MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells
Abstract
1. Introduction
2. Results
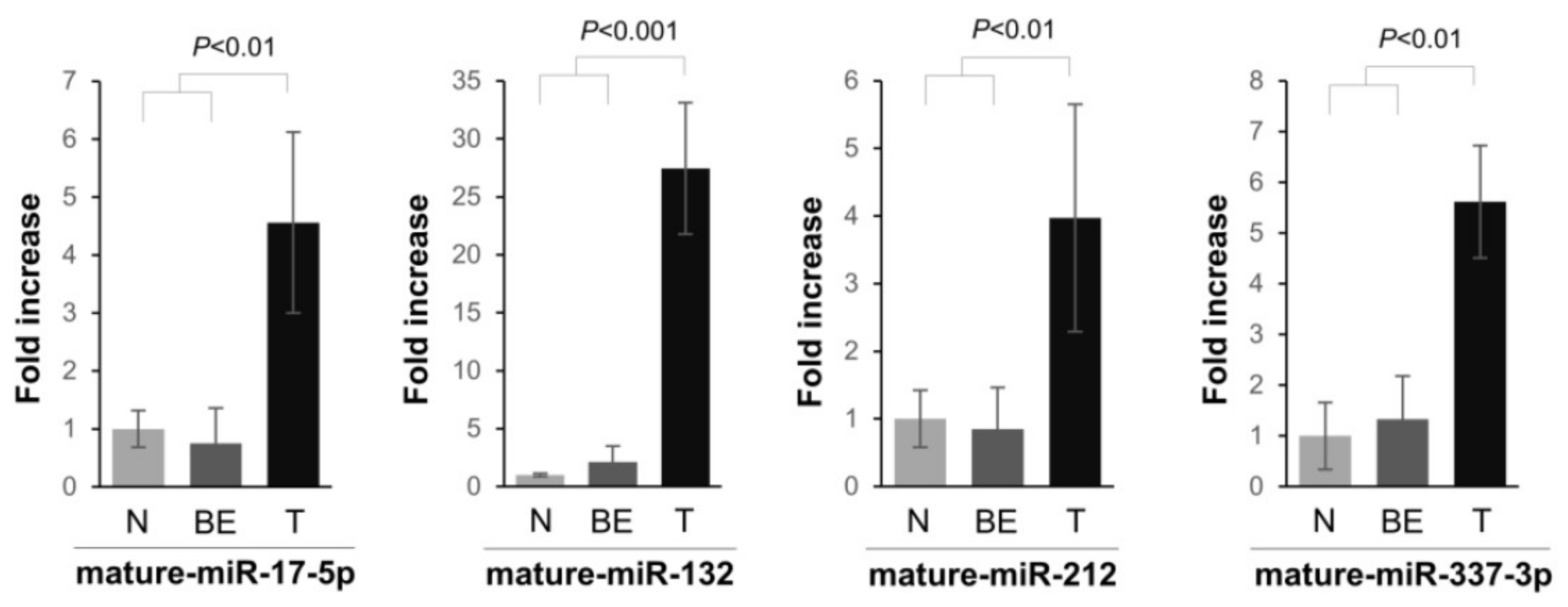
2.1. miRNA Expression in PDAC Specimens
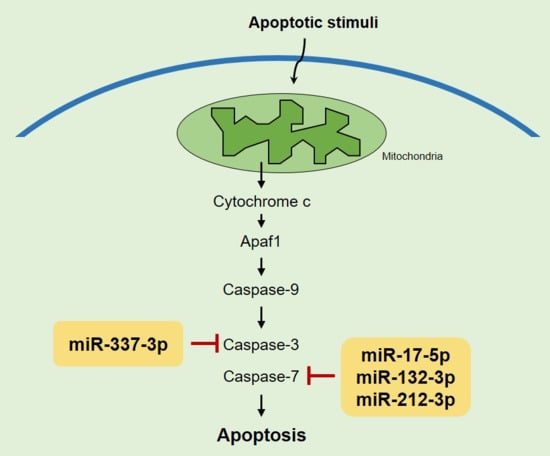
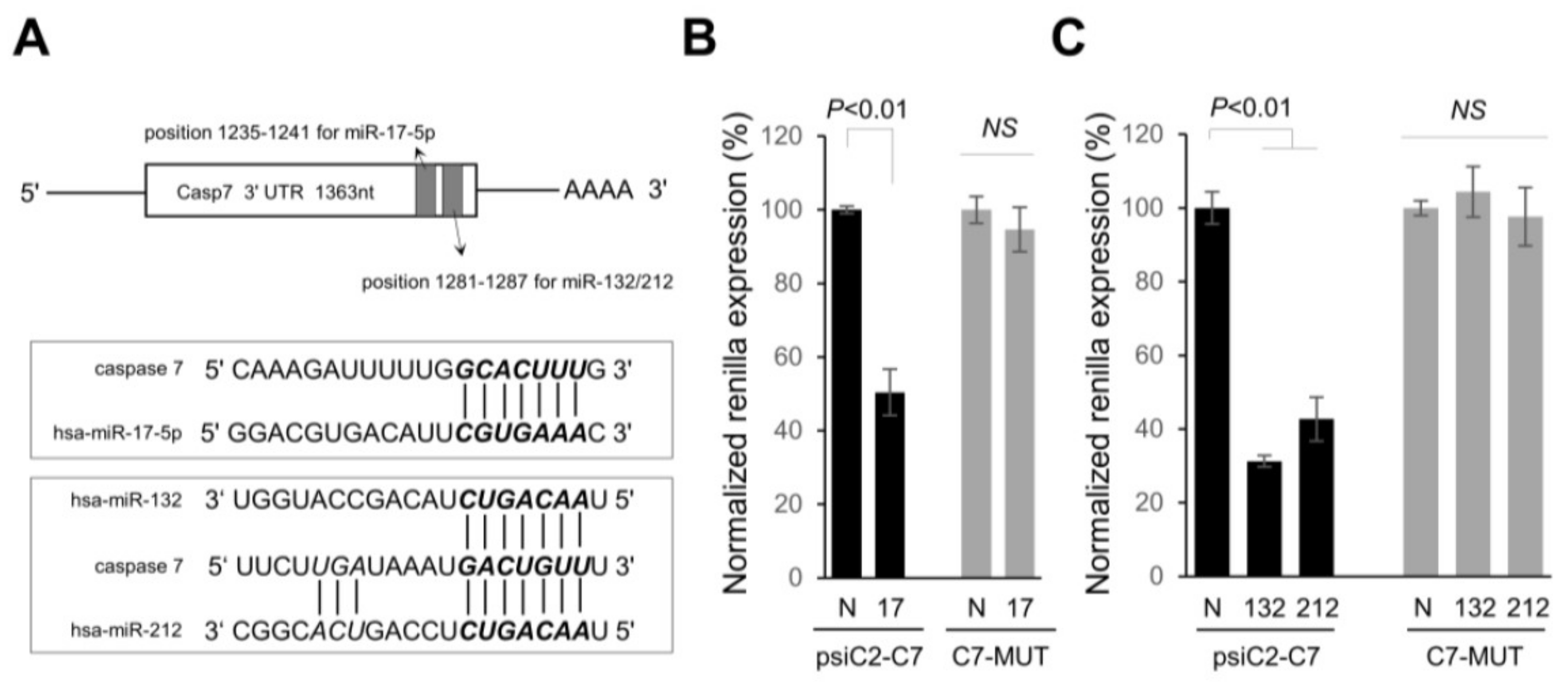
2.2. Identifying Targets of Differentially Expressed miRNAs
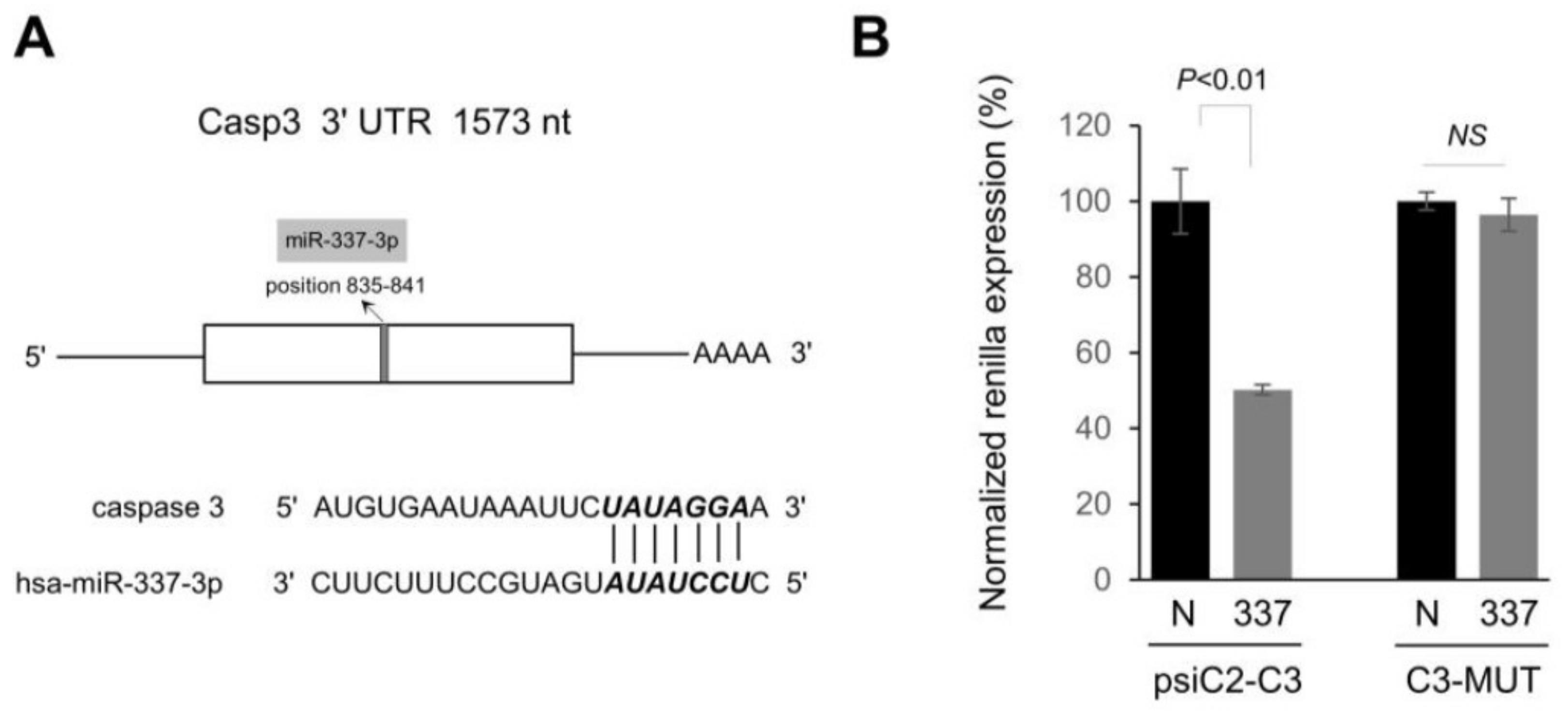
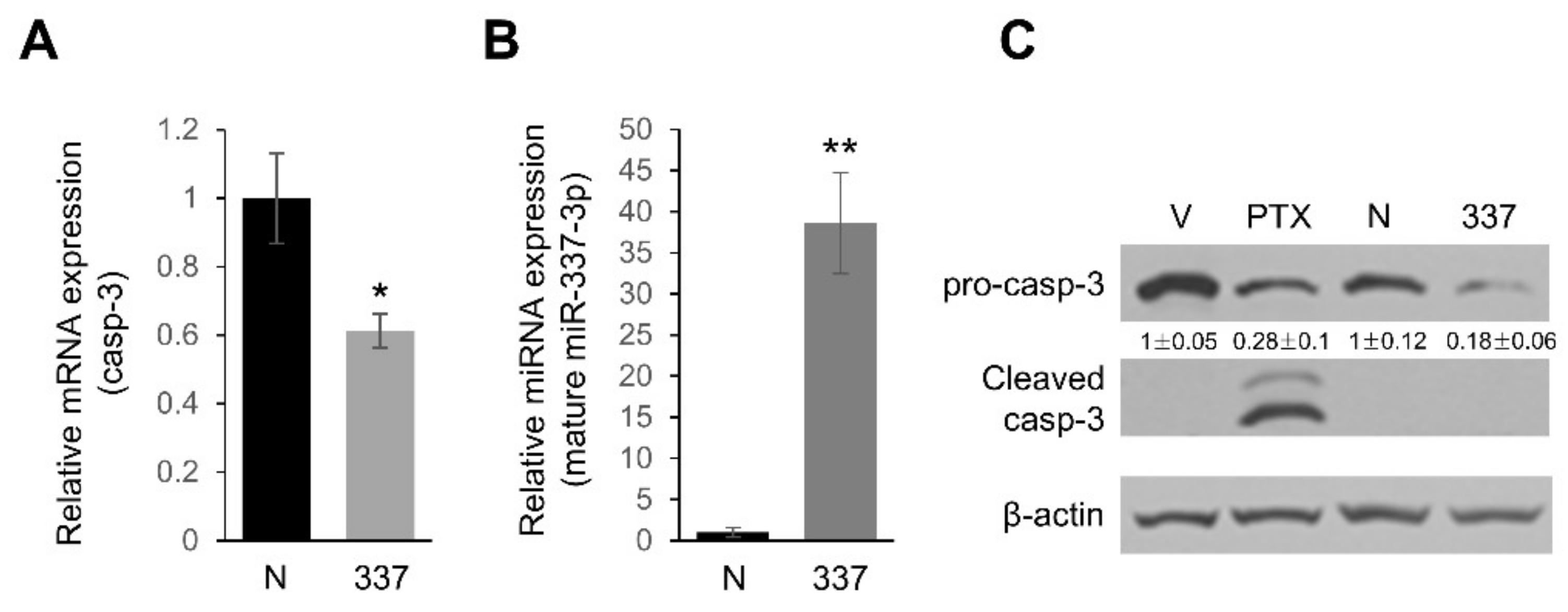
2.3. miR-337-3p Directly Targets Caspase-3
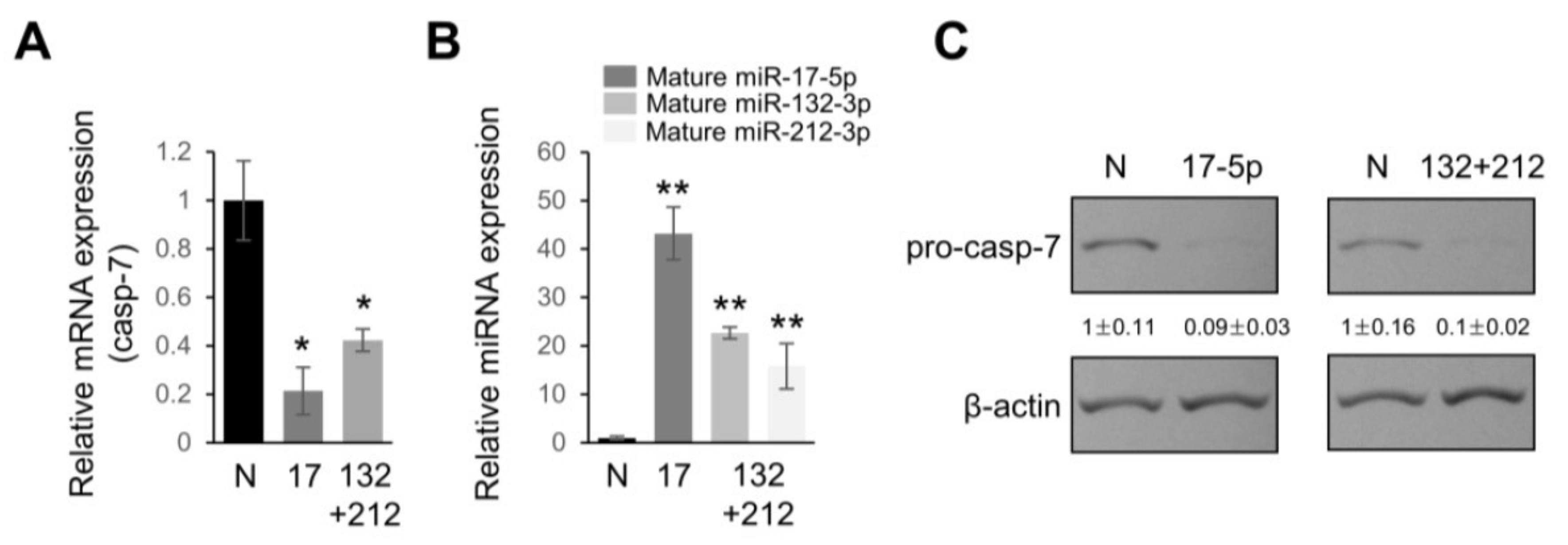
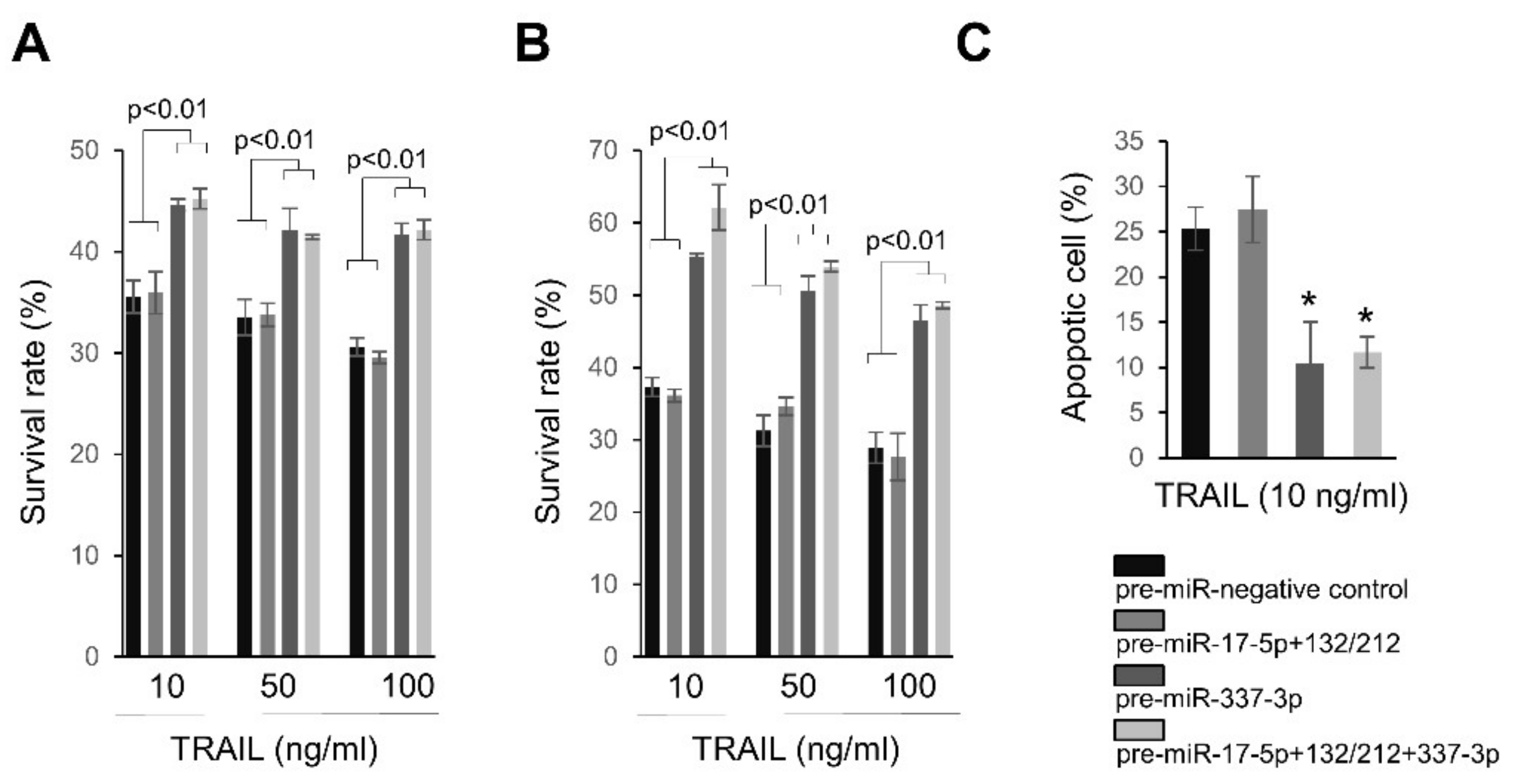
2.4. Overexpression of miRNAs Targeting Caspases-3/7 Diminishes TRAIL Effects on PANC-1 Cells
2.5. Gene Expression Correlation in TCGA Data Set
3. Discussion
4. Materials and Methods
4.1. Tissue Procurement
4.2. Cell Lines and Culture
4.3. qPCR for miRNA and mRNA Expression
4.4. Luciferase Reporter Constructs and Assay
4.5. microRNA Transfections
4.6. Protein Extraction and Immunoblotting
4.7. Cell Proliferation Assay
4.8. Apoptosis Analysis
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PDAC | Pancreatic ductal adenocarcinomas |
| TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| FLIP | FADD-like ICE inhibitory proteins |
| IAP | Inhibitor of apoptosis |
| miRNAs | MicroRNAs |
| FasL | Fas Ligand |
| TNF-α | Tumor Necrosis Factor-Alpha |
