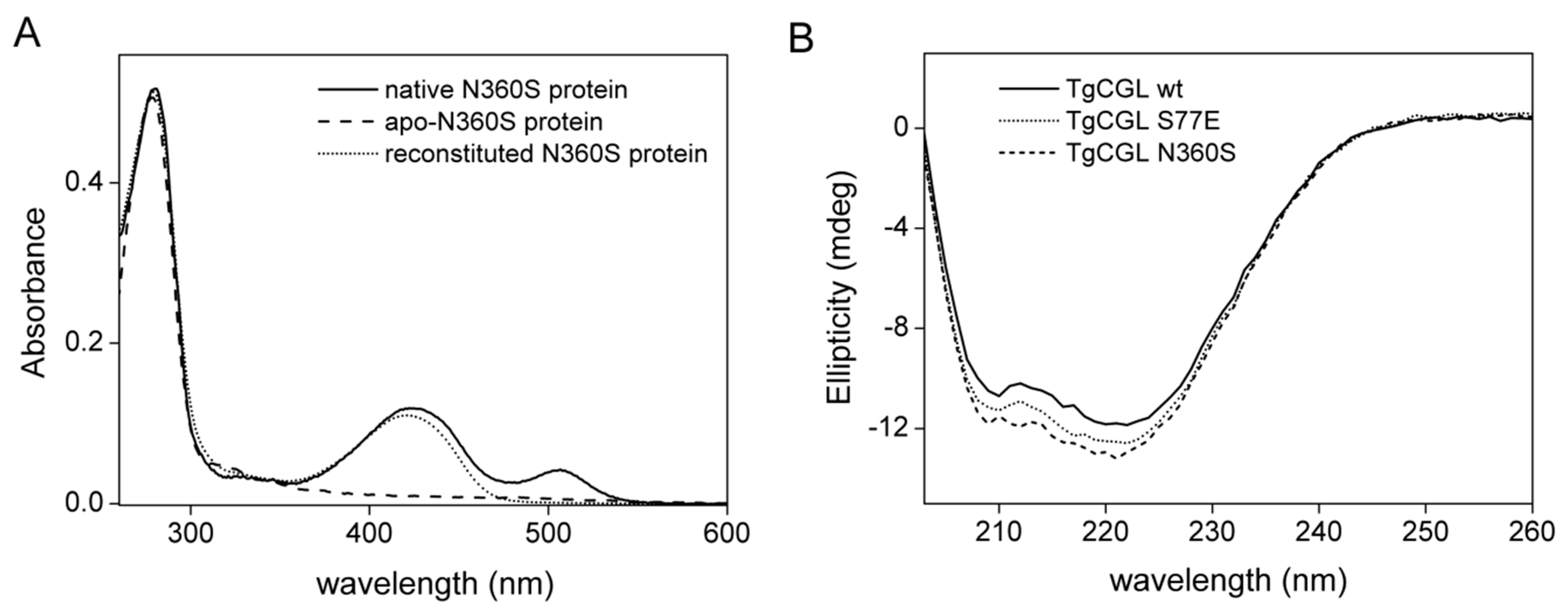
2.5.1. N360S Variant
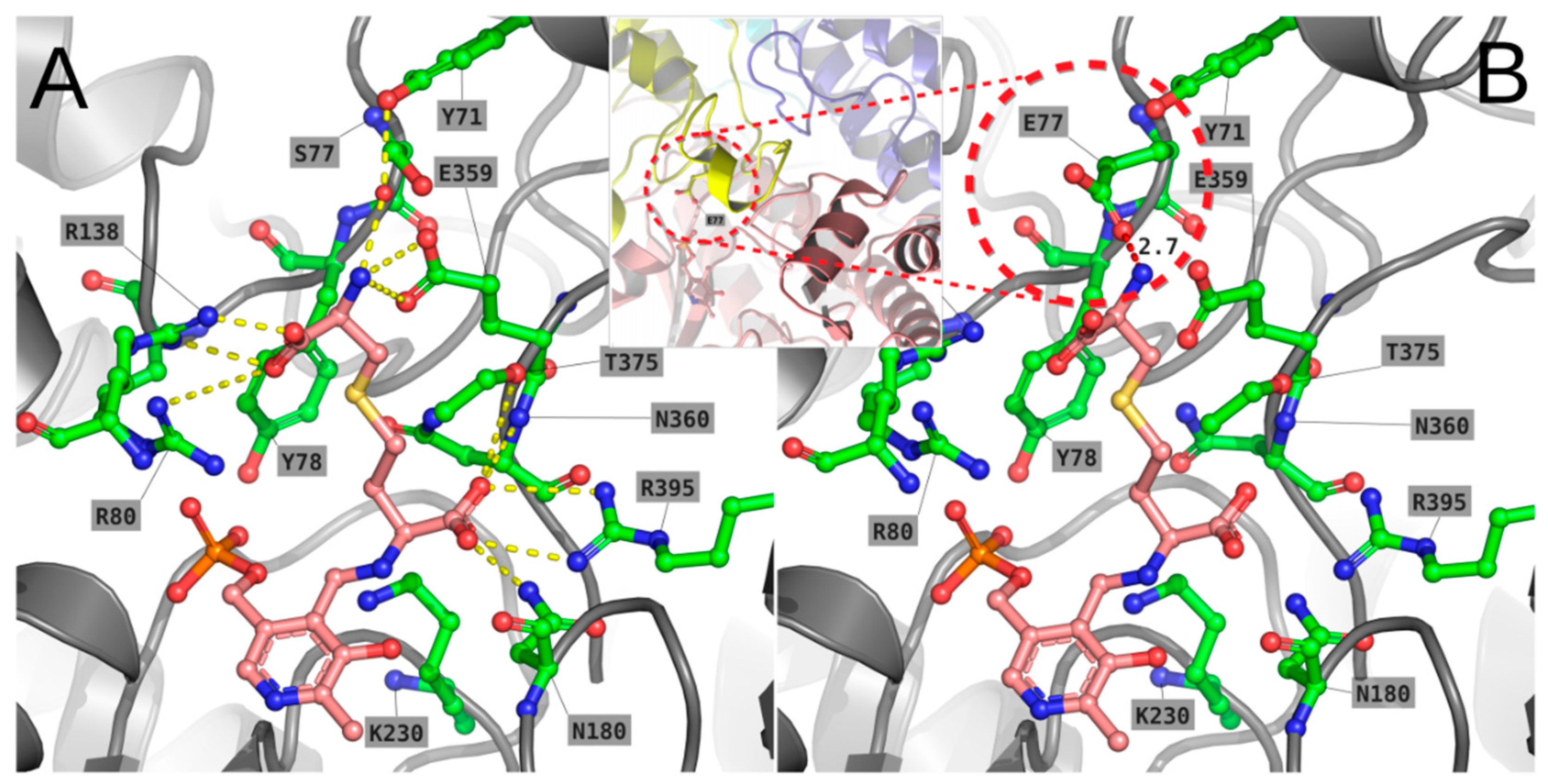
2.5.2. S77E Variant
3. Discussion
4. Materials and Methods
4.1. Protein Production
4.2. Enzyme Activity Assays
4.3. Apo-Protein Preparation
4.4. Size Exclusion Chromatography
4.5. Spectroscopic Measurements
4.6. Limited Proteolysis
4.7. Differential Scanning Calorimetry
4.8. Thin Layer Chromatography
4.9. Molecular Modelling Studies
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CGL | Cystathionine γ-lyase |
| TgCGL | Cystathionine γ-lyase from Toxoplasma gondii |
| CBS | Cystathionine β-synthase |
| CS | Cysteine synthase |
| l-cth | l-Cystathionine |
| l-cys | l-Cysteine |
| l-hcys | l-Homocsyteine |
| DTNB | 5,5′-Dithiobis-2-nitrobenzoic acid |
| LDH | NADH-dependent lactate dehydrogenase |
| TLC | Thin layer chromatography |
| DSC | Differential Scanning Calorimetry |
| CD | Circular Dichroism |
| PLP | Pyridoxal-5′-phosphate |
| PDB | Protein Data Bank |
| Tm | Melting Temperature |
| Km | Michaelis-Menten constant |
| kcat | Turnover rate |
| Ki | Inhibition Constant |
| Kd | Dissociation Constant |
References
- Aitken, S.M.; Lodha, P.H.; Morneau, D.J. The enzymes of the transsulfuration pathways: Active-site characterizations. Biochim. Biophys. Acta 2011, 1814, 1511–1517. [Google Scholar] [CrossRef] [PubMed]
- Tokoro, M.; Asai, T.; Kobayashi, S.; Takeuchi, T.; Nozaki, T. Identification and characterization of two isoenzymes of methionine γ-lyase from entamoeba histolytica: A key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protist that lacks forward and reverse trans-sulfuration pathways. J. Biol. Chem. 2003, 278, 42717–42727. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Asai, T.; Kobayashi, S.; Ikegami, F.; Noji, M.; Saito, K.; Takeuchi, T. Molecular cloning and characterization of the genes encoding two isoforms of cysteine synthase in the enteric protozoan parasite entamoeba histolytica. Mol. Biochem. Parasitol. 1998, 97, 33–44. [Google Scholar] [CrossRef]
- Nozaki, T.; Ali, V.; Tokoro, M. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv. Parasitol. 2005, 60, 1–99. [Google Scholar] [PubMed]
- Ali, V.; Nozaki, T. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin. Microbiol. Rev. 2007, 20, 164–187. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Asai, T.; Sanchez, L.B.; Kobayashi, S.; Nakazawa, M.; Takeuchi, T. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites entamoeba histolytica and entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in entamoeba. J. Biol. Chem. 1999, 274, 32445–32452. [Google Scholar] [PubMed]
