Memory Function in Feeding Habit Transformation of Mandarin Fish (Siniperca chuatsi)
Abstract
1. Introduction
2. Results
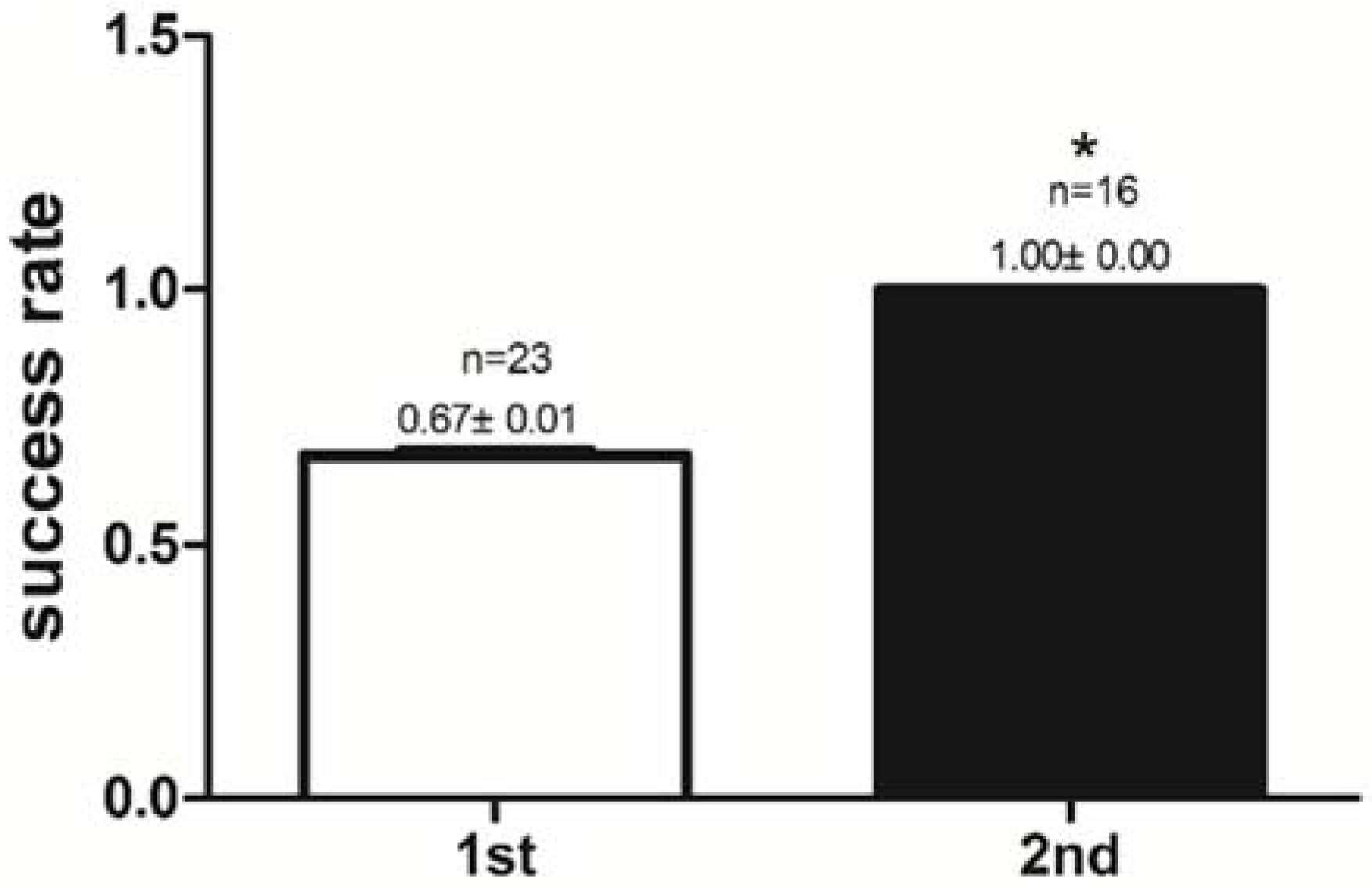
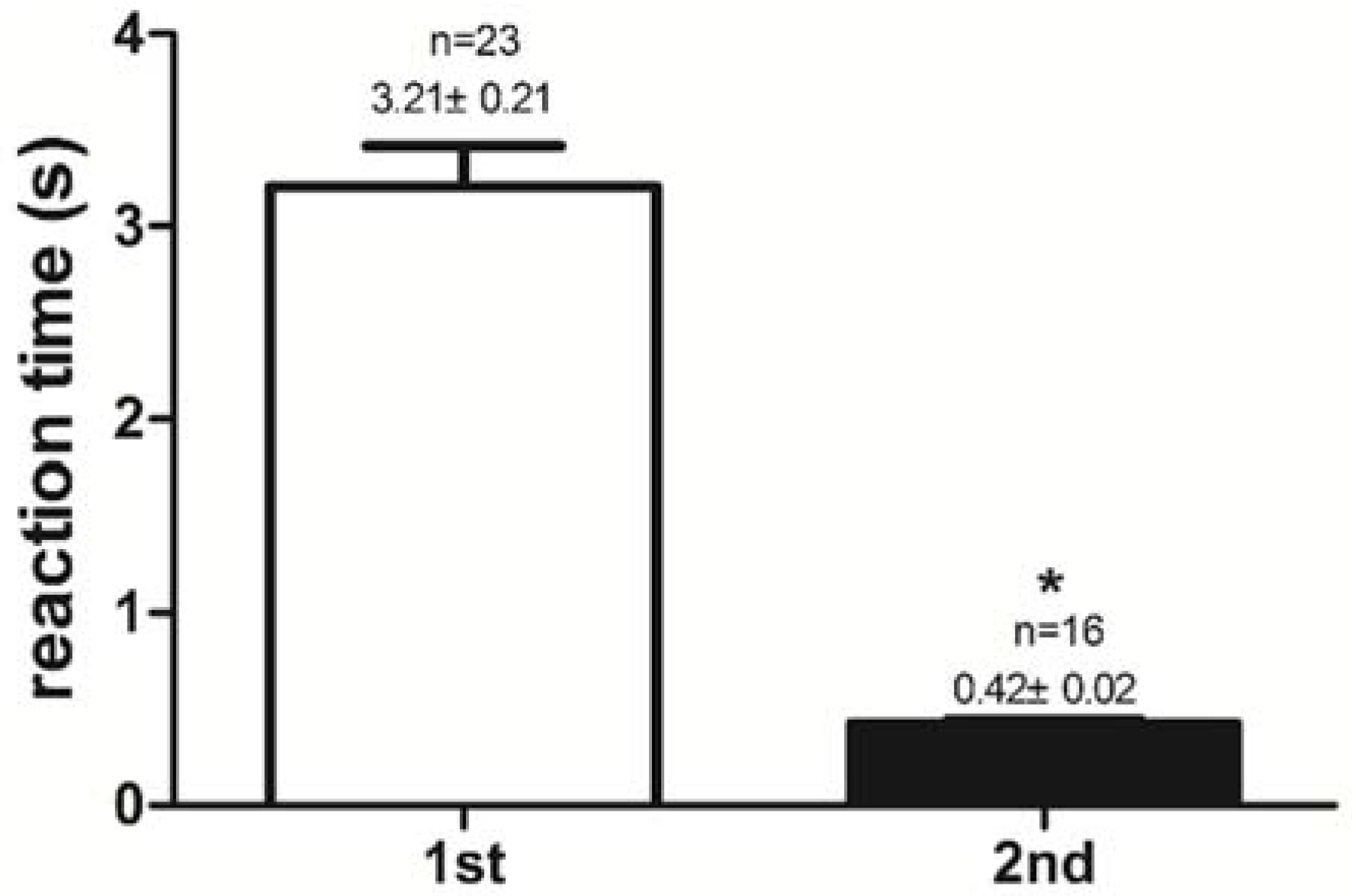
2.1. Success Rate of Feeding Habit Transformation and Reaction Time to Dead Prey Fish
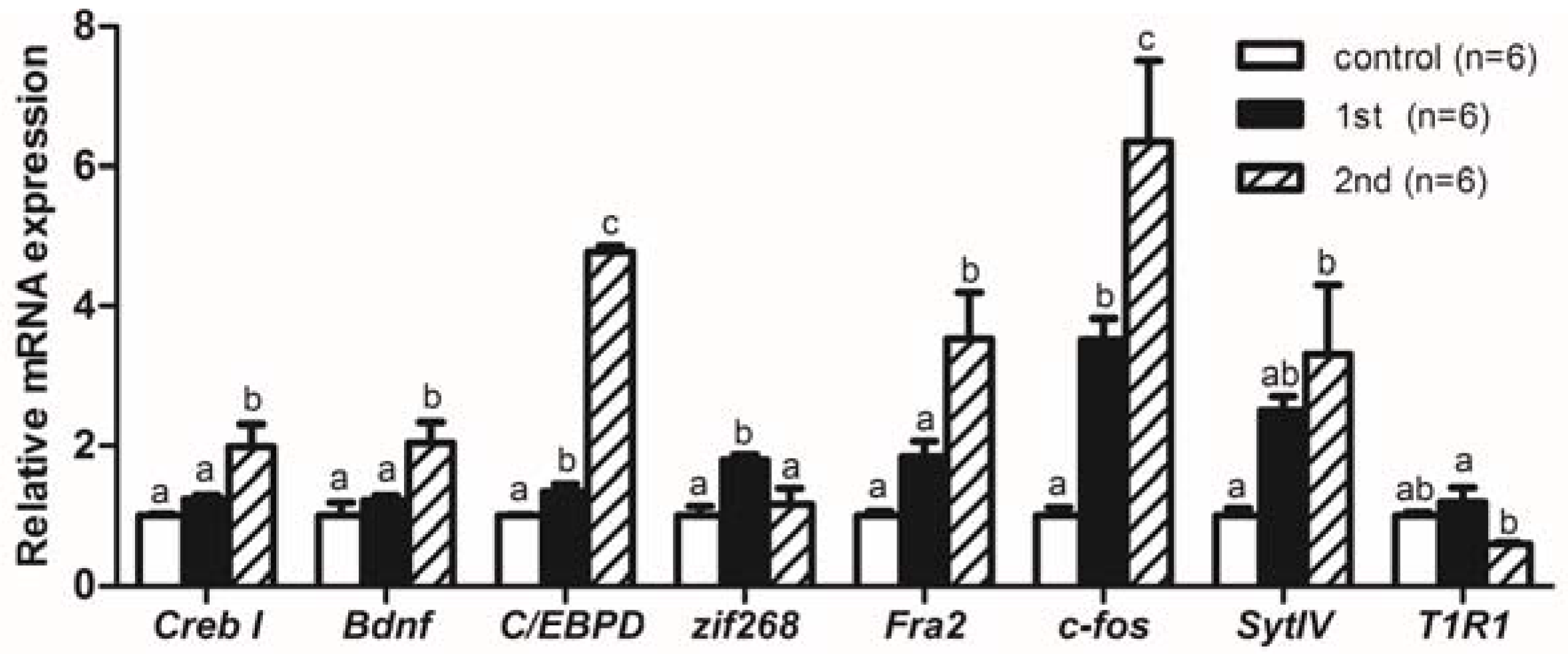
2.2. Gene Expression Levels Analysis of Memory-Relative Genes in Mandarin Fish
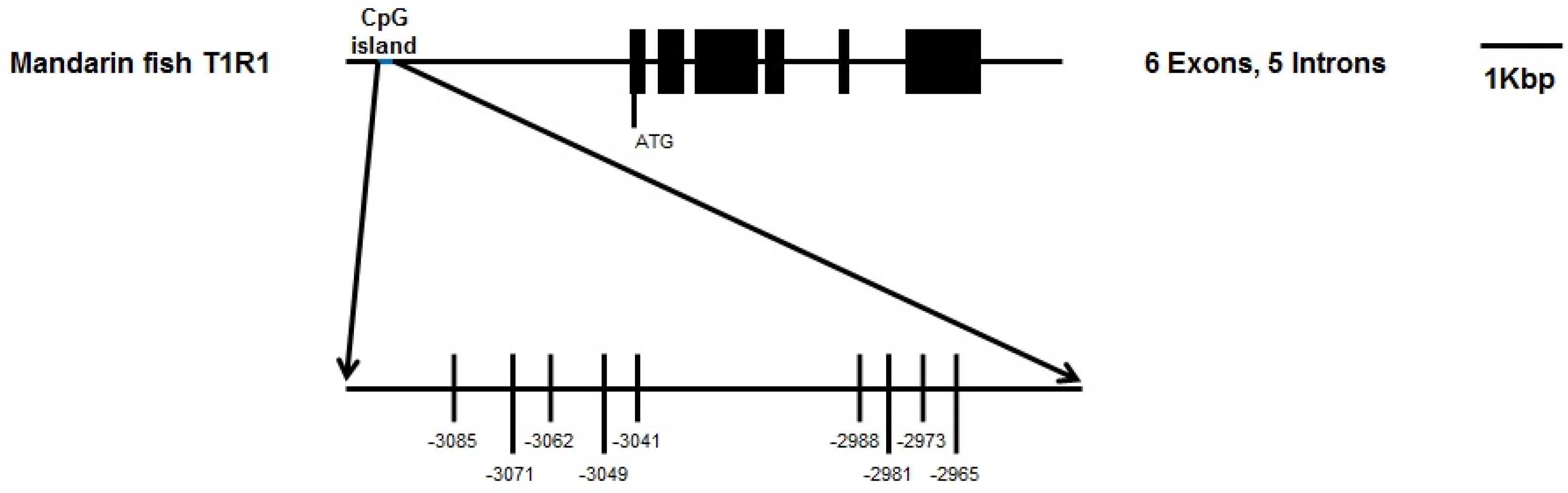
2.3. DNA Methylation Analysis and Bisulphite Sequencing Polymerase Chain Reaction (BSP) of T1R1 Gene
3. Discussion
4. Materials and Methods
4.1. Fish and Sample Preparation
4.2. Experiment Test Phase
4.2.1. Pre-Training
4.2.2. Experiment Training Phase 1
4.2.3. Natural Feed Revert Procedure
4.2.4. Experiment Training Phase 2
4.3. Sample Collection
4.4. RNA Isolation and Reverse Transcription
4.5. Gene Expression Levels Analysis of Memory Relative Genes in Mandarin Fish
4.6. DNA Methylation Analysis and Bisulphite Sequencing Polymerase Chain Reaction (BSP)
4.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chiang, I. On the biology of mandarin fish, Siniperca chuatsi of liang-tze lake. Acta Hydrobiol. Sin. 1959, 3, 375–385. [Google Scholar]
- He, S.; Liang, X.-F.; Sun, J.; Li, L.; Yu, Y.; Huang, W.; Qu, C.-M.; Cao, L.; Bai, X.-L.; Tao, Y.-X. Insights into food preference in hybrid f1 of Siniperca chuatsi (♀) × Siniperca scherzeri (♂) mandarin fish through transcriptome analysis. BMC Genom. 2013, 14, 601. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Oku, H.; Ogata, H.; Liu, J.; He, X. Weaning chinese perch Siniperca chuatsi (Basilewsky) onto artificial diets based upon its specific sensory modality in feeding. Aquac. Res. 2001, 32, 76–82. [Google Scholar] [CrossRef]
- Brown, C.; Laland, K.N. Social learning in fishes: A review. Fish Fish. 2003, 4, 280–288. [Google Scholar] [CrossRef]
- Kelley, J.L.; Magurran, A.E. Learned predator recognition and antipredator responses in fishes. Fish Fish. 2003, 4, 216–226. [Google Scholar] [CrossRef]
- Ajemian, M.; Sohel, S.; Mattila, J. Effects of turbidity and habitat complexity on antipredator behavior of three-spined sticklebacks (Gasterosteus aculeatus). Environ. Biol. Fish. 2015, 98, 45–55. [Google Scholar] [CrossRef]
