Venomics-Accelerated Cone Snail Venom Peptide Discovery
Abstract
1. Introduction
2. Venomics: Heralding a New Era of Venom Bioprospecting
2.1. Cone Snail Venom Duct Transcriptomics
2.2. Proteomics—Exploring the Complexity of Expressed Venom Peptides
2.3. Mass Spectrometry in Venom Characterisation
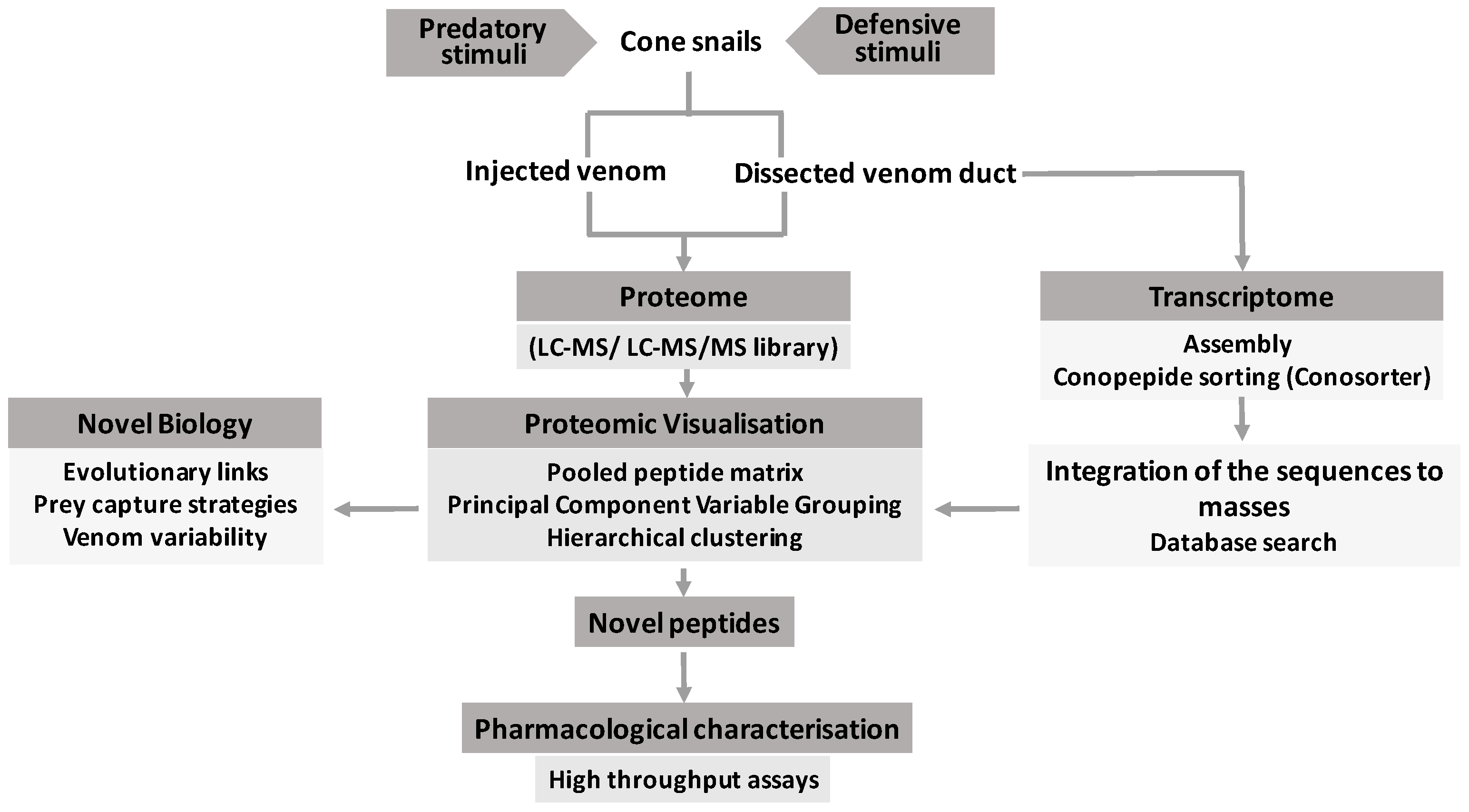
2.4. Omics Data Integration
3. Complex Venom Processing Revealed through Multi-Omics Studies
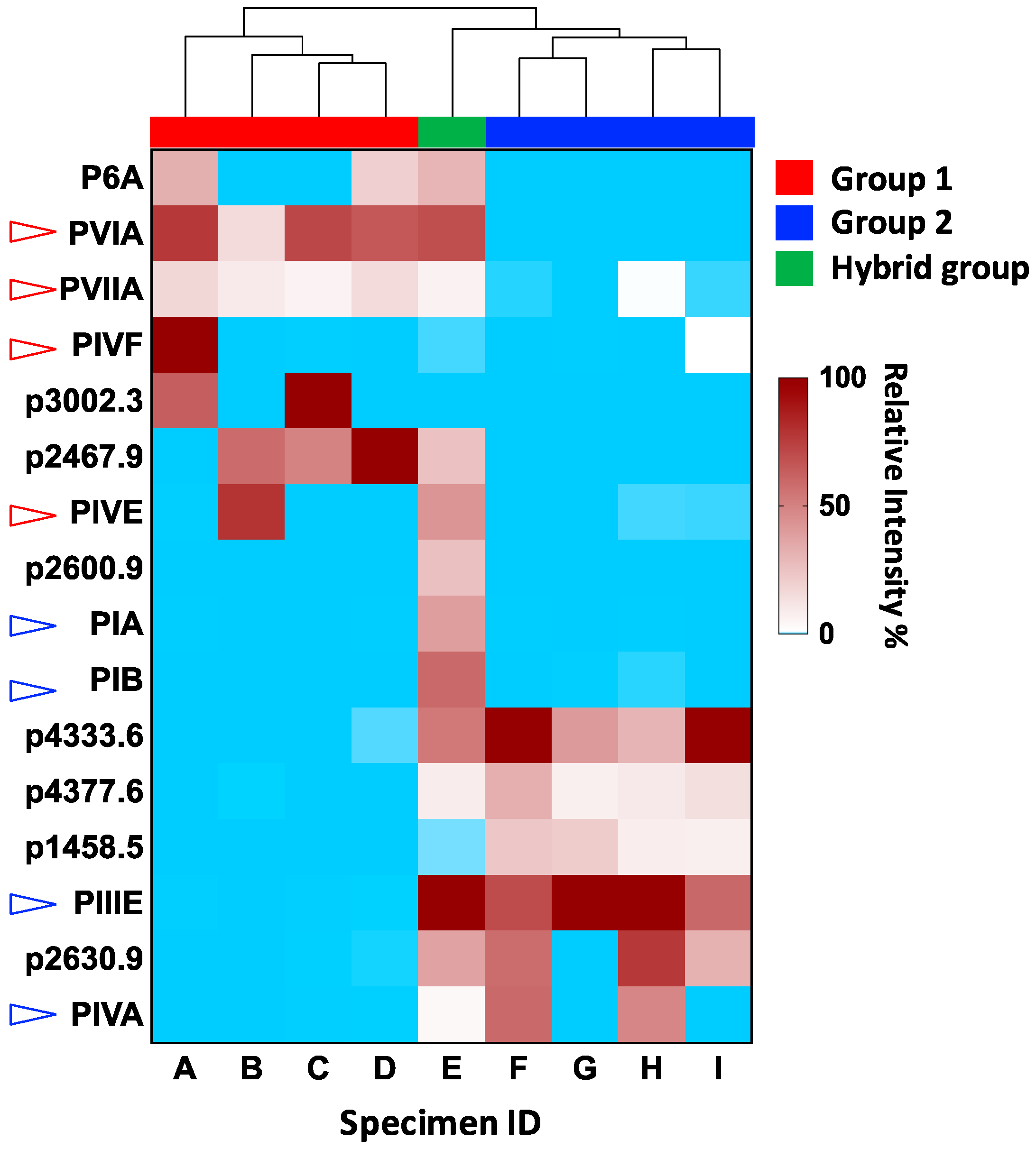
Proteomic Data Visualization Reveals a Remarkable Venom Variability
4. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PTM | Post-translational modification |
| ACE | Angiotensin-converting enzyme |
| LC/MS | Liquid chromatography/mass spectrometry |
| u-HPLC | Ultra-high-performance liquid chromatography |
| Q-TOF-MS | Quadrupole-time-of-flight mass spectrometry |
| DDA | Data-dependent acquisition |
| IDA | Information-Dependent acquisition |
| VPP | Variable peptide processing |
| PCA | Principal component analysis |
References
- Harvey, A.L. Natural products in drug discovery. Drug Discov. Today 2008, 13, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Toxins and drug discovery. Toxins 2014, 92, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Garcia, M.L. Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Pope, J.E.; Deer, T.R. Ziconotide: A clinical update and pharmacologic review. Expert Opin. Pharmacother. 2013, 14, 9957–9966. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.K.; Lewis, R.J.; Alewood, D.; Drinkwater, R.; Palant, E.; Patterson, M.; Yaksh, T.L.; McCumber, D.; Smith, M.T. Anti-allodynic efficacy of the chi-conopeptide, Xen2174, in rats with neuropathic pain. Pain 2005, 118, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.E.; White, H.S.; Wilcox, K.S. The effect of CGX-1007 and CI-1041, novel nmda receptor antagonists, on NMDA receptor-mediated EPSCs. Epilepsy Res. 2004, 59, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lubbers, N.L.; Campbell, T.J.; Polakowski, J.S.; Bulaj, G.; Layer, R.T.; Moore, J.; Gross, G.J.; Cox, B.F. Postischemic administration of CGX-1051, a peptide from cone snail venom, reduces infarct size in both rat and dog models of myocardial ischemia and reperfusion. J. Cardiovasc. Pharmacol. 2005, 46, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Sandall, D.W.; Satkunanathan, N.; Keays, D.A.; Polidano, M.A.; Liping, X.; Pham, V.; Down, J.G.; Khalil, Z.; Livett, B.G.; Gayler, K.R. A novel α-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry 2003, 42, 6904–6911. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Westermann, J.C.; Halai, R.; Wang, C.K.; Craik, D.J. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 2007, 24, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2011, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F. Differentiation of venoms of predatory marine gastropods: Divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J. Mol. Evol. 2008, 67, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V.; Gilly, W.F.; Schulz, J.R. Intraspecific variation of venom injected by fish-hunting Conus snails. J. Exp. Biol. 2005, 208, 2873–2883. [Google Scholar] [CrossRef] [PubMed]