- Nozaki, T.; Shigeta, Y.; Saito-Nakano, Y.; Imada, M.; Kruger, W.D. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, trypanosoma cruzi: Isolation and molecular characterization of cystathionine β-synthase and serine acetyltransferase fromtrypanosoma. J. Biol. Chem. 2001, 276, 6516–6523. [Google Scholar] [CrossRef] [PubMed]
- Marciano, D.; Santana, M.; Nowicki, C. Functional characterization of enzymes involved in cysteine biosynthesis and H2S production in trypanosoma cruzi. Mol. Biochem. Parasitol. 2012, 185, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.K.; Hale, T.I.; Christen, P. Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 1993, 214, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Possenti, A.; Fratini, F.; Fantozzi, L.; Pozio, E.; Dubey, J.P.; Ponzi, M.; Pizzi, E.; Spano, F. Global proteomic analysis of the oocyst/sporozoite of toxoplasma gondii reveals commitment to a host-independent lifestyle. BMC Genom. 2013, 14, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-H.; Wang, Z.-X.; Zhou, C.-X.; He, S.; Elsheikha, H.M.; Zhu, X.-Q. Comparative proteomic analysis of virulent and avirulent strains of toxoplasma gondii reveals strain-specific patterns. Oncotarget 2017, 8, 80481–80491. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Collins, R.; Huang, S.; Holmberg-Schiavone, L.; Anand, G.S.; Tan, C.-H.; van-den-Berg, S.; Deng, L.-W.; Moore, P.K.; Karlberg, T.; et al. Structural basis for the inhibition mechanism of human cystathionine γ-lyase, an enzyme responsible for the production of h2s. J. Biol. Chem. 2009, 284, 3076–3085. [Google Scholar] [CrossRef] [PubMed]
- Messerschmidt, A.; Worbs, M.; Steegborn, C.; Wahl, M.C.; Huber, R.; Laber, B.; Clausen, T. Determinants of enzymatic specificity in the cys-met-metabolism plp-dependent enzymes family: Crystal structure of cystathionine gamma-lyase from yeast and intrafamiliar structure comparison. Biol. Chem. 2003, 384, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Steegborn, C.; Clausen, T.; Sondermann, P.; Jacob, U.; Worbs, M.; Marinkovic, S.; Huber, R.; Wahl, M.C. Kinetics and inhibition of recombinant human cystathionine gamma-lyase. Toward the rational control of transsulfuration. J. Biol. Chem. 1999, 274, 12675–12684. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Two’s company, three’s a crowd: Can H2S be the third endogenous gaseous transmitter? FASEB J. 2002, 16, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.F.; Wedding, R.T. A reaction mechanism from steady state kinetic studies for o-acetylserine sulfhydrylase from salmonella typhimurium lt-2. J. Biol. Chem. 1976, 251, 2023–2029. [Google Scholar] [PubMed]
- Becker, M.A.; Kredich, N.M.; Tomkins, G.M. The purification and characterization of o-acetylserine sulfhydrylase-a from salmonella typhimurium. J. Biol. Chem. 1969, 244, 2418–2427. [Google Scholar] [PubMed]
- Yamagata, S.; Isaji, M.; Yamane, T.; Iwama, T. Substrate inhibition of l-cysteine alpha, beta-elimination reaction catalyzed by l-cystathionine gamma-lyase of saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2002, 66, 2706–2709. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chua, J.H.; Yew, W.S.; Sivaraman, J.; Moore, P.K.; Tan, C.H.; Deng, L.W. Site-directed mutagenesis on human cystathionine-gamma-lyase reveals insights into the modulation of H2S production. J. Mol. Biol. 2010, 396, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Kobylarz, M.J.; Grigg, J.C.; Liu, Y.; Lee, M.S.F.; Heinrichs, D.E.; Murphy, M.E.P. Deciphering the substrate specificity of sbna, the enzyme catalyzing the first step in staphyloferrin b biosynthesis. Biochemistry 2016, 55, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Lowther, J.; Beattie, A.E.; Langridge-Smith, P.R.R.; Clarke, D.J.; Campopiano, D.J. l-penicillamine is a mechanism-based inhibitor of serine palmitoyltransferase by forming a pyridoxal-5 [prime or minute]-phosphate-thiazolidine adduct. MedChemComm 2012, 3, 1003–1008. [Google Scholar] [CrossRef]
- Liu, P.; Torrens-Spence, M.P.; Ding, H.; Christensen, B.M.; Li, J. Mechanism of cysteine-dependent inactivation of aspartate/glutamate/cysteine sulfinic acid alpha-decarboxylases. Amino Acids 2013, 44, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y. Formation of schiff bases of pyridoxal phosphate. Reaction with metal ions1. J. Am. Chem. Soc. 1957, 79, 2011–2015. [Google Scholar] [CrossRef]
- Saiz, C.; Wipf, P.; Manta, E.; Mahler, G. Reversible thiazolidine exchange: A new reaction suitable for dynamic combinatorial chemistry. Org. Lett. 2009, 11, 3170–3173. [Google Scholar] [CrossRef] [PubMed]