- Wund, M.A.; Baker, J.A.; Golub, J.L.; Foster, S.A. The evolution of antipredator behaviour following relaxed and reversed selection in alaskan threespine stickleback fish. Anim. Behav. 2015, 106, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Everley, K.A.; Radford, A.N.; Simpson, S.D. Pile-driving noise impairs antipredator behavior of the european sea bass Dicentrarchus labrax. In The Effects of Noise on Aquatic Life II; Springer: Berlin, Germany, 2016; pp. 273–279. [Google Scholar]
- Broglio, C.; Rodriguez, F.; Salas, C. Spatial cognition and its neural basis in teleost fishes. Fish Fish. 2003, 4, 247–255. [Google Scholar] [CrossRef]
- Odling-Smee, L.; Braithwaite, V.A. The role of learning in fish orientation. Fish Fish. 2003, 4, 235–246. [Google Scholar] [CrossRef]
- Salas, C.; Broglio, C.; Rodríguez, F. Evolution of forebrain and spatial cognition in vertebrates: Conservation across diversity. Brain Behav. Evol. 2003, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Davis, V.; Holbrook, R.; Schumacher, S.; Guilford, T.; de Perera, T.B. Three-dimensional spatial cognition in a benthic fish, Corydoras aeneus. Behav. Process. 2014, 109, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Meckley, T.D.; Gurarie, E.; Miller, J.R.; Wagner, C.M. How fishes find the shore: Evidence for orientation to bathymetry from the non-homing sea lamprey. Can. J. Fish. Aquat. Sci. 2017, 74, 2045–2058. [Google Scholar] [CrossRef]
- Griffiths, S.W. Learned recognition of conspecifics by fishes. Fish Fish. 2003, 4, 256–268. [Google Scholar] [CrossRef]
- Hoare, D.; Krause, J. Social organisation, shoal structure and information transfer. Fish Fish. 2003, 4, 269–279. [Google Scholar] [CrossRef]
- Trompf, L.; Brown, C. Personality affects learning and trade-offs between private and social information in guppies, Poecilia reticulata. Anim. Behav. 2014, 88, 99–106. [Google Scholar] [CrossRef]
- Cui, R.; Delclos, P.J.; Schumer, M.; Rosenthal, G.G. Early social learning triggers neurogenomic expression changes in a swordtail fish. Proc. R. Soc. B 2017, 284, 20170701. [Google Scholar] [CrossRef] [PubMed]
- Milinski, M. Arms races, ornaments and fragrant genes: The dilemma of mate choice in fishes. Neurosci. Biobehav. Rev. 2014, 46, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, S.J.; Heath, D.D.; Devlin, R.H.; Pitcher, T.E. Post-spawning sexual selection in red and white chinook salmon (Oncorhynchus tshawytscha). Behav. Ecol. 2016, 28, 1–10. [Google Scholar] [CrossRef]