- Romeo, C.; Di Francesco, L.; Oliverio, M.; Palazzo, P.; Massilia, G.R.; Ascenzi, P.; Polticelli, F.; Schininà, M.E. Conus ventricosus venom peptides profiling by HPLC-MS: A new insight in the intraspecific variation. J. Sep. Sci. 2008, 31, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable inter-and intra-species complexity of conotoxins revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Biass, D.; Stöcklin, R.; Favreau, P. Dramatic intraspecimen variations within the injected venom of Conus consors: An unsuspected contribution to venom diversity. Toxicon 2010, 55, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Jin, A.H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative venomics reveals the complex prey capture strategy of the piscivorous cone snail Conus catus. J. Proteome Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Dutertre, S.; Lewis, R.J.; Marí, F. Intraspecific variations in Conus purpurascens injected venom using LC/MALDI-TOF-MS and LC-ESI-TripleTOF-MS. Anal. Bioanal. Chem. 2015, 407, 6105–6116. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Mari, F.; Lewis, R.J. Accelerated proteomic visualization of individual predatory venoms of Conus purpurascens reveals separately evolved predation-evoked venom cabals. Sci. Rep. 2018, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Lewis, R.J. Cone snail biology, bioprospecting and conservation. In Snails: Biology, Ecology and Conservation; Nova Science Publisher’s: Hauppauge, NY, USA, 2012; pp. 85–105. [Google Scholar]
- Clark, R.J.; Fischer, H.; Dempster, L.; Daly, N.L.; Rosengren, K.J.; Nevin, S.T.; Meunier, F.A.; Adams, D.J.; Craik, D.J. Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the alpha-conotoxin MII. Proc. Natl. Acad. Sci. USA 2005, 102, 13767–13772. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.J.; Jensen, J.; Nevin, S.T.; Callaghan, B.P.; Adams, D.J.; Craik, D.J. The engineering of an orally active conotoxin for the treatment of neuropathic pain. Angew. Chem. Int. Ed. Engl. 2010, 49, 6545–6548. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, J.; Pi, C.; Zeng, X.; Zhou, M.; Jiang, X.; Chen, S.; Ren, Z.; Xu, A. Identification of a novel M-superfamily conotoxin with the ability to enhance tetrodotoxin sensitive sodium currents. Arch. Toxicol. 2009, 83, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.R.; Haystead, T.A. Molecular biologist’s guide to proteomics. Microbiol. Mol. Biol. Rev. 2002, 66, 39–63. [Google Scholar] [CrossRef] [PubMed]
