Novel C15 Triene Triazole, D-A Derivatives Anti-HepG2, and as HDAC2 Inhibitors: A Synergy Study
Abstract
1. Introduction
2. Results
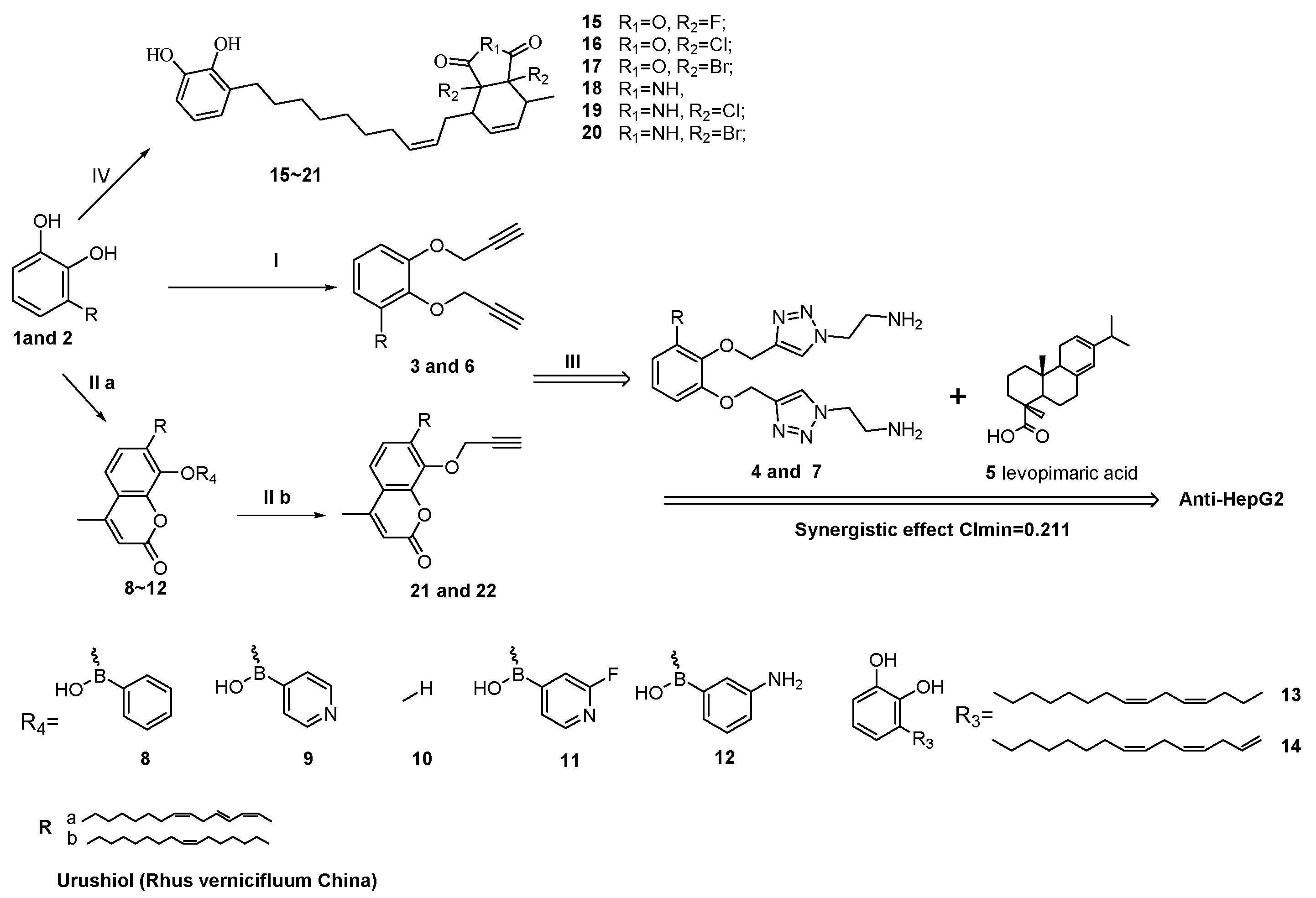
2.1. Synthesis of 3-((8Z,11E,13Z)-Pentadeca-8,11,13-Trien-1-yl) Benzene-1,2-Diol Derivatives
2.2. Anti-Tumor Bioactivity
2.3. QSAR (Quantitative Structure Activity Relationship)
2.4. Molecular Docking
2.4.1. Conjunction Conformation Score
2.4.2. Binding Pattern Analysis
2.5. Western Blot Results
2.6. Flow Cytometry Analysis
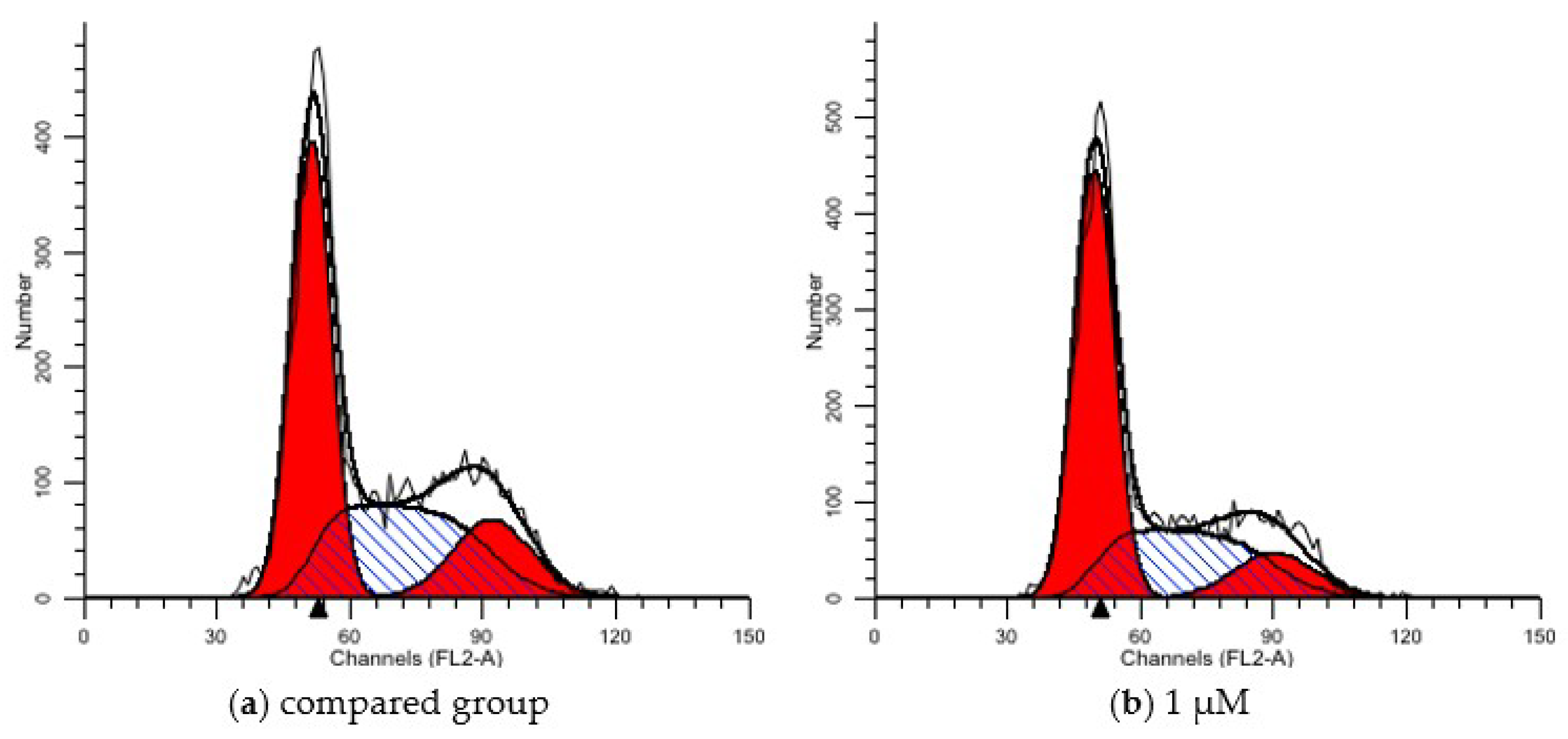
2.6.1. PI Single Staining Assay for Cell Cycle
2.6.2. Annexin-V FITC/PI Double Staining for Apoptosis
2.6.3. JC-1 Staining Assay Mitochondrial Membrane Potential
2.6.4. Calcium Ion Content Detection
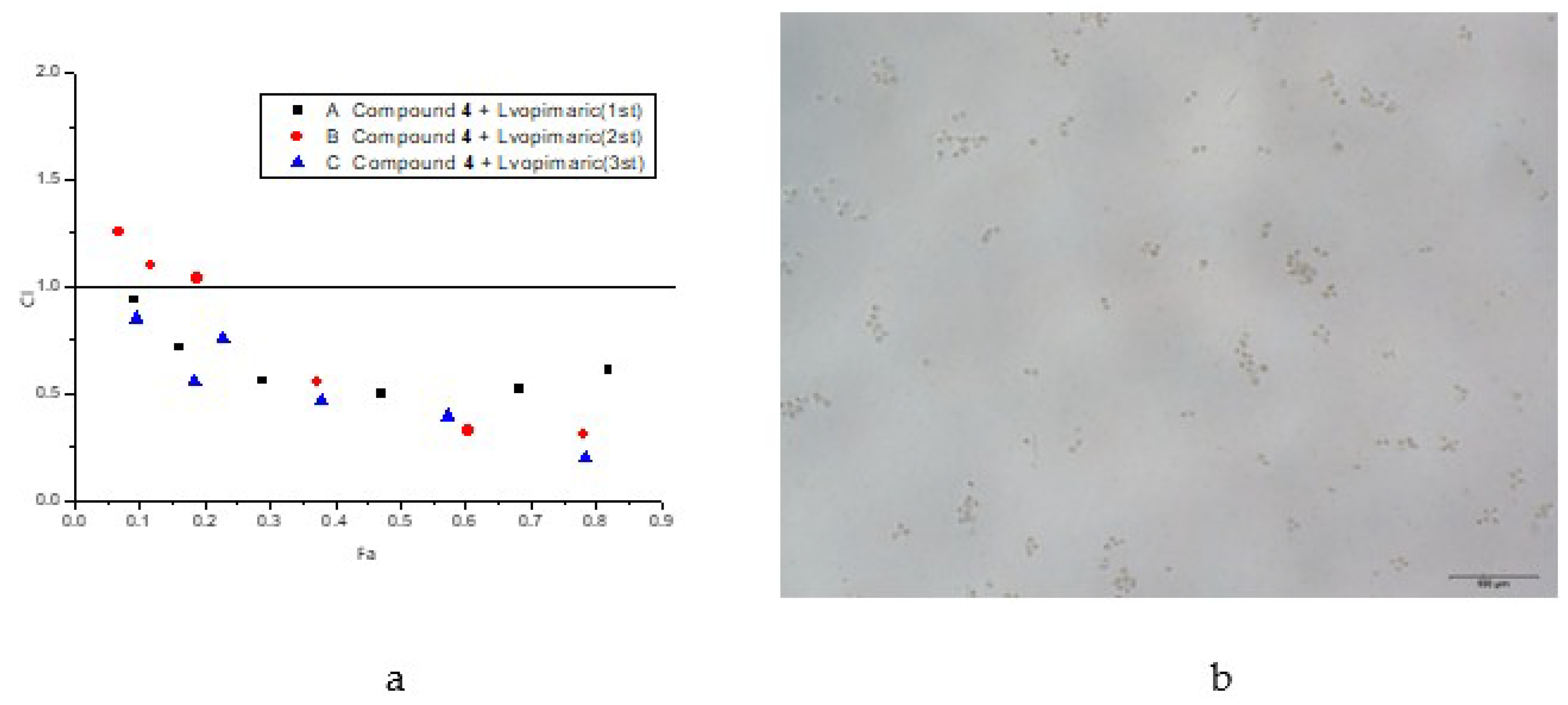
2.7. Synergy
3. Experimental
3.1. Chemistry
3.2. Materials
3.3. Extraction and Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jiang, X. Effects of Docetaxel on HepG2 Proteome in Human Hepatoma Cell; Central South University of China: Changsha, China, 2006. [Google Scholar]
- Fraser, M.; Leung, B.; Jahani-Asl, A.; Yan, X.; Thompson, W.E.; Tsang, B.K. Chemoresi stance in human ovarian cancer: The role of apoptotic regulators. Reprod. Biol. Endocrinol. 2003, 1, 66. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, H.; Tada-Oikawa, S.; Hiraku, Y.; Kojima, M.; Kawanishi, S. Mechanism of apoptosis induced by doxorubicin through the generation of hydrogen peroxide. Life Sci. 2005, 76, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Fujimoto, A.; Takahara, A. Characterization of catechol-containing natural thermosetting polymer “urushiol” thin film. J. Polym. Sci. Part A Polym. Chem. 2013, 51, 3688–3692. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Liu, W.; Xie, N.; Feng, F.; Qu, W. New urushiols with platelet aggregation inhibitory activities from resin of Toxicodendron vernicifluum. Fitoterapia 2016, 112, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Suk, K.T.; Choi, S.H.; Lee, J.W.; Sung, H.T.; Kim, C.H.; Kim, E.J.; Kim, M.J.; Han, S.H.; Kim, M.Y.; et al. Anti-oxidant and natural killer cell activity of Korean red ginseng (Panax ginseng) and urushiol (Rhus vernicifera Stokes) on non-alcoholic fatty liver disease of rat. Food Chem. Toxicol. 2013, 55, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Ryckewaert, L.; Sacconnay, L.; Carrupt, P.A.; Nurisso, A.; Simoes-Pires, C. Non-specific SIRT inhibition as a mechanism for the cytotoxicity of ginkgolic acids and urushiols. Toxicol. Lett. 2014, 229, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.S.; Shin, D.H. Antibacterial capacity of cavity disinfectants against Streptococcus mutans and their effects on shear bond strength of a self-etch adhesive. Dent. Mater. J. 2016, 35, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Park, K.Y.; Kim, S.J.; Oh, S.; Moon, J.H. Antimicrobial activity of the synthesized non-allergenic urushiol derivatives. Biosci. Biotechnol. Biochem. 2015, 79, 1915–1918. [Google Scholar] [CrossRef] [PubMed]
- Halloran, L. Developing Dermatology Detective Powers: Allergic Contact Dermatitis. J. Nurse Pract. 2014, 10, 284–285. [Google Scholar] [CrossRef]
- Hachisuka, J.; Ross, S.E. Understanding the switch from pain-to-itch in dermatitis. Neurosci. Lett. 2014, 579, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Miyakoshi, T. Lacquer Chemistry and Applications; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Coifman, R.; Yang, C. Poster 1019: Novel allergy vaccine delivery system for poison ivy urushiol (PI) and peanut (PN). World Allergy Organ. J. 2014, 7 (Suppl. 1), 10. [Google Scholar] [CrossRef]
