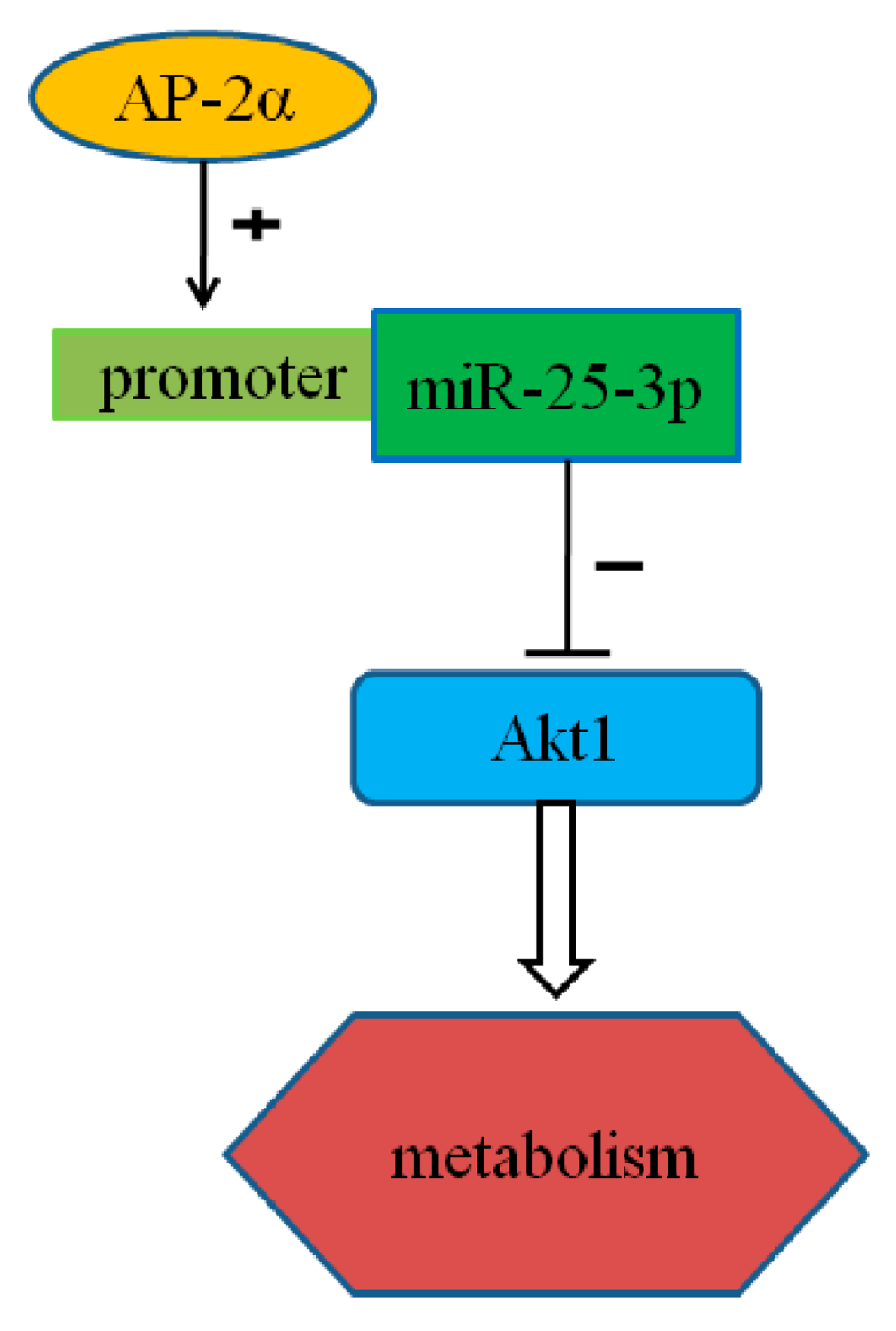
miR-25-3p, Positively Regulated by Transcription Factor AP-2α, Regulates the Metabolism of C2C12 Cells by Targeting Akt1
Abstract
1. Introduction
2. Results
2.1. miR-25 Is Highly Conserved in Mammals
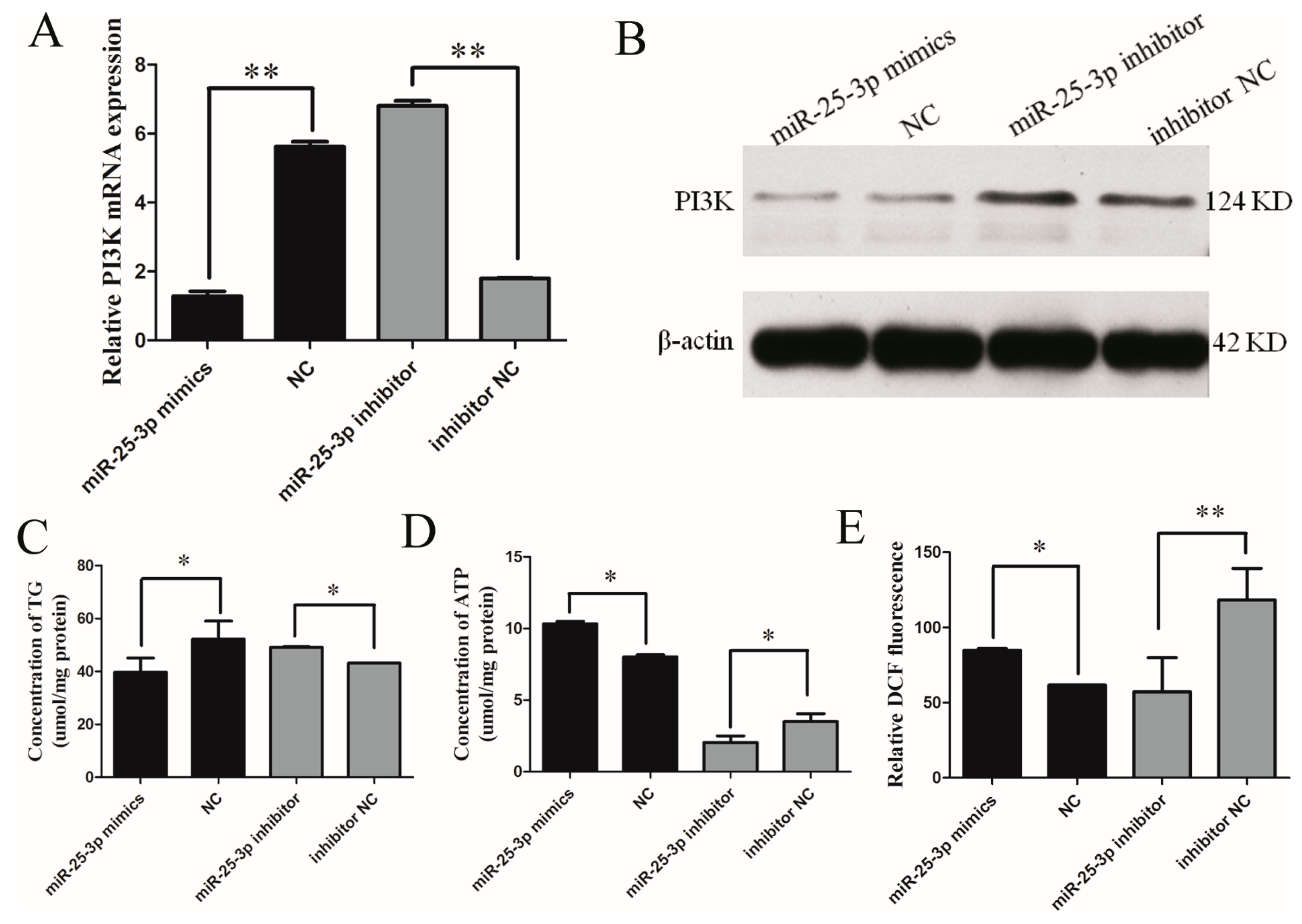
2.2. Effects of miR-25 on the Metabolism of C2C12 Cells
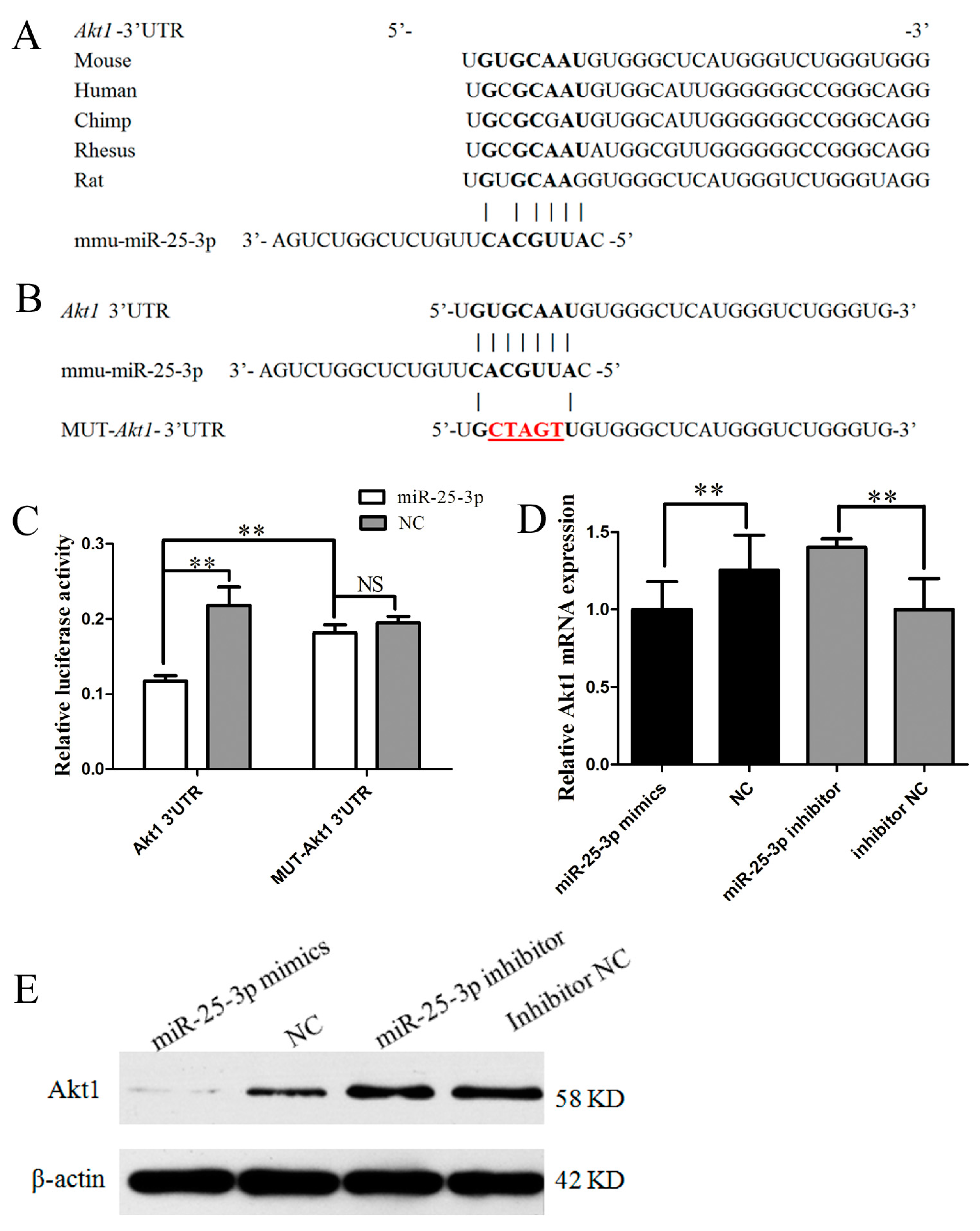
2.3. miR-25-3p Directly Targets Akt1
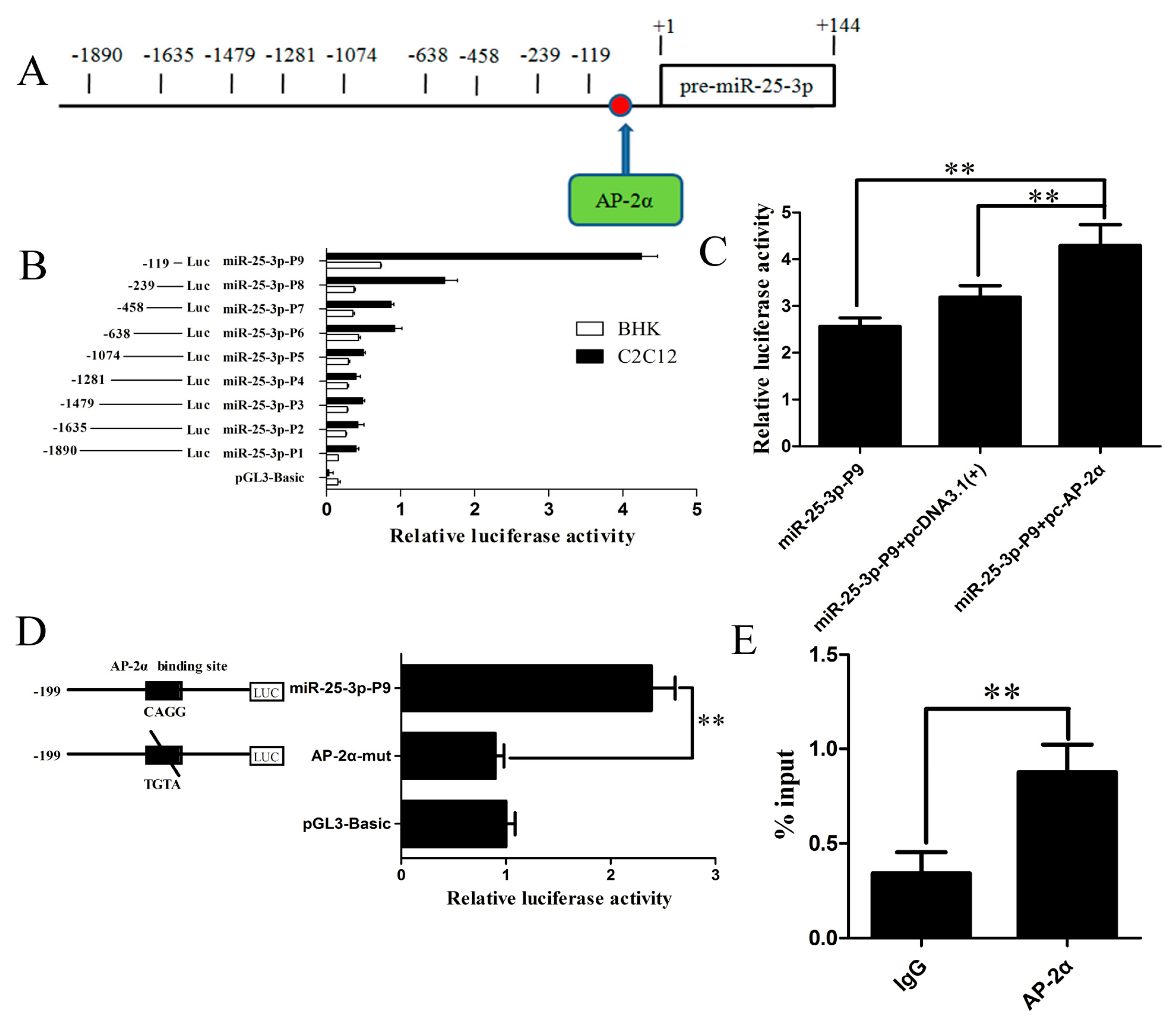
2.4. Identification and Characterization of the Mouse miR-25-3p Promoter
2.5. The Transcription Factor AP-2α Binds to the Core Promoter of Mouse miR-25-3p
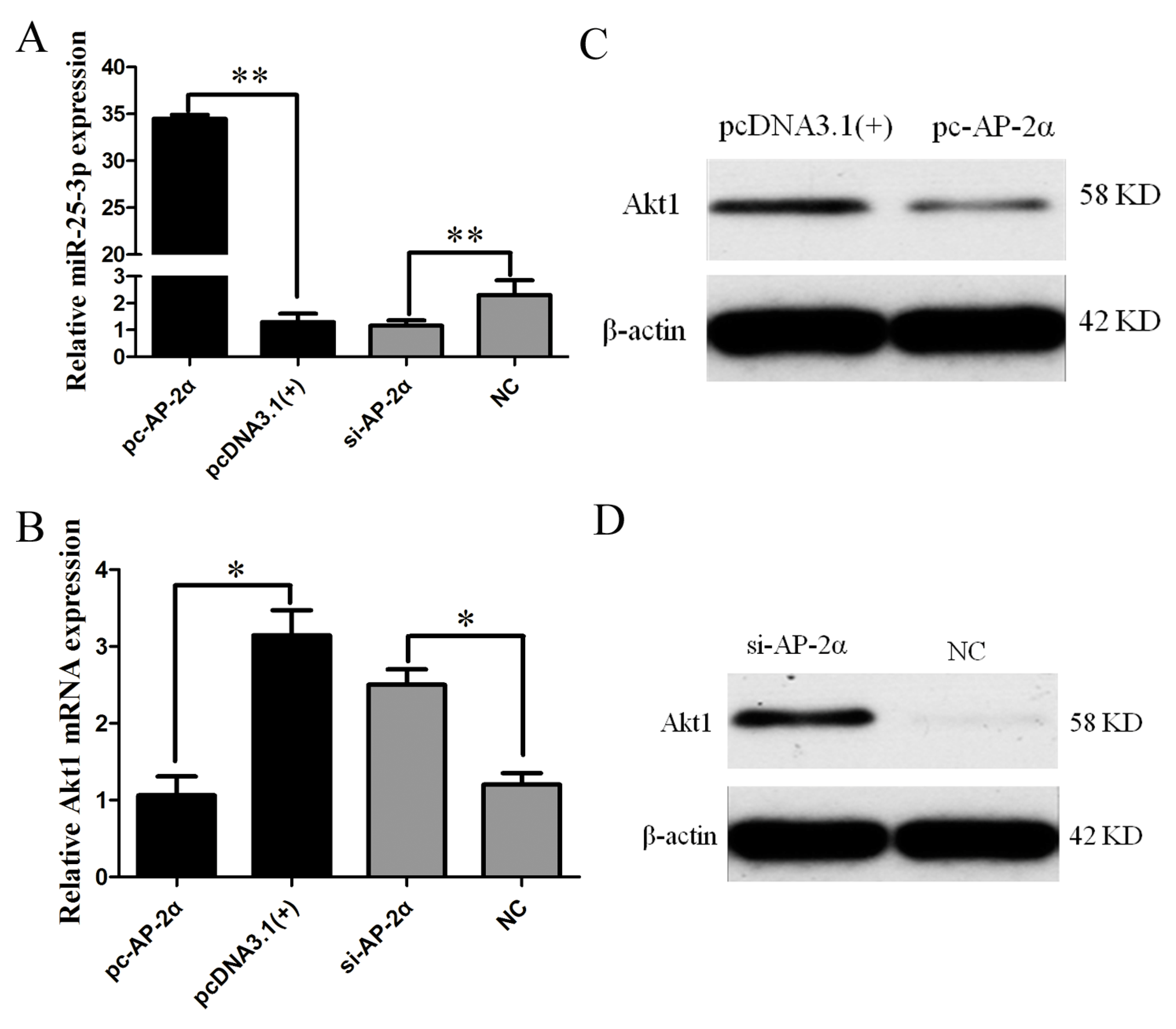
2.6. AP-2α Regulates miR-25-3p and Akt1 Expression
3. Discussion
4. Materials and Methods
4.1. miRNA, Small RNA Oligonucleotide Synthesis, and Plasmid Construction
4.2. Cell Culture and Luciferase Reporter Assays
4.3. Triglyceride Content, ATP, and Reactive Oxygen Species (ROS) Assays
4.4. Chromatin Immunoprecipitation (ChIP)
4.5. RNA Isolation and qRT-PCR
4.6. Protein Isolation and Western Blotting
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Malan-Muller, S.; Hemmings, S.M.; Seedat, S. Big effects of small RNAs: A review of microRNAs in anxiety. Mol. Neurobiol. 2013, 47, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.; Sontheimer, E. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ning, B.; Gao, L.; Liu, R.; Liu, Y.; Zhang, N.; Chen, Z. microRNAs in spinal cord injury: Potential roles and therapeutic implications. Int. J. Biol. Sci. 2014, 10, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.; Murgia, M.; Nagaraj, N.; Treebak, J.; Cox, J.; Mann, M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell Proteom. 2015, 14, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Song, Q.; Li, H.; Lou, Y.; Wang, L. Correction: Circulating miR-25-3p and miR-451a May Be Potential Biomarkers for the Diagnosis of Papillary Thyroid Carcinoma. PLoS ONE 2015, 10, e0135549. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Nagahara, M.; Sato, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Sugihara, K.; Doki, Y.; et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin. Cancer Res. 2012, 18, 3054–3070. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Bronk, S.F.; Smoot, R.L.; Fingas, C.D.; Werneburg, N.W.; Roberts, L.R.; Mott, J.L. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology 2012, 55, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Meng, X.; Li, H.; Liu, W.; Shen, S.; Gao, Z. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin. Transl. Oncol. 2014, 16, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Y.; Yang, L.; Jiang, R.; Li, W. MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol. Cell. Biochem. 2014, 385, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Wahlquist, C.; Jeong, D.; Rojas-Muñoz, A.; Kho, C.; Lee, A.; Mitsuyama, S.; van Mil, A.; Park, W.; Sluijter, J.; Doevendans, P.; et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature 2014, 508, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Huang, B.; Ma, X.; Wang, S.; Feng, J.; Lv, F.; Liu, Y.; Liu, Y.; Li, C.; Liang, D.; et al. MiR-25 protects cardiomyocytes against oxidative damage by targeting the mitochondrial calcium uniporter. Int. J. Mol. Sci. 2015, 16, 5420–5433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Liu, J.; Han, P.; Li, X.; Bai, H.; Zhang, C.; Sun, X.; Teng, Y.; Zhang, Y.; et al. Variations in MicroRNA-25 Expression Influence the Severity of Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2017, 28, 3627–3638. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [PubMed]
- Zhang, Y.; Peng, Z.; Zhao, Y.; Chen, L. microRNA-25 Inhibits Cell Apoptosis of Human Gastric Adenocarcinoma Cell Line AGS via Regulating CCNE1 and MYC. Med. Sci. Monit. 2016, 22, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Yuan, X.; Yuan, J.; Liu, Q.; Dai, M.; Shen, C.; Ma, J.; Liao, Y.; Jiang, W. miR-25 promotes glioblastoma cell proliferation and invasion by directly targeting NEFL. Mol. Cell. Biochem. 2015, 409, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zuo, Z.; Lu, X.; Wang, L.; Wang, H.; Zhu, Z. miR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 2012, 27, 594–598. [Google Scholar] [PubMed]
- Brett, J.O.; Renault, V.M.; Rafalski, V.A.; Webb, A.E.; Brunet, A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging 2011, 3, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lu, X.; Ding, F. microRNA regulatory mechanism by which PLLA aligned nanofibers influence PC12 cell differentiation. J. Neural Eng. 2015, 12, 046010. [Google Scholar] [CrossRef] [PubMed]