| PARP | Poly ADP ribose polymerase |
| MECP2 | Methyl-CpG Binding Protein 2 |
| EGF | Epidermal Growth Factor |
| P120RasGAP | P120 RAS GTPase Activating Protein |
| Rb1 | RB Transcriptional Corepressor 1 |
| HBP1 | HMG-Box Transcription Factor 1 |
| E2F1 | E2F Transcription Factor 1 |
| RBL2 | RB Transcriptional Corepressor Like 2 |
| CBFB | Core binding factor beta subunit |
| PPP3CA | Protein phosphatase 3, catalytic subunit, alpha isozyme |
| CLINT1 | Clathrin interactor 1 |
| HOXB7 | Homeobox B7 |
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Adamska, A.; Domenichini, A.; Falasca, M. Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int. J. Mol. Sci. 2017, 18, 1338. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Simianu, V.V.; Zyromski, N.J.; Nakeeb, A.; Lillemoe, K.D. Pancreatic cancer: Progress made. Acta Oncol. 2010, 49, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, D.W. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999, 6, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Strimpakos, A.; Saif, M.W.; Syrigos, K.N. Pancreatic cancer: From molecular pathogenesis to targeted therapy. Cancer Metastasis Rev. 2008, 27, 495–522. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.A.; Masud, A.; Kuida, K.; Porter, G.A., Jr.; Booth, C.J.; Mehal, W.Z.; Inayat, I.; Flavell, R.A. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science 2006, 311, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Sladek, T.L.; Liu, X.; Butler, B.R.; Froelich, C.J.; Thor, A.D. Reconstitution of caspase 3 sensitizes mcf-7 breast cancer cells to doxorubicin- and etoposide-induced apoptosis. Cancer Res. 2001, 61, 348–354. [Google Scholar] [PubMed]
- Ahmad, M.; Shi, Y. Trail-induced apoptosis of thyroid cancer cells: Potential for therapeutic intervention. Oncogene 2000, 19, 3363–3371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Chen, L.; Jin, H.; Wang, S.; Zhang, Y.; Tang, X.; Tang, G. Screening mirnas for early diagnosis of colorectal cancer by small RNA deep sequencing and evaluation in a Chinese patient population. OncoTargets Ther. 2016, 9, 1159–1166. [Google Scholar]
- Li, C.; Yin, Y.; Liu, X.; Xi, X.; Xue, W.; Qu, Y. Non-small cell lung cancer associated microrna expression signature: Integrated bioinformatics analysis, validation and clinical significance. Oncotarget 2017, 8, 24564–24578. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Kang, J.H.; Lee, K.S.; Nam, S.; Ro, J.; Kim, J.H. Identification and clinical implications of circulating microRNAs for estrogen receptor-positive breast cancer. Tumour Biol. 2014, 