Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors
Abstract
:1. Introduction
2. Optimization of E. coli System for Heterologous Expression of Chloroplast FtsZ
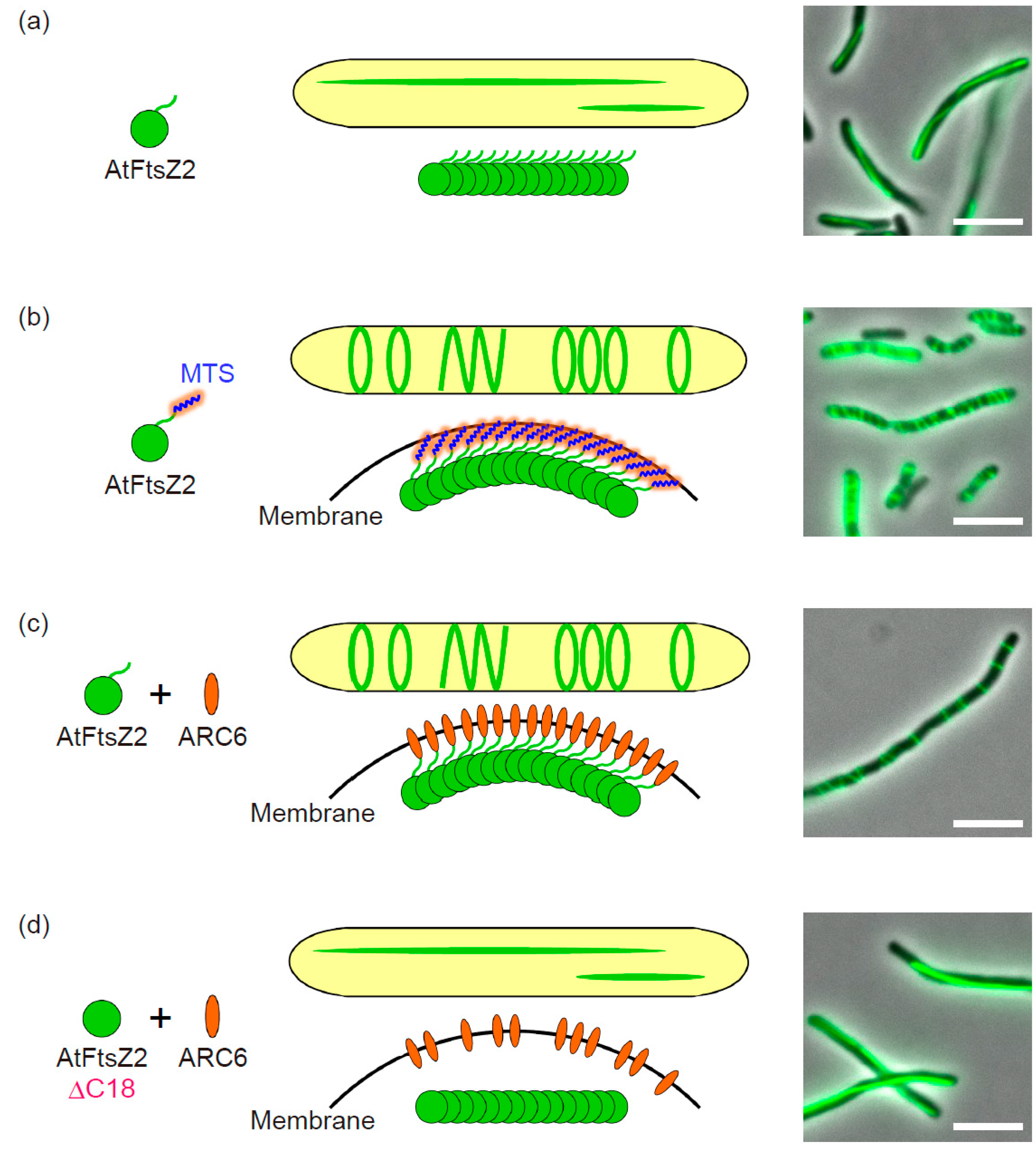
2.1. Fluorescent Tagging of AtFtsZ2 and Culture Condition
2.2. N-Terminal Region of AtFtsZ2
2.3. Membrane-Tethering of AtFtsZ2
3. The Function of Negative and Positive Contributors in Bacterial Reconstitution Systems
3.1. The Negative Regulator ARC3
3.2. The Positive Regulator ARC6
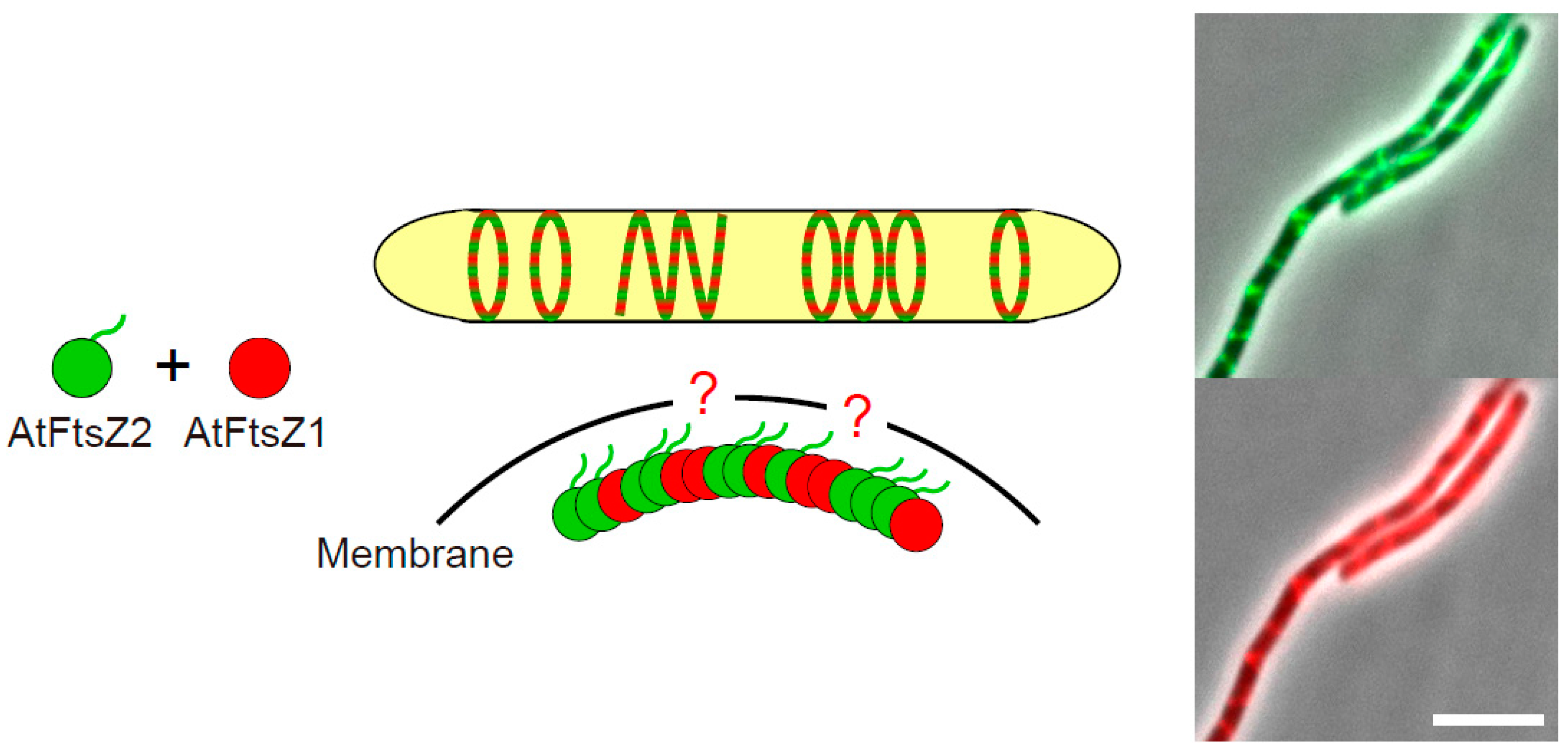
3.3. The Positive Regulator AtFtsZ1
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J. The number, speed, and impact of plastid endosymbiosis in eukaryotic evolution. Annu. Rev. Plant Biol. 2013, 64, 583–607. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nakanishi, H.; Kabeya, Y. Structure, regulation, and evolution of the plastid division machinery. Int. Rev. Cell Mol. Biol. 2011, 291, 115–153. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Pyke, K.A. Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 2014, 65, 443–472. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; MacCready, J.S.; Ducat, D.C.; Osteryoung, K.W. The molecular machinery of chloroplast division. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, T.; Kuroiwa, H.; Sakai, A.; Takahashi, H.; Toda, K.; Itoh, R. The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 1998, 181, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Kuroiwa, H.; Misumi, O.; Yoshida, M.; Ohnuma, M.; Fujiwara, T.; Yagisawa, F.; Hirooka, S.; Imoto, Y.; Matsushita, K.; et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 2010, 329, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.W.; Errington, J. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 2009, 7, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [PubMed]
- Mingorance, J.; Rivas, G.; Vélez, M.; Gómez-Puertas, P.; Vicente, M. Strong FtsZ is with the force: Mechanisms to constrict bacteria. Trends Microbiol. 2010, 18, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Terbush, A.D.; MacCready, J.S.; Chen, C.; Ducat, D.C.; Osteryoung, K.W. Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiol. 2018, 176, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Vierling, E. Conserved cell and organelle division. Nature 1995, 376, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Kuroiwa, H.; Misumi, O.; Nishida, K.; Yagisawa, F.; Fujiwara, T.; Nanamiya, H.; Kawamura, F.; Kuroiwa, T. Isolated chloroplast division machinery can actively constrict after stretching. Science 2006, 313, 1435–1438. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of contractile FtsZ rings in liposomes. Science 2008, 320, 792–794. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Mogi, Y.; TerBush, A.D.; Osteryoung, K.W. Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2016, 2, 16095. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Stokes, K.D.; Rutherford, S.M.; Percival, A.L.; Lee, W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial FtsZ. Plant Cell 1998, 10, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Olson, B.J.; Wang, Q.; Osteryoung, K.W. GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 2010, 285, 20634–20643. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Johnson, C.B.; Vitha, S.; Holzenburg, A. Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett. 2010, 584, 166–172. [Google Scholar] [CrossRef] [PubMed]