- Rottiers, V.; Näär, A. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Zhang, J.; Wu, J.; Xu, Y. Isoflurane promotes glucose metabolism through up-regulation of miR-21 and suppresses mitochondrial oxidative phosphorylation in ovarian cancer cells. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Kokoza, V.; Zhang, C.; Aksoy, E.; Raikhel, A. MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA 2017, 114, E8017–E8024. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.; Lee, Y.; Kita, K.; Ochi, T.; Zeller, K.; De Marzo, A.; van Eyk, J.; Mendell, J.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Pessin, J.E.; Saltiel, A.R. Signaling pathways in insulin action: Molecular targets of insulin resistance. J. Clin. Investig. 2000, 106, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Courtnay, R.; Ngo, D.; Malik, N.; Ververis, K.; Tortorella, S.; Karagiannis, T. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Knight, Z.; Shokat, K. Chemically targeting the PI3K family. Biochem. Soc. Trans. 2007, 35 Pt 2, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Carrière, A.; Fernandez, Y.; Rigoulet, M.; Pénicaud, L.; Casteilla, L. Inhibition of preadipocyte proliferation by mitochondrial reactive oxygen species. FEBS Lett. 2003, 550, 163–167. [Google Scholar] [CrossRef]
- Carrière, A.; Carmona, M.; Fernandez, Y.; Rigoulet, M.; Wenger, R.; Pénicaud, L.; Casteilla, L. Mitochondrial reactive oxygen species control the transcription factor CHOP-10/GADD153 and adipocyte differentiation: A mechanism for hypoxia-dependent effect. J. Biol. Chem. 2004, 279, 40462–40469. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Choi, K.; Kim, J.; Han, X.; Piao, Y.; Jeong, J.; Choe, W.; Kang, I.; Ha, J.; Forman, H.; et al. Endogenous hydrogen peroxide regulates glutathione redox via nuclear factor erythroid 2-related factor 2 downstream of phosphatidylinositol 3-kinase during muscle differentiation. Am. J. Pathol. 2008, 172, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Won, H.; Lim, S.; Jang, M.; Kim, Y.; Rashid, M.; Jyothi, K.; Dashdorj, A.; Kang, I.; Ha, J.; Kim, S. Peroxiredoxin-2 upregulated by NF-κB attenuates oxidative stress during the differentiation of muscle-derived C2C12 cells. Antioxid. Redox Signal. 2012, 16, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Thorvaldsen, J.L.; Chu, Q.; Feng, F.; Birnbaum, M.J. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 2001, 276, 38349–38352. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.; McGraw, T.E. The Akt kinases Isoform specificity in metabolism and cancer. Cell Cycle 2009, 8, 2502–2508. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.S.; Xu, P.-Z.; Gottlob, K.; Chen, M.-L.; Sokol, K.; Shiyanova, T.; Roninson, I.; Weng, W.; Suzuki, R.; Tobe, K.; et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the akt1 gene. Genes Dev. 2001, 15, 2203–2208. [Google Scholar] [CrossRef] [PubMed]