- Hong, M.; Kim, S.W.; Han, S.H.; Kim, D.J.; Suk, K.T.; Kim, Y.S.; Kim, M.J.; Kim, M.Y.; Baik, S.K.; Ham, Y.L. Probiotics (Lactobacillus rhamnosus R0011 and acidophilus R0052) reduce the expression of toll-like receptor 4 in mice with alcoholic liver disease. PLoS ONE 2015, 10, e0117451. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Wang, F.F. Mechanisms underlying the pro-survival pathway of p53 in suppressing mitotic death induced by adriamycin. Cell Signal 2008, 20, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Jantova, S.; Letasiova, S.; Ovadekova, R.; Muckova, M. Cytotoxic/antiproliferative effects of new [1,2,4]triazolo[4,3-c] quinazolines in tum or cell lines H eLa and B16. Neoplasma 2006, 53, 291–300. [Google Scholar] [PubMed]
- Deprez-Poulain, R.; Hennuyer, N.; Bosc, D.; Liang, W.G.; Enee, E.; Marechal, X.; Charton, J.; Totobenazara, J.; Berte, G.; Jahklal, J.; et al. Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice. Nat. Commun. 2015, 6, 8250. [Google Scholar] [CrossRef] [PubMed]
- Marzano, C.; Pellei, M.; Colavito, D.; Alidori, S.; Lobbia, G.G.; Gandin, V.; Tisato, F.; Santini, C. Synthesis, characterization, and invitro antitumor properties of tris(hydroxymethyl) phosphine copper(I) complexes containing the new bis(1,2,4-tri-azol-1-yl) acetate ligand. J. Med. Chem. 2006, 49, 7317–7324. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.S.; Hong, S.H.; Suk, K.T.; Kim, J.B.; Han, S.H.; Sung, H.; Kim, E.J.; Kim, M.J.; Kim, M.Y.; Baik, S.K.; et al. Effects of Korean Red Ginseng (Panax ginseng), urushiol (Rhus vernicifera Stokes), and probiotics (Lactobacillus rhamnosus R0011 and Lactobacillus acidophilus R0052) on the gut-liver axis of alcoholic liver disease. J. Ginseng Res. 2014, 38, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Suk, K.T.; Baik, S.K.; Kim, H.S.; Park, S.M.; Paeng, K.J.; Uh, Y.; Jang, I.H.; Cho, M.Y.; Choi, E.H.; Kim, M.J.; et