- TerBush, A.D.; Osteryoung, K.W. Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z ring structure and remodeling. J. Cell Biol. 2012, 199, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nozaki, H.; Nishida, K.; Nishida, K.; Matsuzaki, M.; Kuroiwa, T. Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: The duplication of FtsZ is implicated in endosymbiosis. J. Mol. Evol. 2004, 58, 291–303. [Google Scholar] [CrossRef] [PubMed]
- TerBush, A.D.; Yoshida, Y.; Osteryoung, K.W. FtsZ in chloroplast division: Structure, function and evolution. Curr. Opin. Cell Biol. 2013, 25, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Glynn, J.M.; Olson, B.J.; Stokes, K.D.; Osteryoung, K.W. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2009, 2, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Karamoko, M.; El-Kafafi, E.S.; Mandaron, P.; Lerbs-Mache, S.; Falconet, D. Multiple FtsZ2 isoforms involved in chloroplast division and biogenesis are developmentally associated with thylakoid membranes in Arabidopsis. FEBS Lett. 2011, 585, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Koizumi, M.; Kuroki, K.; Mochizuki, M.; Fujimoto, H.; Ohta, H.; Masuda, T.; Takamiya, K. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004, 45, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Maple, J.; Vojta, L.; Soll, J.; Møller, S.G. ARC3 is a stromal Z ring accessory protein essential for plastid division. EMBO Rep. 2007, 8, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Schmitz, A.J.; Kadirjan-Kalbach, D.K.; TerBush, A.D.; Osteryoung, K.W. Chloroplast division protein ARC3 regulates chloroplast FtsZ ring assembly and positioning in Arabidopsis through interaction with FtsZ2. Plant Cell 2013, 25, 1787–1802. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Mukherjee, A.; Pichoff, S.; Lutkenhaus, J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 1999, 96, 14819–14824. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; Froehlich, J.E.; Koksharova, O.; Pyke, K.A.; van Erp, H.; Osteryoung, K.W. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 2003, 15, 1918–1933. [Google Scholar] [CrossRef] [PubMed]
- Maple, J.; Aldridge, C.; Møller, S.G. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 2005, 43, 811–823. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, R.S.; Olson, B.J.; Kadirjan-Kalbach, D.K.; Chi-Ham, C.L.; Vitha, S.; Froehlich, J.E.; Osteryoung, K.W. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 2008, 412, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Maple, J.; Chua, N.H.; Møller, S.G. The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 2002, 31, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.M.; Yang, Y.; Vitha, S.; Schmitz, A.J.; Hemmes, M.; Miyagishima, S.Y.; Osteryoung, K.W. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 2009, 59, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, Y.; Jia, J.; Li, D.; Zhang, R.; Gao, H.; He, Y. CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 2009, 19, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, C.; Froehlich, J.E.; TerBush, A.D.; Osteryoung, K.W. Roles of Arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol. 2016, 170, 250–262. [Google Scholar] [CrossRef] [PubMed]