- Jensen, O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006, 7, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 131, 341. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Next-generation snake venomics: Protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteom. 2014, 11, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I. Development and optimization of FLIPR high throughput calcium assays for ion channels and GPCRs. In Calcium Signaling; Springer: Amsterdam, The Netherlands, 2012; pp. 45–82. [Google Scholar]
- Calvete, J.J. Venomics: Integrative venom proteomics and beyond. Biochem. J. 2017, 474, 611–634. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Dutertre, S.; Jin, A.H.; Lewis, R.J.; Taft, R.J.; Alewood, P.F. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genom. 2013, 14, 708. [Google Scholar] [CrossRef] [PubMed]
- Koua, D.; Brauer, A.; Laht, S.; Kaplinski, L.; Favreau, P.; Remm, M.; Lisacek, F.; Stöcklin, R. ConoDictor: A tool for prediction of conopeptide superfamilies. Nucleic Acids Res. 2012, 40, W238–W241. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Lewis, R.J. An efficient transcriptome analysis pipeline to accelerate venom peptide discovery and characterisation. Toxicon 2015, 107, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol. Cell. Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Aman, J.W.; Imperial, J.S.; Ueberheide, B.; Zhang, M.M.; Aguilar, M.; Taylor, D.; Watkins, M.; Yoshikami, D.; Showers-Corneli, P.; Safavi-Hemami, H.; et al. Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc. Natl. Acad. Sci. USA 2015, 112, 5087–5092. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Hemami, H.; Gajewiak, J.; Karanth, S.; Robinson, S.D.; Ueberheide, B.; Douglass, A.D.; Schlegel, A.; Imperial, J.S.; Watkins, M.; Bandyopadhyay, P.K.; et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. USA 2015, 112, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.; Kelley, W.P.; Rubakhin, S.S.; Bingham, J.P.; Sweedler, J.V.; Gilly, W.F. Anatomical correlates of venom production in Conus californicus. Biol. Bull. 2002, 203, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Oldrati, V.; Arrell, M.; Violette, A.; Perret, F.; Sprüngli, X.; Wolfender, J.L.; Stöcklin, R. Advances in venomics. Mol. Biosyst. 2016, 12, 3530–3543. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Hemami, H.; Young, N.D.; Williamson, N.A.; Purcell, A.W. Proteomic interrogation of venom delivery in marine cone snails: Novel insights into the role of the venom bulb. J. Proteome Res. 2010, 9, 5610–5619. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.J.; Reid-Bayliss, K.S.; Emond, M.J.; Loeb, L.A. Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl. 2014, 1. [Google Scholar] [CrossRef]