- Lehnert, S.J.; Pitcher, T.E.; Devlin, R.H.; Heath, D.D. Red and white Chinook salmon: Genetic divergence and mate choice. Mol. Ecol. 2016, 25, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- Warburton, K. Learning of foraging skills by fish. Fish Fish. 2003, 4, 203–215. [Google Scholar] [CrossRef]
- Pike, T.W.; Laland, K.N. Conformist learning in nine-spined sticklebacks’ foraging decisions. Biol. Lett. 2010. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, J.D.; Colgan, P.W. The role of learning in fish behaviour. Rev. Fish Biol. Fish. 1992, 2, 125–143. [Google Scholar] [CrossRef]
- Provenza, F.D.; Pfister, J.A.; Cheney, C.D. Mechanisms of learning in diet selection with reference to phytotoxicosis in herbivores. J. Range Manag. 1992, 45, 36–45. [Google Scholar] [CrossRef]
- Liu, H.; Li, S.; Bai, J. Research advances in gene regulation and genetic improvement of fish feeding. Agric. Sci. Technol. 2015, 16, 2277–2282. [Google Scholar]
- Gräff, J.; Joseph, N.F.; Horn, M.E.; Samiei, A.; Meng, J.; Seo, J.; Rei, D.; Bero, A.W.; Phan, T.X.; Wagner, F. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell 2014, 156, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Stafford, J.M.; Raybuck, J.D.; Ryabinin, A.E.; Lattal, K.M. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol. Psychiatry 2012, 72, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Han, C.; Luo, W.; Li, H.; Luo, H.; Qiang, M.; Su, T.; Wu, B.; Liu, Y.; Yang, X. Aging-associated excess formaldehyde leads to spatial memory deficits. Sci. Rep. 2013, 3, 1807. [Google Scholar] [CrossRef] [PubMed]
- Roth, E.D.; Roth, T.L.; Money, K.M.; SenGupta, S.; Eason, D.E.; Sweatt, J.D. DNA methylation regulates neurophysiological spatial representation in memory formation. Neuroepigenetics 2015, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sweatt, J.D. Experience-dependent epigenetic modifications in the central nervous system. Biol. Psychiatry 2009, 65, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Erlenbach, I.; Ryba, N.J.; Zuker, C.S. The receptors for mammalian sweet and umami taste. Cell 2003, 115, 255–266. [Google Scholar] [CrossRef]