- TerBush, A.D.; Porzondek, C.A.; Osteryoung, K.W. Functional analysis of the chloroplast division complex using Schizosaccharomyces pombe as a heterologous expression system. Microsc. Microanal. 2016, 22, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Mishra, M.; Murata-Hori, M.; Balasubramanian, M.K. Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr. Biol. 2007, 17, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Mishra, M.; Wu, L.; Yin, Z.; Balasubramanian, M.K. The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 2008, 22, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Irieda, H.; Shiomi, D. ARC6-mediated Z ring-like structure formation of prokaryote-descended chloroplast FtsZ in Escherichia coli. Sci. Rep. 2017, 7, 3492. [Google Scholar] [CrossRef] [PubMed]
- Haeusser, D.P.; Margolin, W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 2016, 14, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Bi, E.F.; Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Levin, P.A.; Losick, R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996, 10, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ehrhardt, D.W.; Margolin, W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 1996, 93, 12998–13003. [Google Scholar] [CrossRef] [PubMed]
- Addinall, S.G.; Bi, E.; Lutkenhaus, J. FtsZ ring formation in fts mutants. J. Bacteriol. 1996, 178, 3877–3884. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Margolin, W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol. 1998, 180, 2050–2056. [Google Scholar] [PubMed]
- McAndrew, R.S.; Froehlich, J.E.; Vitha, S.; Stokes, K.D.; Osteryoung, K.W. Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 2001, 127, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Kuroiwa, H.; Takahara, M.; Miyagishima, S.Y.; Kuroiwa, T. Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 2001, 42, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; McAndrew, R.S.; Osteryoung, K.W. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001, 153, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; de Boer, P.A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 2005, 18, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Erickson, H.P. Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 2005, 151, 4033–4043. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Margolin, W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999, 181, 7531–7544. [Google Scholar] [PubMed]
- Johnson, C.B.; Shaik, R.; Abdallah, R.; Vitha, S.; Holzenburg, A. FtsZ1/FtsZ2 turnover in chloroplasts and the role of ARC3. Microsc. Microanal. 2015, 21, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Yoshida, S. Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 2001, 287, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.A.; de Boer, P.A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 1997, 88, 175–185. [Google Scholar] [CrossRef]