- Terzuoli, L.; Leoncini, R.; Pagani, R.; Guerranti, R.; Vannoni, D.; Ponticelli, F.; Marinello, E. Some chemical properties and biological role of thiazolidine compounds. Life Sci. 1998, 63, 1251–1267. [Google Scholar] [CrossRef]
- Zhu, W.; Lin, A.; Banerjee, R. Kinetic properties of polymorphic variants and pathogenic mutants in human cystathionine gamma-lyase. Biochemistry 2008, 47, 6226–6232. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Shirley, B.A.; Thompson, J.A. Measuring the Conformational Stability of a Protein. In Protein Structure: A Practical Approach, 2nd ed.; Creighton, T.E., Ed.; IRL Press: Oxford, UK, 2002. [Google Scholar]
- Hopwood, E.M.; Ahmed, D.; Aitken, S.M. A role for glutamate-333 of saccharomyces cerevisiae cystathionine gamma-lyase as a determinant of specificity. Biochim. Biophys. Acta 2014, 1844, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Cherest, H.; Thomas, D.; Surdin-Kerjan, Y. Cysteine biosynthesis in saccharomyces cerevisiae occurs through the transsulfuration pathway which has been built up by enzyme recruitment. J. Bacteriol. 1993, 175, 5366–5374. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Stone, E.; Zhang, Y.J. Structural snapshots of an engineered cystathionine-gamma-lyase reveal the critical role of electrostatic interactions in the active site. Biochemistry 2017, 56, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.; Huber, R.; Messerschmidt, A.; Pohlenz, H.D.; Laber, B. Slow-binding inhibition of escherichia coli cystathionine beta-lyase by l-aminoethoxyvinylglycine: A kinetic and X-ray study. Biochemistry 1997, 36, 12633–12643. [Google Scholar] [CrossRef] [PubMed]
- Gut, H.; Dominici, P.; Pilati, S.; Astegno, A.; Petoukhov, M.V.; Svergun, D.I.; Grutter, M.G.; Capitani, G. A common structural basis for ph- and calmodulin-mediated regulation in plant glutamate decarboxylase. J. Mol. Biol. 2009, 392, 334–351. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; Maresi, E.; Bertoldi, M.; La Verde, V.; Paiardini, A.; Dominici, P. Unique substrate specificity of ornithine aminotransferase from Toxoplasma gondii. Biochem. J. 2017, 474, 939–955. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; Giorgetti, A.; Allegrini, A.; Cellini, B.; Dominici, P. Characterization of c-s lyase from c. Diphtheriae: A possible target for new antimicrobial drugs. Biomed Res. Int. 2013, 2013, 701536. [Google Scholar] [CrossRef] [PubMed]