- Yasuo, T.; Kusuhara, Y.; Yasumatsu, K.; Ninomiya, Y. Multiple receptor systems for glutamate detection in the taste organ. Biol. Pharm. Bull. 2008, 31, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Mace, O.J.; Lister, N.; Morgan, E.; Shepherd, E.; Affleck, J.; Helliwell, P.; Bronk, J.R.; Kellett, G.L.; Meredith, D.; Boyd, R. An energy supply network of nutrient absorption coordinated by calcium and t1r taste receptors in rat small intestine. J. Physiol. 2009, 587, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Nagasawa, M.; Yamada, S.; Hara, A.; Mogami, H.; Nikolaev, V.O.; Lohse, M.J.; Shigemura, N.; Ninomiya, Y.; Kojima, I. Sweet taste receptor expressed in pancreatic beta-cells activates the calcium and cyclic amp signaling systems and stimulates insulin secretion. PLoS ONE 2009, 4, e5106. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhou, L.; Terwilliger, R.; Newton, S.S.; Araujo, I.E.D. Sweet taste signaling functions as a hypothalamic glucose sensor. Front. Integr. Neurosci. 2009, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Treesukosol, Y.; Smith, K.R.; Spector, A.C. The functional role of the t1r family of receptors in sweet taste and feeding. Physiol. Behav. 2011, 105, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.; Bojahr, J.; Narukawa, M.; Hübner, S.; Boehm, U.; Meyerhof, W. Transsynaptic tracing from taste receptor cells reveals local taste receptor gene expression in gustatory ganglia and brain. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 9717–9729. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, J.-R.; Xu, H.; Zhang, J. Pseudogenization of the umami taste receptor gene tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol. Boil. Evol. 2010, 27, 2669–2673. [Google Scholar] [CrossRef] [PubMed]
- Raliou, M.; Grauso, M.; Hoffmann, B.; Schlegel-Le-Poupon, C.; Nespoulous, C.; Débat, H.; Belloir, C.; Wiencis, A.; Sigoillot, M.; Bano, S.P. Human genetic polymorphisms in T1R1 and T1R3 taste receptor subunits affect their function. Chem. Senses 2011, 36, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Alarcon, S.; Tharp, A.; Ahmed, O.M.; Estrella, N.L.; Greene, T.A.; Rucker, J.; Breslin, P.A.S.; Fernstrom, J.D.; Beauchamp, G.K. Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes. Am. J. Clin. Nutr. 2009, 90, 770S–779S. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.F.; Liu, J.K.; Huang, B.Y. The role of sense organs in the feeding behaviour of chinese perch. J. Fish Biol. 1998, 52, 1058–1067. [Google Scholar] [CrossRef]