- Liu, Z.; Mukherjee, A.; Lutkenhaus, J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 1999, 31, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Mosyak, L.; Zhang, Y.; Glasfeld, E.; Haney, S.; Stahl, M.; Seehra, J.; Somers, W.S. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000, 19, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002, 21, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Lutkenhaus, J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005, 55, 1722–1734. [Google Scholar] [CrossRef] [PubMed]
- Koksharova, O.A.; Wolk, C.P. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 2002, 184, 5524–5528. [Google Scholar] [CrossRef] [PubMed]
- Mazouni, K.; Domain, F.; Cassier-Chauvat, C.; Chauvat, F. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 2004, 52, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Saguez, C.; Cassier-Chauvat, C.; Chauvat, F. ZipN, an FtsA-like orchestrator of divisome assembly in the model cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 2009, 74, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Erickson, H.P. Liposome division by a simple bacterial division machinery. Proc. Natl. Acad. Sci. USA 2013, 110, 11000–11004. [Google Scholar] [CrossRef] [PubMed]
- Pyke, K.A.; Rutherford, S.M.; Robertson, E.J.; Leech, R.M. arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol. 1994, 106, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.B.; Tang, L.K.; Smith, A.G.; Ravichandran, A.; Luo, Z.; Vitha, S.; Holzenburg, A. Single particle tracking analysis of the chloroplast division protein FtsZ anchoring to the inner envelope membrane. Microsc. Microanal. 2013, 19, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Porter, K.; Osawa, M.; Augustus, A.M.; Milam, S.L.; Joshi, C.; Osteryoung, K.W.; Erickson, H.P. The chloroplast tubulin homologs FtsZA and FtsZB from the red alga Galdieria sulphuraria co-assemble into dynamic filaments. J. Biol. Chem. 2017, 292, 5207–5215. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Kabeya, Y.; Suzuki, K.; Mori, T.; Ichikawa, T.; Matsui, M.; Nakanishi, H.; Miyagishima, S. The PLASTID DIVISION1 and 2 Components of the Chloroplast Division Machinery Determine the Rate of Chloroplast Division in Land Plant Cell Differentiation. Plant Cell 2009, 21, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Lutkenhaus, J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 2007, 76, 539–562. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, V.W.; Margolin, W. The bacterial Min system. Curr. Biol. 2013, 23, R553–R556. [Google Scholar] [CrossRef] [PubMed]
- MacCready, J.S.; Schossau, J.; Osteryoung, K.W.; Ducat, D.C. Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol. Microbiol. 2017, 103, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Glynn, J.M.; Olson, B.J.; Schmitz, A.J.; Osteryoung, K.W. Plastid division: Across time and space. Curr. Opin. Plant Biol. 2008, 11, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Kabeya, Y. Chloroplast division: Squeezing the photosynthetic captive. Curr. Opin. Microbiol. 2010, 13, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Pyke, K.A.; Leech, R.M. Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 1992, 99, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.M.; Miyagishima, S.Y.; Yoder, D.W.; Osteryoung, K.W.; Vitha, S. Chloroplast division. Traffic 2007, 8, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Shaik, R.S.; Sung, M.W.; Vitha, S.; Holzenburg, A. Chloroplast division protein ARC3 acts on FtsZ2 by preventing filament bundling and enhancing GTPase activity. Biochem. J. 2018, 475, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Cao, C.; Lutkenhaus, J. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 1996, 178, 5080–5085. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.M.; Froehlich, J.E.; Osteryoung, K.W. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 2008, 20, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, J.; Sun, Q.; Yu, X.; Zhang, W.; Jia, N.; An, C.; Li, Y.; Dong, Y.; Han, F.; et al. Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat. Plants 2017, 3, 17011. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Mukherjee, A.; Cao, C.; Lutkenhaus, J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 1997, 179, 5551–5559. [Google Scholar] [CrossRef] [PubMed]
- Feucht, A.; Lucet, I.; Yudkin, M.D.; Errington, J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 2001, 40, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Krupka, M.; Rowlett, V.W.; Morado, D.; Vitrac, H.; Schoenemann, K.; Liu, J.; Margolin, W. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat. Commun. 2017, 8, 15957. [Google Scholar] [CrossRef] [PubMed]
- Yoder, D.W.; Kadirjan-Kalbach, D.; Olson, B.J.; Miyagishima, S.Y.; Deblasio, S.L.; Hangarter, R.P.; Osteryoung, K.W. Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 2007, 48, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Suzuki, K.; Kabeya, Y.; Miyagishima, S.Y. Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 2009, 19, 151–156. [Google Scholar] [CrossRef] [PubMed]
- De Boer, P.A.; Crossley, R.E.; Rothfield, L. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 1989, 56, 641–649. [Google Scholar] [CrossRef]
- Gao, H.; Kadirjan-Kalbach, D.; Froehlich, J.E.; Osteryoung, K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 4328–4333. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Froehlich, J.E.; Osteryoung, K.W. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 2006, 18, 2517–2530. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irieda, H.; Shiomi, D. Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors. Int. J. Mol. Sci. 2018, 19, 544. https://doi.org/10.3390/ijms19020544
Irieda H, Shiomi D. Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors. International Journal of Molecular Sciences. 2018; 19(2):544. https://doi.org/10.3390/ijms19020544
Chicago/Turabian StyleIrieda, Hiroki, and Daisuke Shiomi. 2018. "Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors" International Journal of Molecular Sciences 19, no. 2: 544. https://doi.org/10.3390/ijms19020544
APA StyleIrieda, H., & Shiomi, D. (2018). Bacterial Heterologous Expression System for Reconstitution of Chloroplast Inner Division Ring and Evaluation of Its Contributors. International Journal of Molecular Sciences, 19(2), 544. https://doi.org/10.3390/ijms19020544