al. Antibacterial Effects of the Urushiol Component in the Sap of the Lacquer Tree (Rhus verniciflua Stokes) on Helicobacter pylori. Helicobacter 2011, 16, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Tretyakova, E.V.; Smirnova, I.E.; Kazakova, O.B.; Tolstikov, G.A.; Yavorskaya, N.P.; Golubeva, I.S.; Pugacheva, R.B.; Apryshko, G.N.; Poroikov, V.V. Synthesis and anticancer activity of quinopimaric and maleopimaric acids’ derivatives. Bioorg. Med. Chem. 2014, 22, 6481–6489. [Google Scholar] [CrossRef] [PubMed]
- Tretyakova, E.V.; Smirnova, I.E.; Salimova, E.V.; Odinokov, V.N. Synthesis and antiviral activity of maleopimaric and quinopimaric acids’ derivatives. Bioorg. Med. Chem. 2015, 23, 6543–6550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, C.; Ye, J.; Chen, H.; Tao, R. Design, virtual screening, molecular docking and molecular dynamics studies of novel urushiol derivatives as potential HDAC2 selective inhibitors. Gene 2017, 637, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, C.; Deng, T.; Tao, R.; Li, W. Novel urushiol derivatives as, H.D.AC8 inhibitors: Rational design, virtual screening, molecular docking and molecular dynamics studies. J. Biomol. Struct. Dyn. 2018, 36, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Bi, Y.; He, Y.; Huang, W.; Liu, L.; Li, Y.; Zhang, S.; Liu, X.; Yu, N. Design, synthesis and biological evaluation of di-substituted cinnamic hydroxamic acids bearing urea/thiourea unit as potent histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 6432–6435. [Google Scholar] [CrossRef] [PubMed]
- Hildmann, C.; Riester, D.; Schwienhorst, A. Histone deacetylases—An important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 2007, 75, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Drummond, D.C.; Noble, C.O.; Kirpotin, D.B.; Guo, Z.; Scott, G.K.; Benz, C.C. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 495–528. [Google Scholar] [CrossRef] [PubMed]