- McGaugh, J.L. Memory—A century of consolidation. Science 2000, 287, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Nader, K.; Schafe, G.E.; Le Doux, J.E. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature 2000, 406, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. Mechanisms of memory stabilization: Are consolidation and reconsolidation similar or distinct processes? Trends Neurosci. 2005, 28, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Brightwell, J.J.; Smith, C.A.; Neve, R.L.; Colombo, P.J. Long-term memory for place learning is facilitated by expression of cAMP response element-binding protein in the dorsal hippocampus. Learn. Mem. 2007, 14, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Bourtchuladze, R.; Frenguelli, B.; Blendy, J.; Cioffi, D.; Schutz, G.; Silva, A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 1994, 79, 59–68. [Google Scholar] [CrossRef]
- Tao, X.; Finkbeiner, S.; Arnold, D.B.; Shaywitz, A.J.; Greenberg, M.E. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 1998, 20, 709–726. [Google Scholar] [CrossRef]
- Yamada, K.; Mizuno, M.; Nabeshima, T. Role for brain-derived neurotrophic factor in learning and memory. Life Sci. 2002, 70, 735–744. [Google Scholar] [CrossRef]
- Tyler, W.J.; Alonso, M.; Bramham, C.R.; Pozzo-Miller, L.D. From acquisition to consolidation: On the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learn. Mem. 2002, 9, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M.; Ghirardi, M.; Huang, Y.Y.; Nguyen, P.V.; Kandel, E.R. A molecular switch for the consolidation of long-term memory: cAMP—inducible gene expression. Ann. N. Y. Acad. Sci. 1995, 758, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E.R. The molecular biology of memory storage: A dialogue between genes and synapses. Science 2001, 294, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
- Neale, J.H.; Klinger, P.; Agranoff, B. Camptothecin blocks memory of conditioned avoidance in the goldfish. Science 1973, 179, 1243–1246. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Countryman, R.A.; Kaban, N.L.; Colombo, P.J. Hippocampal c-fos is necessary for long-term memory of a socially transmitted food preference. Neurobiol. Learn. Mem. 2005, 84, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Bozon, B.; Davis, S.; Laroche, S. Regulated transcription of the immediate-early gene Zif268: Mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 2002, 12, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Bozon, B.; Davis, S.; Laroche, S. A requirement for the immediate early gene Zif268 in reconsolidation of recognition memory after retrieval. Neuron 2003, 40, 695–701. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Matsubara, J.A.; Cynader, M.S. Neuronal activity in primate visual cortex assessed by immunostaining for the transcription factor Zif268. Vis. Neurosci. 1995, 12, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Worley, P.F.; Christy, B.A.; Nakabeppu, Y.; Bhat, R.V.; Cole, A.J.; Baraban, J.M. Constitutive expression of Zif268 in neocortex is regulated by synaptic activity. Proc. Natl. Acad. Sci. USA 1991, 88, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.M. Gene targeting of presynaptic proteins in synaptic plasticity and memory: Across the great divide. Neurobiol. Learn. Mem. 2006, 85, 2–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dean, C.; Liu, H.; Dunning, F.M.; Chang, P.Y.; Jackson, M.B.; Chapman, E.R. Synaptotagmin-iv modulates synaptic function and long-term potentiation by regulating BDNF release. Nat. Neurosci. 2009, 12, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Oike, H.; Nagai, T.; Furuyama, A.; Okada, S.; Aihara, Y.; Ishimaru, Y.; Marui, T.; Matsumoto, I.; Misaka, T.; Abe, K. Characterization of ligands for fish taste receptors. J. Neurosci. 2007, 27, 5584–5592. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, J. Mismatches between feeding ecology and taste receptor evolution: An inconvenient truth. Proc. Natl. Acad. Sci. USA 2012, 109, E1464. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Gao, X.; Xie, A.; Wu, X.; Ma, F. Interaction between umami peptide and taste receptor T1R1/T1R3. Cell Biochem. Biophys. 2014, 70, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.N. Understanding safety of glutamate in food and brain. Indian J. Physiol. Pharmacol. 2007, 51, 216–234. [Google Scholar] [PubMed]
- Niki, M.; Takai, S.; Kusuhara, Y.; Ninomiya, Y.; Yoshida, R. Responses to apical and basolateral application of glutamate in mouse fungiform taste cells with action potentials. Cell. Mol. Neurobiol. 2011, 31, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.W.; French, P.J.; Bliss, T.; Rosenblum, K. Molecular mechanisms of long-term potentiation in the insular cortex in vivo. J. Neurosci. 1999, 19, RC36. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leung, F.C. Gc content increased at CpG flanking positions of fish genes compared with sea squirt orthologs as a mechanism for reducing impact of DNA methylation. PLoS ONE 2008, 3, e3612. [Google Scholar] [CrossRef] [PubMed]