- Allegrini, A.; Astegno, A.; La Verde, V.; Dominici, P. Characterization of c-s lyase from lactobacillus delbrueckii subsp. Bulgaricus atcc baa-365 and its potential role in food flavour applications. J. Biochem. 2017, 161, 349–360. [Google Scholar] [PubMed]
- Copeland, R.A. Kinetics of single-substrate enzyme reactions. In Enzymes; John Wiley & Sons, Inc.: New York, NY, USA, 2002; pp. 109–145. [Google Scholar]
- Bertoldi, M.; Cellini, B.; Clausen, T.; Voltattorni, C.B. Spectroscopic and kinetic analyses reveal the pyridoxal 5′-phosphate binding mode and the catalytic features of treponema denticola cystalysin. Biochemistry 2002, 41, 9153–9164. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; Capitani, G.; Dominici, P. Functional roles of the hexamer organization of plant glutamate decarboxylase. Biochim. Biophys. Acta 2015, 1854, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; Allegrini, A.; Piccoli, S.; Giorgetti, A.; Dominici, P. Role of active-site residues Tyr55 and Tyr114 in catalysis and substrate specificity of Corynebacterium diphtheriae C-S lyase. Proteins Struct. Funct. Bioinform. 2015, 83, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; La Verde, V.; Marino, V.; Dell’Orco, D.; Dominici, P. Biochemical and biophysical characterization of a plant calmodulin: Role of the N- and C-lobes in calcium binding, conformational change, and target interaction. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2016, 1864, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Vallone, R.; La Verde, V.; D’Onofrio, M.; Giorgetti, A.; Dominici, P.; Astegno, A. Metal binding affinity and structural properties of calmodulin-like protein 14 from arabidopsis thaliana. Protein Sci. 2016, 25, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Oppici, E.; Fodor, K.; Paiardini, A.; Williams, C.; Voltattorni, C.B.; Wilmanns, M.; Cellini, B. Crystal structure of the S187F variant of human liver alanine: Glyoxylate [corrected] aminotransferase associated with primary hyperoxaluria type i and its functional implications. Proteins 2013, 81, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Janson, G.; Zhang, C.; Prado, M.G.; Paiardini, A. Pymod 2.0: Improvements in protein sequence-structure analysis and homology modeling within pymol. Bioinformatics 2017, 33, 444–446. [Google Scholar] [CrossRef] [PubMed]
- Schuttelkopf, A.W.; van Aalten, D.M. Prodrg: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. Moldock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]