- Mardis, E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Yao, G.; Gao, B.M.; Fan, C.X.; Bian, C.; Wang, J.; Cao, Y.; Wen, B.; Zhu, Y.; Ruan, Z.; et al. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multi-transcriptome sequencing. GigaScience 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Lluisma, A.O.; Milash, B.A.; Moore, B.; Olivera, B.M.; Bandyopadhyay, P.K. Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar. Genom. 2012, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Harliwong, I.; Jones, A.; Miller, D.; Taft, R.J.; Alewood, P.F. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl. Acad. Sci. USA 2015, 112, E3782–E3791. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Safavi-Hemami, H.; Raghuraman, S.; Imperial, J.S.; Papenfuss, A.T.; Teichert, R.W.; Purcell, A.W.; Olivera, B.M.; Norton, R.S. Discovery by proteogenomics and characterization of an RF-amide neuropeptide from cone snail venom. J. Proteom. 2015, 114, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. Comparison of the venom peptides and their expression in closely related Conus species: Insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biol. Evol. 2015, 7, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Decaestecker, T.N.; Vande Casteele, S.R.; Wallemacq, P.E.; Van Peteghem, C.H.; Defore, D.L.; Van Bocxlaer, J.F. Information-dependent acquisition-mediated LC−MS/MS screening procedure with semiquantitative potential. Anal. Chem. 2004, 76, 6365–6373. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dekan, Z.; Smout, M.J.; Wilson, D.; Dutertre, S.; Vetter, I.; Lewis, R.J.; Loukas, A.; Daly, N.L.; Alewood, P.F. Conotoxin Φ-MiXXVIIA from the superfamily G2 employs a novel cysteine framework that mimics granulin and displays anti-apoptotic activity. Angew. Chem. 2017, 129, 15169–15172. [Google Scholar] [CrossRef]
- Reimers, C.; Lee, C.H.; Kalbacher, H.; Tian, Y.; Hung, C.H.; Schmidt, A.; Prokop, L.; Kauferstein, S.; Mebs, D.; Chen, C.C.; et al. Identification of a cono-RFamide from the venom of Conus textile that targets ASIC3 and enhances muscle pain. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Lu, A.; Bandyopadhyay, P.K.; Yandell, M.; Olivera, B.M.; Safavi-Hemami, H. The Venom Repertoire of Conus gloriamaris (Chemnitz, 1777), the Glory of the Sea. Mar. Drugs 2017, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Sanchez, J.C.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1995, 13, 19–50. [Google Scholar] [CrossRef]
- Mann, M. Origins of mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 678. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, Y.; Subramanian, R. Comparison of information-dependent acquisition, SWATH, and MSAll techniques in metabolite identification study employing ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal. Chem. 2014, 86, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Fanali, S. An overview to nano-scale analytical techniques: Nano-liquid chromatography and capillary electrochromatography. Electrophoresis 2017, 38, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Macek, B.; Lange, O.; Makarov, A.; Horning, S.; Mann, M. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 2007, 4, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Syka, J.E.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Yang, L.; Xu, S.; Wang, C. Various conotoxin diversifications revealed by a venomic study of Conus flavidus. Mol. Cell. Proteom. 2014, 13, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Ivosev, G.; Burton, L.; Bonner, R. Dimensionality reduction and visualization in principal component analysis. Anal. Chem. 2008, 80, 4933–4944. [Google Scholar] [CrossRef] [PubMed]
- Ringner, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B.; Dumas, E.; Piec, I.; Béchet, D.; Hébraud, M.; Hocquette, J.F. Assessment of hierarchical clustering methodologies for proteomic data mining. J. Proteome Res. 2007, 6, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Verhoeckx, K.; Bijlsma, S.; de Groene, E.M.; Witkamp, R.F.; van der Greef, J.; Rodenburg, R.J. A combination of proteomics, principal component analysis and transcriptomics is a powerful tool for the identification of biomarkers for macrophage maturation in the U937 cell line. Proteomics 2004, 4, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Turchetto, J.; Sequeira, A.F.; Ramond, L.; Peysson, F.; Brás, J.L.; Saez, N.J.; Duhoo, Y.; Blemont, M.; Guerreiro, C.I.P.D.; Quinton, L.; et al. High-throughput expression of animal venom toxins in Escherichia coli to generate a large library of oxidized disulphide-reticulated peptides for drug discovery. Microb. Cell Fact. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
| Developer | System | Read Length (bp) | Error Rate (%) |
|---|---|---|---|
| Illumina | Illumina Hi-Seq2000 | 2 × 150 bp | 0.1 |
| Illumina Hi-Seq3000/4000 | 2 × 150 bp | 0.1 | |
| Illumina Mi-Seq (Version 3) | 2 × 300 bp | 0.1 | |
| Thermo Fisher Scientific | Ion Torrent PGM 318 | 400 bp | 1 |
| Applied Biosystems | ABI 3730/ABI 3730xl | 500 bp | 0.4 |
| Pacific Biosciences | PacBio RS II | 10–20 kb | 10–15 |
| PacBio Sequel | >20 kb | 10–15 | |
| Oxford | Nanopore PromethION | 900 kb | 4 |
| Roche | 454 (discontinued in 2013) | 700 bp | 1 |
| Species | New Superfamily | Cysteine Framework | Sequencing | Reference |
|---|---|---|---|---|
| Conus arenatus Conus coronatus Conus rattus | MRFYM- | VI/VII | Illumina HiSeq 2000 | [13] |
| Conus arenatus Conus imperialis Conus lividus Conus quercinus Conus sponsalis Conus virgo | MKISL- | VI/VII | Illumina HiSeq 2000 | [13] |
| Conus marmoreus | N, B, H, E, F, H2, I4, M2, N2, O4, Q, R, U, W, X, Y2, Y3, Z | XV, VIIII, VI/VII, N, N, III, C1, C7 *, N, VI/VII, N, N, VI/VII, N, C2, C4 *, C2, C2 | Roche 454 | [15] |
| Conus catus | Cat-NSF1 | VI/VII | Roche 454 | [20] |
| Conus miles | SF-mi 1–8 | XIII, C8-novel, VI/VII, N, N, XV, NA, XIII | Roche 454 | [38] |