- Pierron, F.; Baillon, L.; Sow, M.; Gotreau, S.; Gonzalez, P. Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ. Sci. Technol. 2013, 48, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Song, C.; Liu, S.; Tao, M.; Hu, J.; Wang, J.; Liu, W.; Zeng, M.; Liu, Y. DNA methylation analysis of allotetraploid hybrids of red crucian carp (Carassius auratus red var.) and common carp (Cyprinus carpio L.). PLoS ONE 2013, 8, e56409. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.; You, F.; Sun, P.; Li, J.; Xu, D.; Wu, Z.; Ma, D.; Zhang, P. CpG methylation of dmrt1 and cyp19a promoters in relation to their sexual dimorphic expression in the japanese flounder Paralichthys olivaceus. J. Fish Boil. 2014, 84, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, C.; Wu, H.; Chen, D.; Li, S.; Xu, S. Effects of atrazine and chlorpyrifos on DNA methylation in the brain and gonad of the common carp. Comp. Biochem. Physiol. Pt. C 2015, 168, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent Bdnf gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P.; Wolffe, A.P. Methylation-induced repression—Belts, braces, and chromatin. Cell 1999, 99, 451–454. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D.; Jones, P.A. DNA methylation: Past, present and future directions. Carcinogenesis 2000, 21, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 2004, 15, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007, 35, 1245. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. The DNA methylation paradox. Trends Genet. 1999, 15, 34–37. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Selker, E.U. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997, 11, 2383–2395. [Google Scholar] [CrossRef] [PubMed]
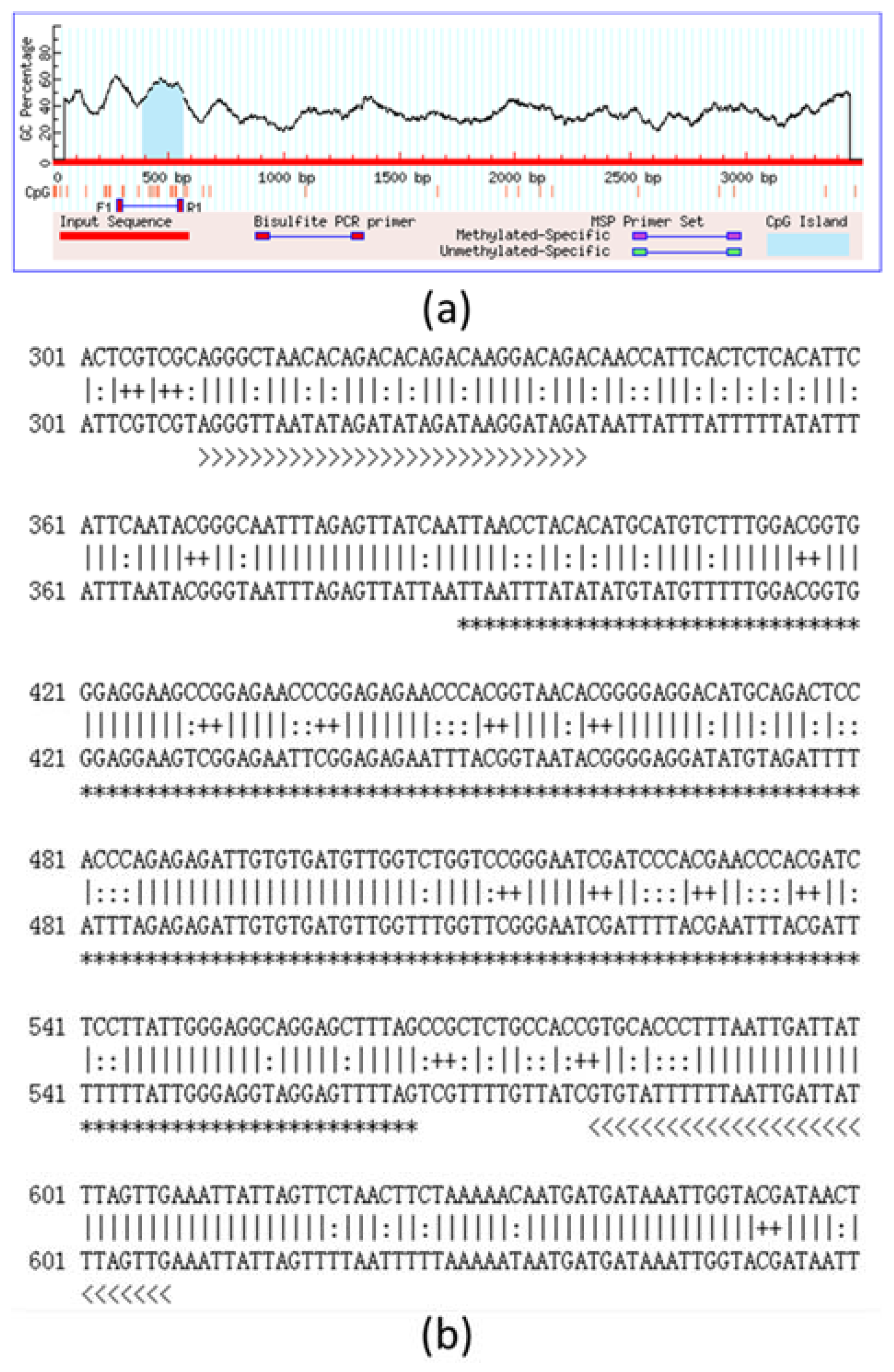

| CpG Position | −3085 | −3071 | −3062 | −3049 | −3041 | −2988 | −2981 | −2973 | −2965 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Me-CpG 1st 1 (%) | 29/30 | 30/30 | 29/30 | 29/30 | 29/30 | 30/30 | 30/30 | 29/30 | 27/30 | 262/270 |
| 96.70 | 100.00 | 96.70 | 96.70 | 96.70 | 100.00 | 100.00 | 96.70 | 90.00 | 97.00 | |
| Me-CpG 2nd 2 (%) | 18/30 | 27/30 | 22/30 | 19/30 | 20/30 | 27/30 | 21/30 | 28/30 | 24/30 | 206/270 |