- Ewa Augustin a, A.M.-R. Induction of G2/M phase arrest and apoptosis of human leukemia cells by potent antitumor triazoloacridinone C-1305. Biochem. Pharmacol. 2006, 72, 1668–1679. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, Z.S.; Wang, H.L. Carboxyamido-triazole inhibits proliferation of human breast cancer cells via G2/M cell cycle arrest and apoptosis. Eur. J. Pharmacol. 2006, 538, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Rezayati, S.; Sheikholeslami-Farahani, F.; Rostami-Charati, F.; Abad, S.A. One-pot synthesis of coumarine derivatives using butylenebispyridinium hydrogen sulfate as novel ionic liquid catalyst. Res. Chem. Intermed. 2015, 42, 4097–4107. [Google Scholar] [CrossRef]
- Potdar, M.K.; Mohile, S.S.; Salunkhe, M.M. Coumarin syntheses via Pechmann condensation in Lewis acidic chloroaluminate ionic liquid. Tetrahedron Lett. 2001, 42, 9285–9287. [Google Scholar] [CrossRef]
- Khandekar, A.C.; Khadilkar, B.M. Pechmann Reaction in Chloroaluminate Ionic Liquid. Synlett 2002, 1, 152–154. [Google Scholar] [CrossRef]
- Futagawa, M.; Wedge, D.E.; Dayan, F.E. Physiological factors influencing the antifungal activity of zopfiellin. FEBS Lett. 2002, 73, 87–93. [Google Scholar] [CrossRef]
- Hosoe, T.; Fukushima, K.; Itabashi, T. The abeolute structures of dihydroepiheveadride, as characteristic antifungal agent filamentous fimi, and its related compounds from unidentified fungus, I.F.M52672. Eterocycles 2004, 63, 2581–2589. [Google Scholar]
- Dörwald, F.Z. Side Reactions in Organic Syntheis, I.I.: Aromatic Substitutions; John Wiley & Sons: Visp, Switzerland, 2014. [Google Scholar]