| Conus vexillum | NSVx1-4 | XIV, XXIV, VI/VII, XXVIII (novel) | Roche 454 | [40] |
| Conus betulinus | NSF-bt01–09 | IX, XV, VI/VII, XIV, VI/VII, IX, VIII, VI/VII, IX | ABI 3730 | [47] |
| Conus episcopatus | SF-Epi 1–16 | variable, III a, V, C7 *, V, V, V, XI, C7 *, N, C5 *, NA, XVIII, VI/VII, IV, C5 * | Illumina MiSeq | [48] |
| Conus tribblei and Conus lenavati | SF-01-04 | C12 *, C12 *, IX, XIII, | Illumina HiSeq 2000 | [49] |
| Conus pullicarius | J2 | XIV | Roche GS-FLX | [52] |
| Conus gloriamaris | MKAVA-, MSRLF-, MMLFM-, MLSML- | XXII, N, VIII, C12 * | Illumina HiSeq 2000 | [53] |
| Conus lenavati | SF-05-06 | XIV, C12 * | Illumina HiSeq 2000 | [54] |
| Conus andremenezi | Put.MGGRF, Put.MKAVA | N, C8 * | Illumina HiSeq 2000 | [55] |
| Conus andremenezi Conus praecellens | Put.MSGLR, Put.MUSGK | VI/VII, C6 * | Illumina HiSeq 2000 | [55] |
| Tool | Function |
|---|---|
| Database search algorithms | |
| PEAKS DB | Database search engine, run in parallel with de novo sequencing, to automatically validate the search results |
| ProteinPilot Software | Enables peptide identification while considering PTMs, non-tryptic cleavages, and amino acid substitutions. Supports quantification using iTRAQ, mTRAQ, and SILAC |
| Mascot | Peptide mass fingerprinting and MS/MS database searching |
| Protein Prospector | Proteomic analysis tools including “Batch-Tag” for instrument- and fragmentation mode-optimised analysis |
| MassMatrix | Search algorithm for tandem MS data that ranks peptide and protein matches |
| Byonic | MS/MS data search for fragment identifications to produce protein scores and identification probabilities |
| MS/MS peptide quantification and visualisation software | |
| MarkerView Software | Statistical analysis and visualisation of quantitative mass spec data sets from proteomic profiling applications |
| MultiQuant Software | Quantitation and targeted visualisation of TripleTOF or QTRAP data, including MRM and SWATH acquisition |
| MaxQuant | Quantitative proteomics tool for analysis of label-free and SILAC-based proteomics data |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Himaya, S.W.A.; Lewis, R.J. Venomics-Accelerated Cone Snail Venom Peptide Discovery. Int. J. Mol. Sci. 2018, 19, 788. https://doi.org/10.3390/ijms19030788
Himaya SWA, Lewis RJ. Venomics-Accelerated Cone Snail Venom Peptide Discovery. International Journal of Molecular Sciences. 2018; 19(3):788. https://doi.org/10.3390/ijms19030788
Chicago/Turabian StyleHimaya, S. W. A., and Richard J. Lewis. 2018. "Venomics-Accelerated Cone Snail Venom Peptide Discovery" International Journal of Molecular Sciences 19, no. 3: 788. https://doi.org/10.3390/ijms19030788
APA StyleHimaya, S. W. A., & Lewis, R. J. (2018). Venomics-Accelerated Cone Snail Venom Peptide Discovery. International Journal of Molecular Sciences, 19(3), 788. https://doi.org/10.3390/ijms19030788