| 60.00 | 90.00 | 73.30 | 63.30 | 66.70 | 90.00 | 70.00 | 93.30 | 80.00 | 76.30 | |
| significance | 0.007 * | 0.236 | 0.030 * | 0.004 * | 0.008 * | 0.236 | 0.004 * | 1.000 | 0.470 | 0.000 * |
| Primers Name | Sequences (5′-3′) |
|---|---|
| RTsc Creb I F | ATACACCCTCCCACTTCA |
| RTsc Creb I R | TCTCCTCCACATCCGTTC |
| RTsc Bdnf F | AACTGCCCTCACTCACA |
| RTsc Bdnf R | ACCTCCCTGGCTCTTAT |
| RTsc C/EBPD F | GCAGGAGAAGGCGGATTT |
| RTsc C/EBPD R | CTGGGAAGGCAGGGATGA |
| RTsc zif268 F | GGATCTTGCCGTGCCTCTTG |
| RTsc zif268 R | TTGCGACCGCCGTTTCTC |
| RTsc Fra2 F | CAACCAGGACCTCCAGTG |
| RTsc Fra2 R | TCTACGCCTTTCAATCTC |
| RTsc c-fos F | CGATGATGTTTACCGCTTTC |
| RTsc c-fos R | TAGTATCCCAGATTGTCCC |
| RTsc SytIV F | TGTCGGAGGATTAGAACG |
| RTsc SytIV R | CTGAAAGTCCAATGGGTAC |
| RTsc T1R1 F | TGTATTTTGTTTGATAGAATAAGAGT |
| RTsc T1R1 R | TAAAAAAACTTAATATAATACTTTTTAAAA |
| RTsc RPL13A F | TATCCCCCCACCCTATGACA |
| RTsc RPL13A R | ACGCCCAAGGAGAGCGAACT |
| Primers | Sequences (5′-3′) |
|---|---|
| Primers for genomic DNA amplicon | |
| CPG1 T1R1 F | AGGGCTAACACAGACACAGACAAGGACAGA |
| CPG1 T1R1 R | CAACTAAATAATCAATTAAAGGGTGCAC |
| Primers for BSP amplicon | |
| BSP1 T1R1 F | AGGGTTAATATAGATATAGATAAGGATAGA |
| BSP1 T1R1 R | CAACTAAATAATCAATTAAAAAATACAC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, Y.; He, S.; Liang, X.-F.; Cai, W.; Wang, J.; Shi, L.; Li, J. Memory Function in Feeding Habit Transformation of Mandarin Fish (Siniperca chuatsi). Int. J. Mol. Sci. 2018, 19, 1254. https://doi.org/10.3390/ijms19041254
Dou Y, He S, Liang X-F, Cai W, Wang J, Shi L, Li J. Memory Function in Feeding Habit Transformation of Mandarin Fish (Siniperca chuatsi). International Journal of Molecular Sciences. 2018; 19(4):1254. https://doi.org/10.3390/ijms19041254
Chicago/Turabian StyleDou, Yaqi, Shan He, Xu-Fang Liang, Wenjing Cai, Jie Wang, Linjie Shi, and Jiao Li. 2018. "Memory Function in Feeding Habit Transformation of Mandarin Fish (Siniperca chuatsi)" International Journal of Molecular Sciences 19, no. 4: 1254. https://doi.org/10.3390/ijms19041254
APA StyleDou, Y., He, S., Liang, X.-F., Cai, W., Wang, J., Shi, L., & Li, J. (2018). Memory Function in Feeding Habit Transformation of Mandarin Fish (Siniperca chuatsi). International Journal of Molecular Sciences, 19(4), 1254. https://doi.org/10.3390/ijms19041254