35, 12173–12180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, Y.W.; Wang, Y.Y.; Song, Y.; Zhu, J.; Si, P.C.; Ma, R. Identification of microrna biomarkers in the blood of breast cancer patients based on microrna profiling. Gene 2017, 619, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.G.; Bhatt, D.; Taccioli, C.; Croce, C.M. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007, 297, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.E.; Davison, T.S.; John, J.; Cannon, T.; Sipos, B.; Maghnouj, A.; Labourier, E.; Hahn, S.A. microRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007, 26, 4442–4452. [Google Scholar] [CrossRef] [PubMed]
- Slee, E.A.; Harte, M.T.; Kluck, R.M.; Wolf, B.B.; Casiano, C.A.; Newmeyer, D.D.; Wang, H.G.; Reed, J.C.; Nicholson, D.W.; Alnemri, E.S.; et al. Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 1999, 144, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Korfali, N.; Ruchaud, S.; Loegering, D.; Bernard, D.; Dingwall, C.; Kaufmann, S.H.; Earnshaw, W.C. Caspase-7 gene disruption reveals an involvement of the enzyme during the early stages of apoptosis. J. Biol. Chem. 2004, 279, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Belaguli, N.; Berger, D.H. microRNA and colorectal cancer. World J. Surg. 2009, 33, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. microRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Akiyama, Y.; Hashimoto, Y.; Fukamachi, H.; Yuasa, Y. Mir-212 is downregulated and suppresses methyl-cpg-binding protein MECP2 in human gastric cancer. Int. J. Cancer 2010, 127, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Cheng, H.; Wirth, P.; Counsell, A.; Marcrom, S.R.; Wood, C.B.; Pohlmann, P.R.; Gilbert, J.; Murphy, B.; Yarbrough, W.G.; et al. Regulation of heparin-binding egf-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS ONE 2010, 5, e12702. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Majeti, B.K.; Acevedo, L.M.; Murphy, E.A.; Mukthavaram, R.; Scheppke, L.; Huang, M.; Shields, D.J.; Lindquist, J.N.; Lapinski, P.E.; et al. microRNA-132-mediated loss of p120rasgap activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010, 16, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Henry, J.C.; Jiang, J.; Esau, C.; Gusev, Y.; Lerner, M.R.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem. Biophys. Res. Commun. 2011, 406, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.K.; Veremeyko, T.; Patel, N.; Lemere, C.A.; Walsh, D.M.; Esau, C.; Vanderburg, C.; Krichevsky, A.M. De-repression of foxo3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer’s disease. Hum. Mol. Genet. 