- Qi, Z.; Wang, C.; Jiang, J. Synthesis and Evaluation of C15 Triene Urushiol Derivatives as Potential Anticancer Agents and, H.D.AC2 Inhibitor. Molecules 2018, 23, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Study of the influence of taxol on hepatocytes of rats in vitro. J. Jinan Univ. 2015, 36, 146–149. [Google Scholar]
- Schiff, P.B.; Fant, J.; Horwitz, S.B. Promotion of microtubule assembly in vitro by taxol. Nature 1979, 277, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Parness, J.; Horwitz, S.B. Taxol binds to polymerized tubulinin vitro. J. Cell Biol. 1981, 91, 479–481. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, J.J.; Parness, J.; Horwitz, S.B. Taxol binds to cellular microtubules. J. Cell Biol. 1982, 94, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Schiff, P.; Horwitz, S.B. Taxol stabilizes microtubules in mouse fibmblaSt cells. Proc. Natl. Acad. Sci. USA 1980, 77, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining techniques to model, RNA—Small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Balius, T.E.; Rizzo, R.C. Docking Validation Resources: Protein Family and Ligand Flexibility Experiments. J. Chem. Inf. Model. 2010, 50, 1986–2000. [Google Scholar] [CrossRef] [PubMed]
- Tret’yakova, E.V.; Salimova, E.V.; Parfenova, L.V.; Odinokov, V.N. Synthesis and modifications of alkyne derivatives of dihydroquinopimaric, maleopimaric, and fumaropimaric acids. Russ. J. Organ. Chem. 2016, 52, 1496–1502. [Google Scholar] [CrossRef]
- Shen, Q.-K.; Liu, C.-F.; Zhang, H.-J.; Tian, Y.-S.; Quan, Z.-S. Design and synthesis of new triazoles linked to xanthotoxin for potent and highly selective anti-gastric cancer agents. Bioorg. Med. Chem. Lett. 2017, 27, 4871–4875. [Google Scholar] [CrossRef] [PubMed]
- Zareyee, D.; Serehneh, M. Recyclable CMK-5 supported sulfonic acid as an environmentally benign catalyst for solvent-free one-pot construction of coumarin through Pechmann condensation. J. Mol. Catal. A Chem. 2014, 391, 88–91. [Google Scholar] [CrossRef]