- Beurel, E.; Grieco, S.; Jope, R. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Agius, L. Role of glycogen phosphorylase in liver glycogen metabolism. Mol. Asp. Med. 2015, 46, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, L.; Ma, C.; Huang, Y.; Zhu, G.; Chen, Q. MiR-25-3p attenuates the proliferation of tongue squamous cell carcinoma cell line Tca8113. Asian Pac. J. Trop. Med. 2013, 6, 743–747. [Google Scholar] [CrossRef]
- Zheng, H.; Dong, X.; Liu, N.; Xia, W.; Zhou, L.; Chen, X.; Yang, Z.; Chen, X. Regulation and mechanism of mouse miR-130a/b in metabolism-related inflammation. Int. J. Biochem. Cell Biol. 2016, 74, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ye, J.; Ashraf, U.; Li, Y.; Chen, H.; Song, Y.; Cao, S. Transcriptional regulation of miR-15b by c-Rel and CREB in Japanese encephalitis virus infection. Sci. Rep. 2016, 6, 22581. [Google Scholar] [CrossRef] [PubMed]
- Bueno, M.; Gómez de Cedrón, M.; Laresgoiti, U.; Fernández-Piqueras, J.; Zubiaga, A.; Malumbres, M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol. Cell. Biol. 2010, 30, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Wenke, A.; Bosserhoff, A. Roles of AP-2 transcription factors in the regulation of cartilage and skeletal development. FEBS J. 2010, 277, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Eckert, D.; Buhl, S.; Weber, S.; Jäger, R.; Schorle, H. The AP-2 family of transcription factors. Genome Biol. 2005, 6, 246. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Wang, C.; Tjian, R. Positive and negative regulation of transcription in vitro: Enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell 1987, 50, 847–861. [Google Scholar] [CrossRef]
- Holt, E.; Lane, M. Downregulation of repressive CUP/AP-2 isoforms during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2001, 288, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Vujanovic, N. Soluble TNF Regulates TACE via AP-2α Transcription Factor in Mouse Dendritic Cells. J. Immunol. 2017, 198, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Li, C.; Zhang, A.; Hou, L.; Yang, J.; Hu, H. Regulation of DEK expression by AP-2α and methylation level of DEK promoter in hepatocellular carcinoma. Oncol. Rep. 2016, 36, 2382–2390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, C.; Geng, J.; Wei, X.; Zhang, R.; Jiang, S. MiR-144-3p regulates osteogenic differentiation and proliferation of murine mesenchymal stem cells by specifically targeting Smad4. FEBS Lett. 2016, 590, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Deng, B.; Zhang, F.; Chen, K.; Wen, J.; Huang, H.; Liu, W.; Ye, S.; Wang, L.; Yang, Y.; Gong, P.; et al. MyoD promotes porcine PPARgamma gene expression through an E-box and a MyoD-binding site in the PPARgamma promoter region. Cell Tissue Res. 2016, 365, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Cheng, X.; Peng, Y.; Zheng, R.; Chai, J.; Jiang, S. STAT5a promotes the transcription of mature mmu-miR-135a in 3T3-L1 cells by binding to both miR-135a-1 and miR-135a-2 promoter elements. Int. J. Biochem. Cell Biol. 2016, 77 Pt A, 109–119. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Chen, K.; Tao, H.; Kang, T.; Xiong, Q.; Zeng, Q.; Liu, Y.; Jiang, S.; Chen, M. miR-25-3p, Positively Regulated by Transcription Factor AP-2α, Regulates the Metabolism of C2C12 Cells by Targeting Akt1. Int. J. Mol. Sci. 2018, 19, 773. https://doi.org/10.3390/ijms19030773
Zhang F, Chen K, Tao H, Kang T, Xiong Q, Zeng Q, Liu Y, Jiang S, Chen M. miR-25-3p, Positively Regulated by Transcription Factor AP-2α, Regulates the Metabolism of C2C12 Cells by Targeting Akt1. International Journal of Molecular Sciences. 2018; 19(3):773. https://doi.org/10.3390/ijms19030773
Chicago/Turabian StyleZhang, Feng, Kun Chen, Hu Tao, Tingting Kang, Qi Xiong, Qianhui Zeng, Yang Liu, Siwen Jiang, and Mingxin Chen. 2018. "miR-25-3p, Positively Regulated by Transcription Factor AP-2α, Regulates the Metabolism of C2C12 Cells by Targeting Akt1" International Journal of Molecular Sciences 19, no. 3: 773. https://doi.org/10.3390/ijms19030773
APA StyleZhang, F., Chen, K., Tao, H., Kang, T., Xiong, Q., Zeng, Q., Liu, Y., Jiang, S., & Chen, M. (2018). miR-25-3p, Positively Regulated by Transcription Factor AP-2α, Regulates the Metabolism of C2C12 Cells by Targeting Akt1. International Journal of Molecular Sciences, 19(3), 773. https://doi.org/10.3390/ijms19030773