2013, 22, 3077–3092. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bian, C.; Liao, L.; Li, J.; Zhao, R.C. Mir-17-5p promotes human breast cancer cell migration and invasion through suppression of HPH1. Breast Cancer Res. Treat. 2011, 126, 565–575. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. C-myc-regulated micrornas modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Thomson, J.M.; Wong, H.Y.; Hammond, S.M.; Hogan, B.L. Transgenic over-expression of the microrna miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007, 310, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Ostenfeld, M.S.; Bramsen, J.B.; Lamy, P.; Villadsen, S.B.; Fristrup, N.; Sorensen, K.D.; Ulhoi, B.; Borre, M.; Kjems, J.; Dyrskjot, L.; et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in ta bladder tumors. Oncogene 2010, 29, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 2008, 13, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Lu, Y.; Pan, Z.; Chu, W.; Luo, X.; Lin, H.; Xiao, J.; Shan, H.; Wang, Z.; Yang, B. The muscle-specific micrornas miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell Sci. 2007, 120, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C.; Harland, R.M. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009, 23, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Slee, E.A.; Adrain, C.; Martin, S.J. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 2001, 276, 7320–7326. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Madhukrishna, B.; Adhya, A.K.; Keshari, S.; Mishra, S.K. Overexpression of caspase 7 is eralpha dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(cip). Oncogenesis 2016, 5, e219. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.G.; Cullen, S.P.; Sheridan, C.; Luthi, A.U.; Gerner, C.; Martin, S.J. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. USA 2008, 105, 12815–12819. [Google Scholar] [CrossRef] [PubMed]
- Svandova, E.; Lesot, H.; Vanden Berghe, T.; Tucker, A.S.; Sharpe, P.T.; Vandenabeele, P.; Matalova, E. Non-apoptotic functions of caspase-7 during osteogenesis. Cell Death Dis. 2014, 5, e1366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Leng, H.; Huang, J.; Du, Y.; Wang, Y.; Zang, W.; Chen, X.; Zhao, G. miR-337 regulates the proliferation and invasion in pancreatic ductal adenocarcinoma by targeting hoxb7. Diagn. Pathol. 