- Chen, D.Z.; Jing, C.X.; Cai, J.Y.; Wu, J.B.; Wang, S.; Yin, J.L.; Li, X.N.; Li, L.; Hao, X.J. Design, Synthesis, and Structural Optimization of Lycorine-Derived Phenanthridine Derivatives as Wnt/beta-Catenin Signaling Pathway Agonists. J. Nat. Prod. 2016, 79, 180–188. [Google Scholar] [CrossRef] [PubMed]
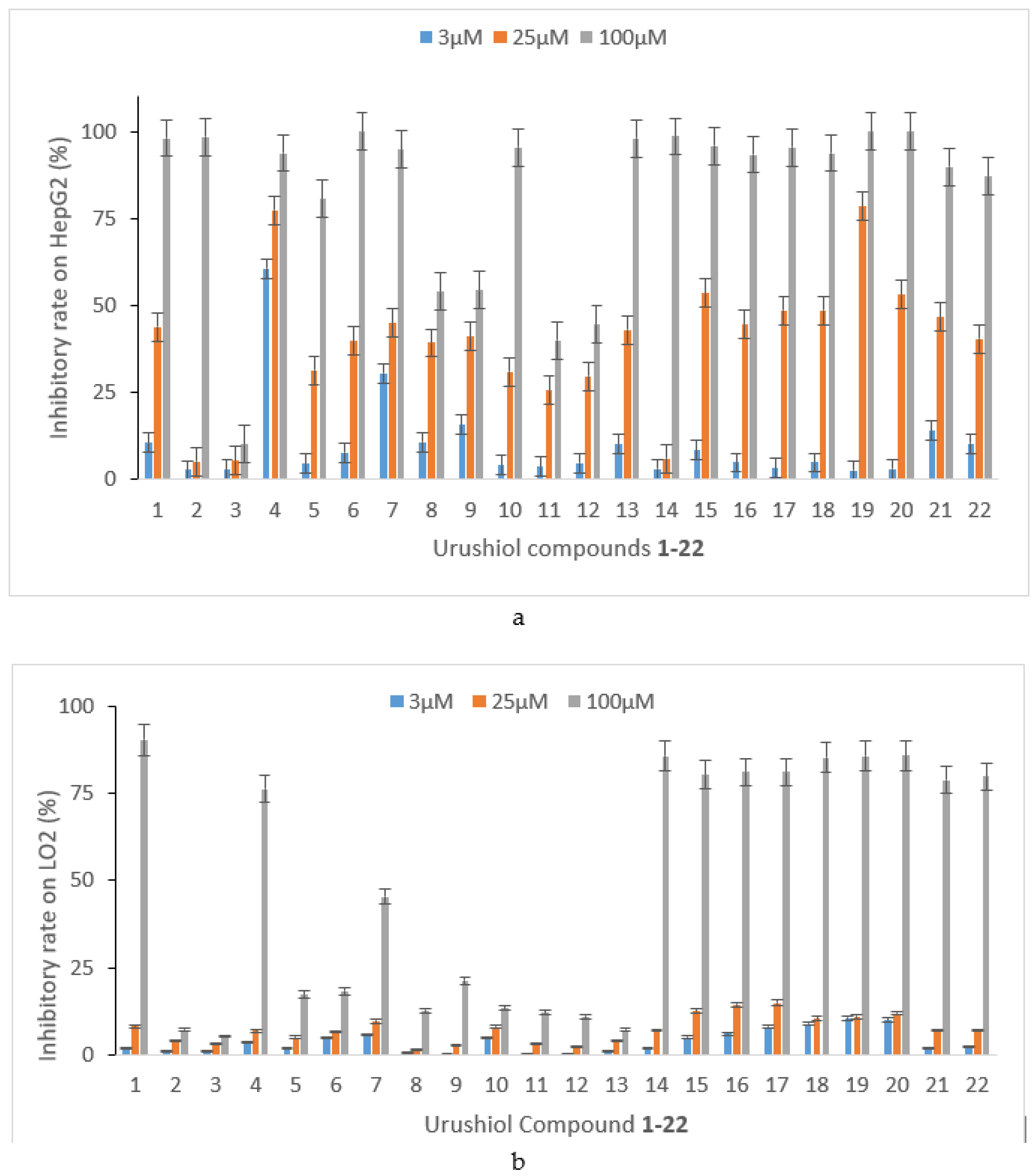
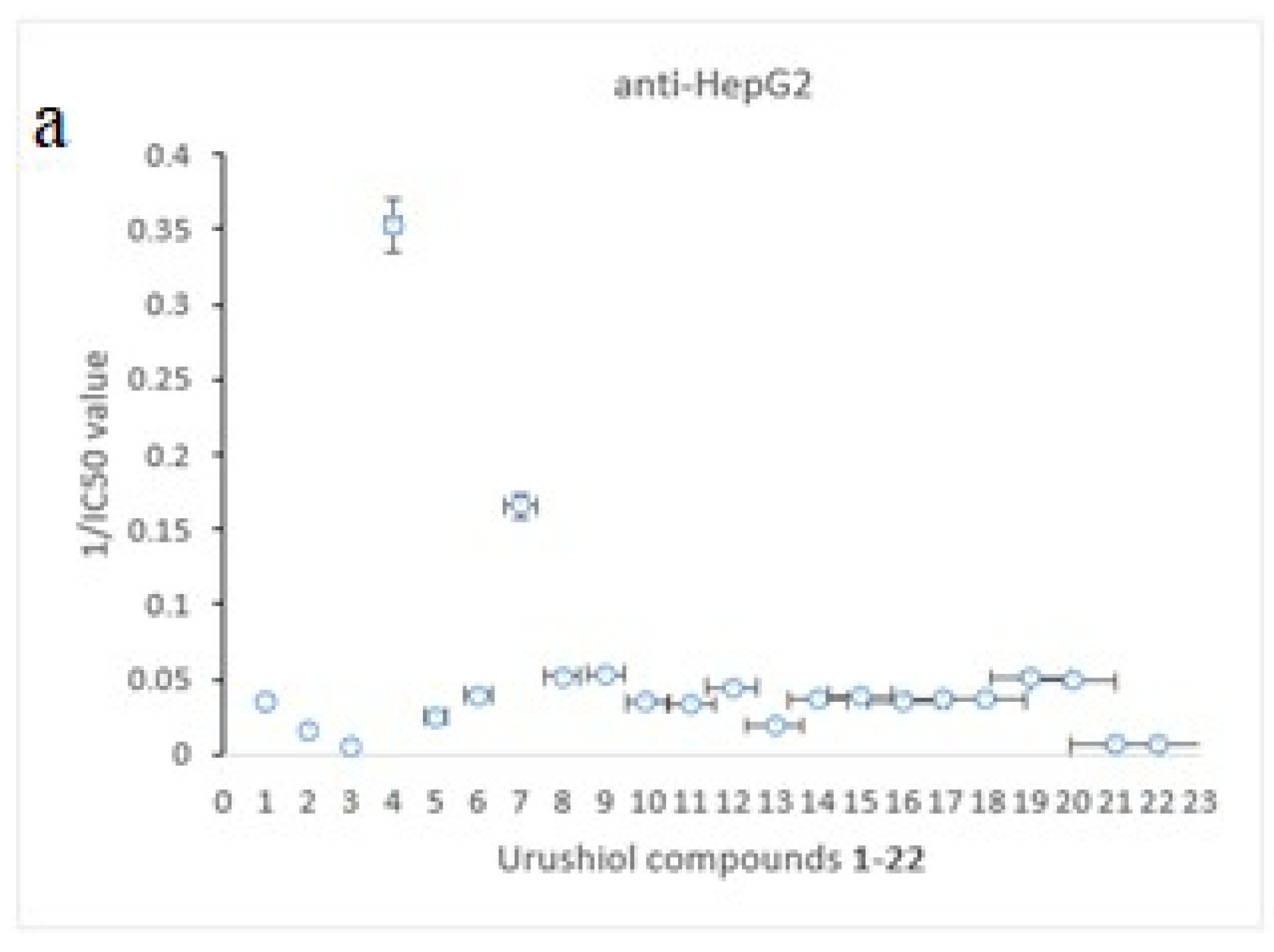

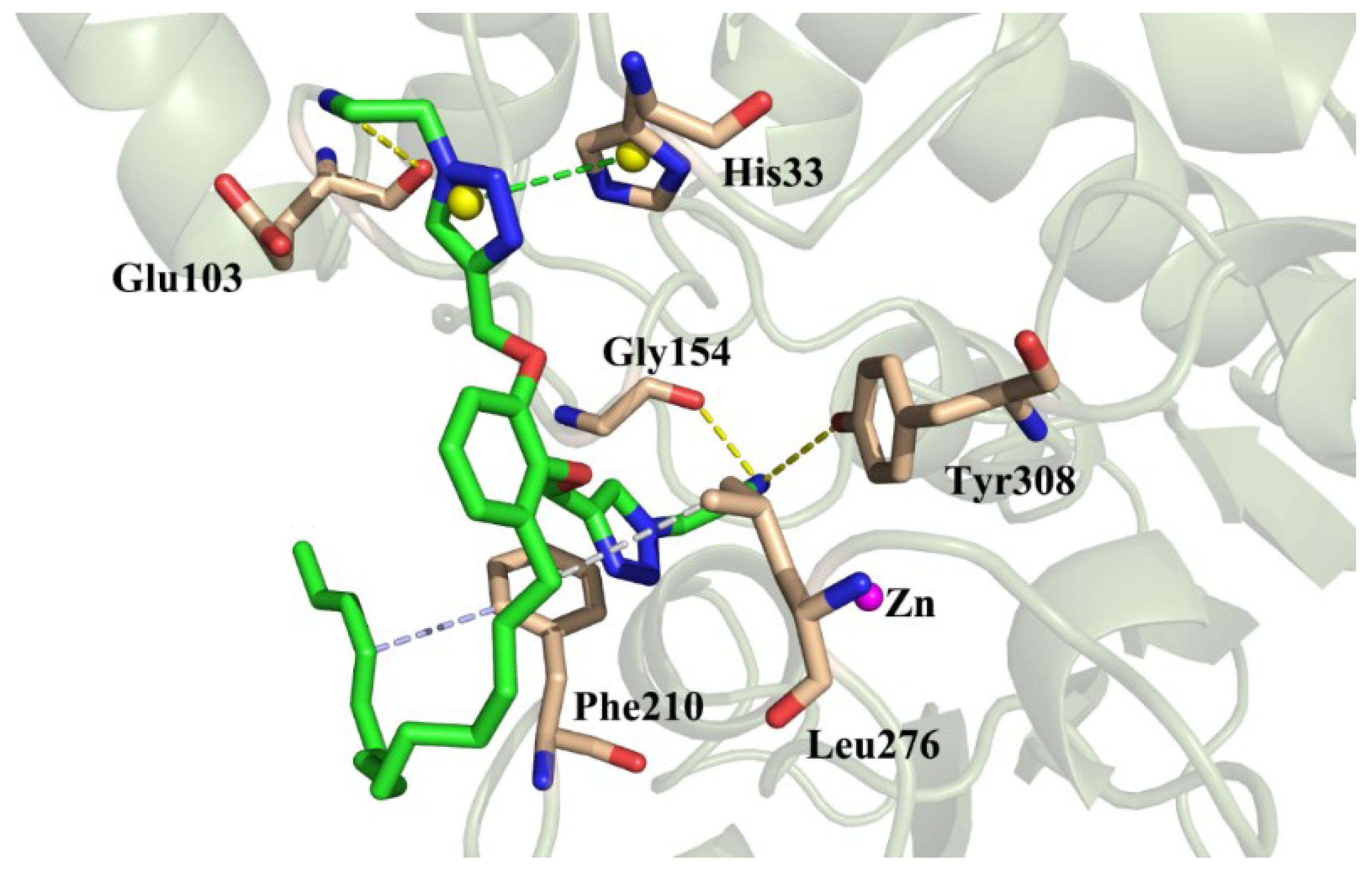


| Cpd. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 10 | 15 | 16 | 22 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HepG2 a | 28.6 | 63.2 | >200 | 2.8 | 39.5 | 25.0 | 6.0 | 28.9 | 29.4 | 23.9 | 65.8 |
| LO2 b | 66.1 | 130.6 | >200 | 80.9 | >200 | 64.0 | 100.1 | 198.0 | >200 | >200 | 150.5 |
| Compound | Pose | Grid Score | Grid_vdw | Grid_es | Int_energy |
|---|---|---|---|---|---|
| 1 | 1 | −46.492264 | −40.641602 | −5.850661 | 4.597153 |
| 2 | 1 | −44.553402 | −40.142418 | −4.410985 | 9.792090 |
| 2 | −41.821983 | −36.967918 | −4.854066 | 7.414151 | |
| 3 | 1 | −47.741497 | −46.867558 | −0.873939 | 11.230281 |
| 4 | 1 | −70.507187 | −55.308952 | −15.198232 | 19.825512 |
| 2 | −68.278595 | −56.138966 | −12.139633 | 18.417746 | |
| 3 | −66.865288 | −53.072853 | −13.792435 | 11.982115 | |
| 5 | 1 | −50.337765 | −44.250916 | −6.086848 | 96.381271 |
| 6 | 1 | −45.127968 | −42.331528 | −2.796439 | 9.279520 |
| 7 | 1 | −72.713211 | −51.605087 | −21.108124 | 15.293597 |
| 10 | 1 | −44.027946 | −34.921326 | −9.106619 | 8.631289 |
| 2 | −42.797626 | −35.534809 | −7.262817 | 7.740744 | |
| 22 | 1 | −46.489151 | −41.222374 | −5.266776 | 9.209332 |
| Compound | Pose | Grid Score | Grid_vdw | Grid_es | Int_energy |
|---|---|---|---|---|---|
| Ligand2 | 1 | −67.1829 | −44.7886 | −22.3944 | 13.1161 |
| 2 | −62.4968 | −48.8023 | −13.69444 | 16.3890 | |