| Assay | kcat (s−1) | Km (mM) | Kil-cys (mM) | kcat/Km (M−1·s−1) | |
|---|---|---|---|---|---|
| Hydrolysis of l-cystathionine | |||||
| wild type | DTNB | 2.0 ± 0.1 | 0.9 ± 0.1 | - | (2.2 ± 0.4) × 103 |
| N360S | DTNB | 0.24 ± 0.02 | 6.1 ± 1.2 | - | (3.9 ± 1.1) × 10 |
| LDH | 0.08 ± 0.01 | 1.1 ± 0.1 | - | (7.3 ± 1.6) × 10 | |
| S77E | LDH/DTNB | n.d.b | n.d. b | - | - |
| S77A | DTNB | 0.7 ± 0.1 | 2.4 ± 0.3 | - | (2.9 ± 0.8) × 102 |
| Hydrolysis of djenkolic acid | |||||
| wild type | LDH | 0.24 ± 0.01 | 0.51 ± 0.01 | - | (4.7 ± 0.3) × 102 |
| N360S | LDH | 0.19 ± 0.01 | 0.86 ± 0.03 | - | (2.2 ± 0.2) × 102 |
| S77E | LDH | n.s. c | n.s. c | - | 2.2 ± 0.6 |
| S77A | LDH | 0.22 ± 0.02 | 1.5 ± 0.2 | - | (1.5 ± 0.3) × 102 |
| Hydrolysis of l-cysteine | |||||
| wild type | LDH | 0.009 ± 0.001 | 0.21 ± 0.1 | 0.53 ± 0.02 | (4.3 ± 2.1) × 10 |
| N360S | LDH | 0.007 ± 0.001 | 0.4 ± 0.1 | 1.11 ± 0.04 | (1.8 ± 0.7) ×10 |
| S77E | LDH | n.d. b | n.d. b | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maresi, E.; Janson, G.; Fruncillo, S.; Paiardini, A.; Vallone, R.; Dominici, P.; Astegno, A. Functional Characterization and Structure-Guided Mutational Analysis of the Transsulfuration Enzyme Cystathionine γ-Lyase from Toxoplasma gondii. Int. J. Mol. Sci. 2018, 19, 2111. https://doi.org/10.3390/ijms19072111
Maresi E, Janson G, Fruncillo S, Paiardini A, Vallone R, Dominici P, Astegno A. Functional Characterization and Structure-Guided Mutational Analysis of the Transsulfuration Enzyme Cystathionine γ-Lyase from Toxoplasma gondii. International Journal of Molecular Sciences. 2018; 19(7):2111. https://doi.org/10.3390/ijms19072111
Chicago/Turabian StyleMaresi, Elena, Giacomo Janson, Silvia Fruncillo, Alessandro Paiardini, Rosario Vallone, Paola Dominici, and Alessandra Astegno. 2018. "Functional Characterization and Structure-Guided Mutational Analysis of the Transsulfuration Enzyme Cystathionine γ-Lyase from Toxoplasma gondii" International Journal of Molecular Sciences 19, no. 7: 2111. https://doi.org/10.3390/ijms19072111
APA StyleMaresi, E., Janson, G., Fruncillo, S., Paiardini, A., Vallone, R., Dominici, P., & Astegno, A. (2018). Functional Characterization and Structure-Guided Mutational Analysis of the Transsulfuration Enzyme Cystathionine γ-Lyase from Toxoplasma gondii. International Journal of Molecular Sciences, 19(7), 2111. https://doi.org/10.3390/ijms19072111