2014, 9, 171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, Y.J.; Lee, C.H.; Lin, Y.P.; Huang, J.Y.; Su, C.C.; Chang, W.T.; Yang, B.C. Caspase-3 enhances lung metastasis and cell migration in a protease-independent mechanism through the ERK pathway. Int. J. Cancer 2008, 123, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Giles, C.B.; Girija-Devi, R.; Dozmorov, M.G.; Wren, J.D. Mircox: A database of miRNA-mRNA expression correlations derived from rna-seq meta-analysis. BMC Bioinform. 2013, 14 (Suppl. S14), S17. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Azevedo-Pouly, A.C.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.K.; Doseff, A.I.; Schmittgen, T.D. MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells. Int. J. Mol. Sci. 2018, 19, 1206. https://doi.org/10.3390/ijms19041206
Park JK, Doseff AI, Schmittgen TD. MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells. International Journal of Molecular Sciences. 2018; 19(4):1206. https://doi.org/10.3390/ijms19041206
Chicago/Turabian StylePark, Jong Kook, Andrea I. Doseff, and Thomas D. Schmittgen. 2018. "MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells" International Journal of Molecular Sciences 19, no. 4: 1206. https://doi.org/10.3390/ijms19041206
APA StylePark, J. K., Doseff, A. I., & Schmittgen, T. D. (2018). MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells. International Journal of Molecular Sciences, 19(4), 1206. https://doi.org/10.3390/ijms19041206