| 3 | −62.2309 | −37.3300 | −24.90084 | 35.4859 |
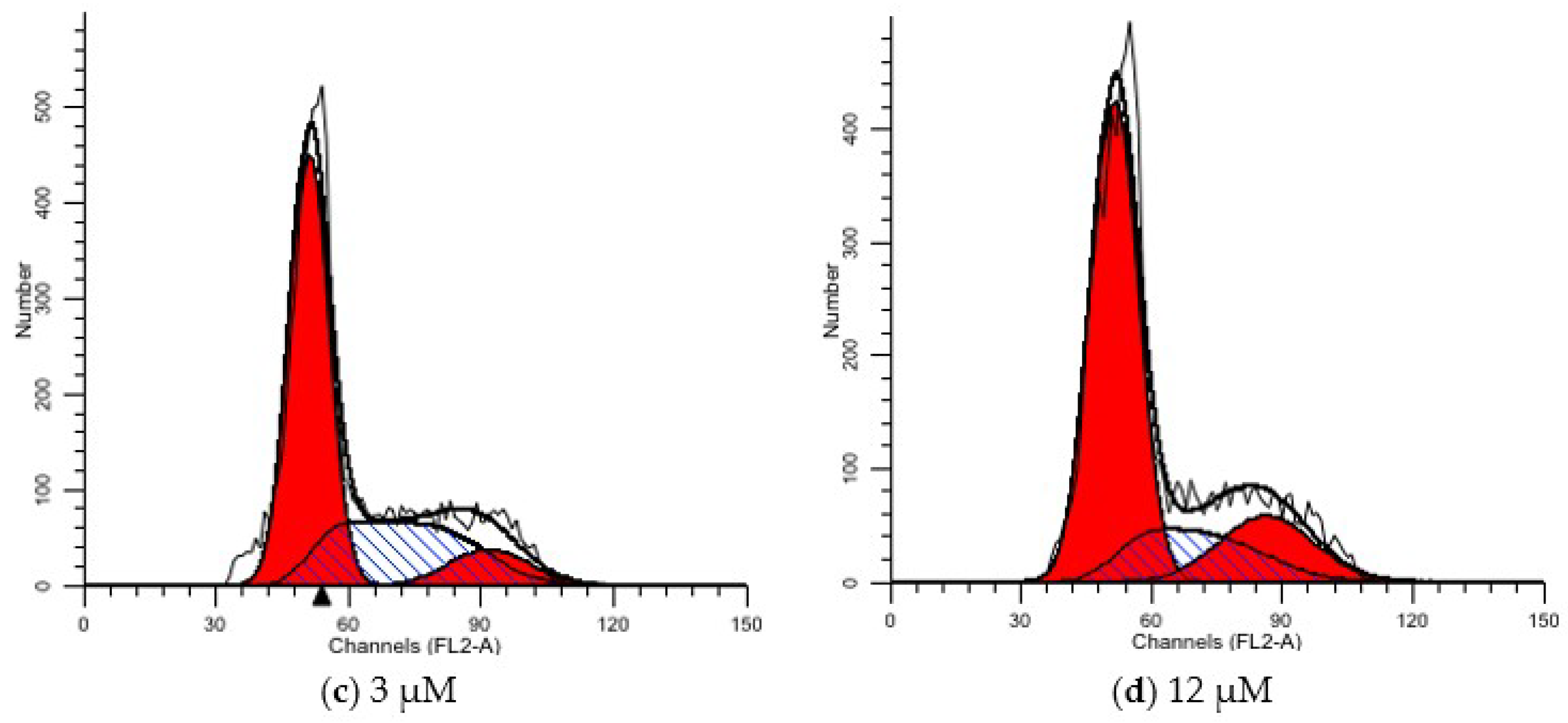
| Group | G1 (%) | S (%) | G2 (%) |
|---|---|---|---|
| CON | 48.06 | 35.8 | 16.14 |
| 1 μM | 55.92 | 32.46 | 11.62 |
| 3 μM | 58.06 | 32.36 | 9.57 |
| 12 μM | 63.84 | 18.76 | 17.4 |
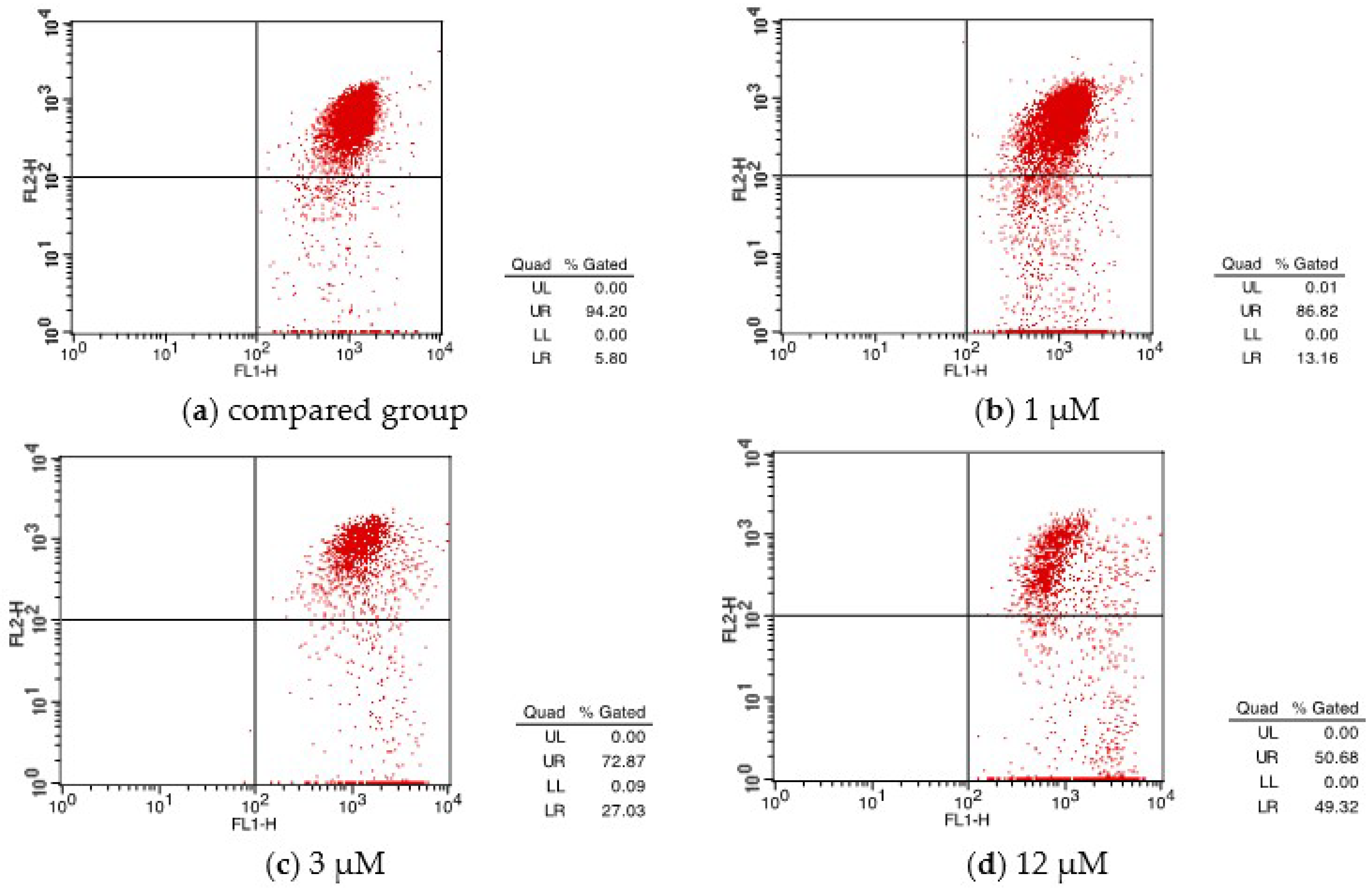
| Group | UL (%) | UR (%) | LL (%) | LR (%) | Apoptosis (%) |
|---|---|---|---|---|---|
| CON | 0.63 | 2.44 | 94.78 | 2.16 | 4.6 |
| 1 μM | 0.09 | 7.80 | 81.93 | 10.17 | 17.97 |
| 3 μM | 0.12 | 13.92 | 70.57 | 15.39 | 29.31 |
| 12 μM | 0.08 | 20.00 | 57.52 | 22.40 | 42.40 |
| Group | Green Fluorescence (%) |
|---|---|
| CON | 5.80 |
| 1 μM | 13.16 |
| 3 μM | 27.03 |
| 12 μM | 49.32 |
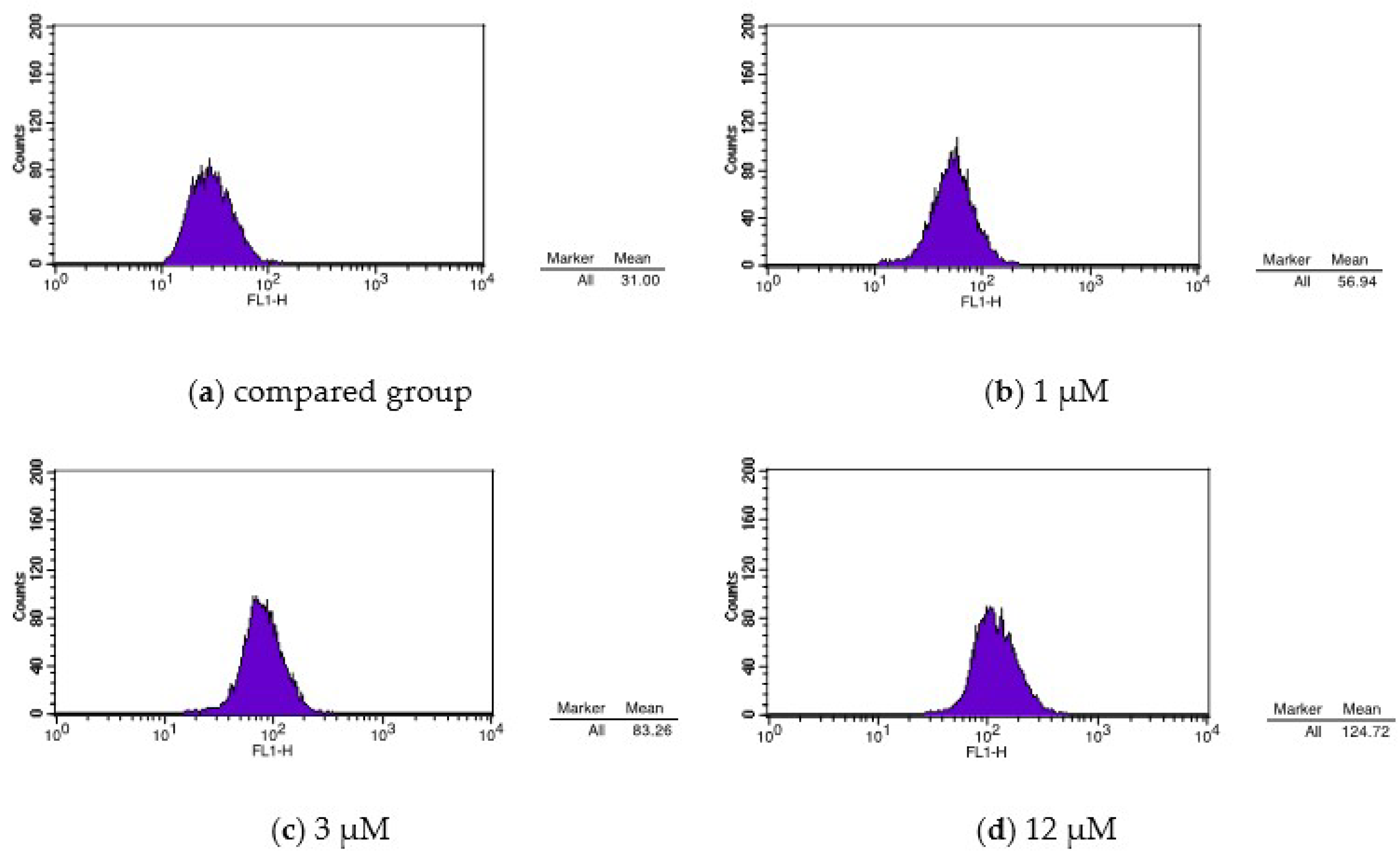
| Group | Mean |
|---|---|
| CON | 31.00 |
| 1 μM | 56.94 |
| 3 μM | 83.26 |
| 12 μM | 124.72 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Z.; Wang, C.; Jiang, J.; Wu, C. Novel C15 Triene Triazole, D-A Derivatives Anti-HepG2, and as HDAC2 Inhibitors: A Synergy Study. Int. J. Mol. Sci. 2018, 19, 3184. https://doi.org/10.3390/ijms19103184
Qi Z, Wang C, Jiang J, Wu C. Novel C15 Triene Triazole, D-A Derivatives Anti-HepG2, and as HDAC2 Inhibitors: A Synergy Study. International Journal of Molecular Sciences. 2018; 19(10):3184. https://doi.org/10.3390/ijms19103184
Chicago/Turabian StyleQi, Zhiwen, Chengzhang Wang, Jianxin Jiang, and Caie Wu. 2018. "Novel C15 Triene Triazole, D-A Derivatives Anti-HepG2, and as HDAC2 Inhibitors: A Synergy Study" International Journal of Molecular Sciences 19, no. 10: 3184. https://doi.org/10.3390/ijms19103184
APA StyleQi, Z., Wang, C., Jiang, J., & Wu, C. (2018). Novel C15 Triene Triazole, D-A Derivatives Anti-HepG2, and as HDAC2 Inhibitors: A Synergy Study. International Journal of Molecular Sciences, 19(10), 3184. https://doi.org/10.3390/ijms19103184







