Site-Specific Cleavage by Topoisomerase 2: A Mark of the Core Centromere
Abstract
1. Introduction
2. Results
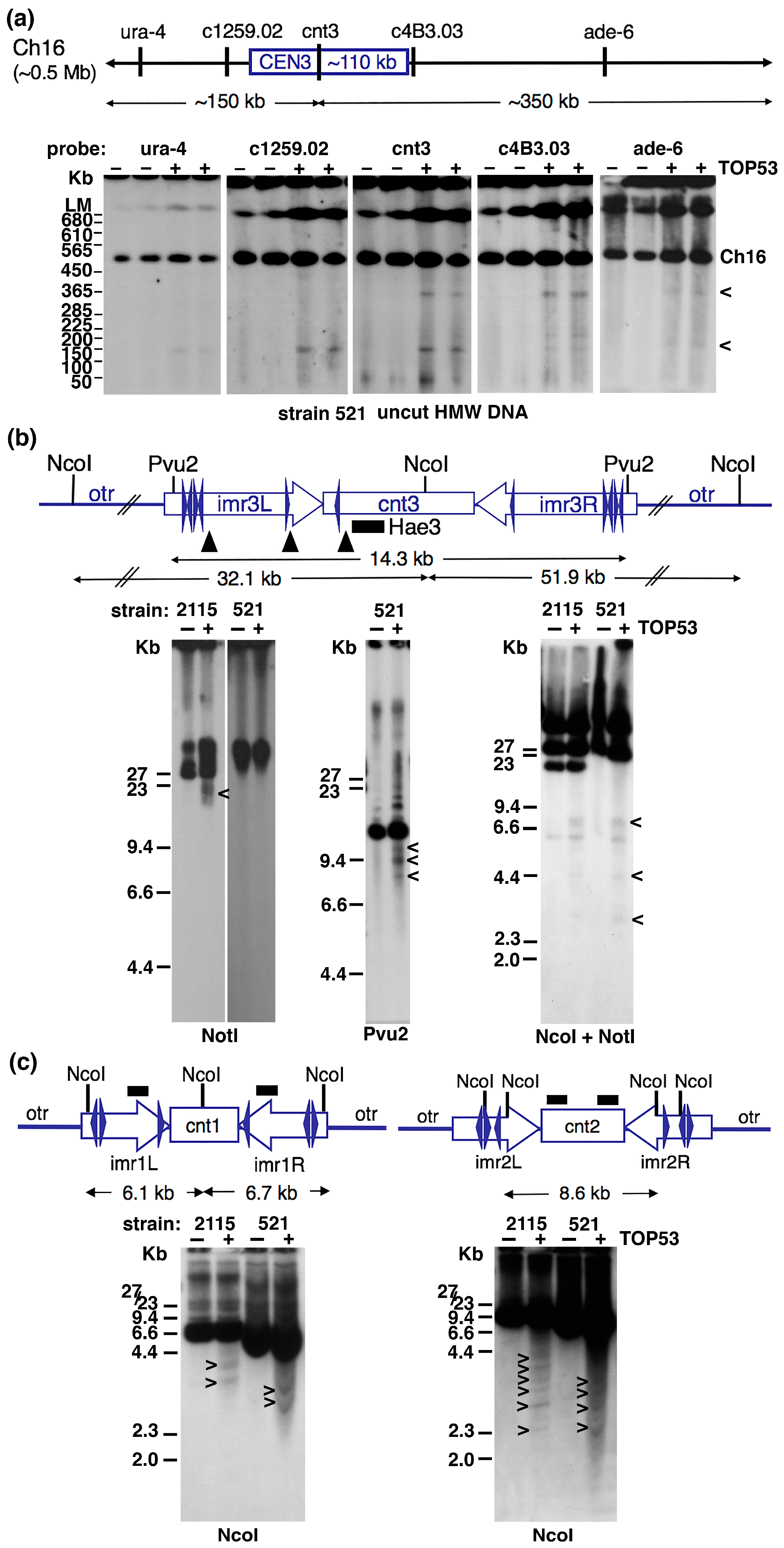
2.1. Topo 2 Cleavage within Centromeres of S. pombe
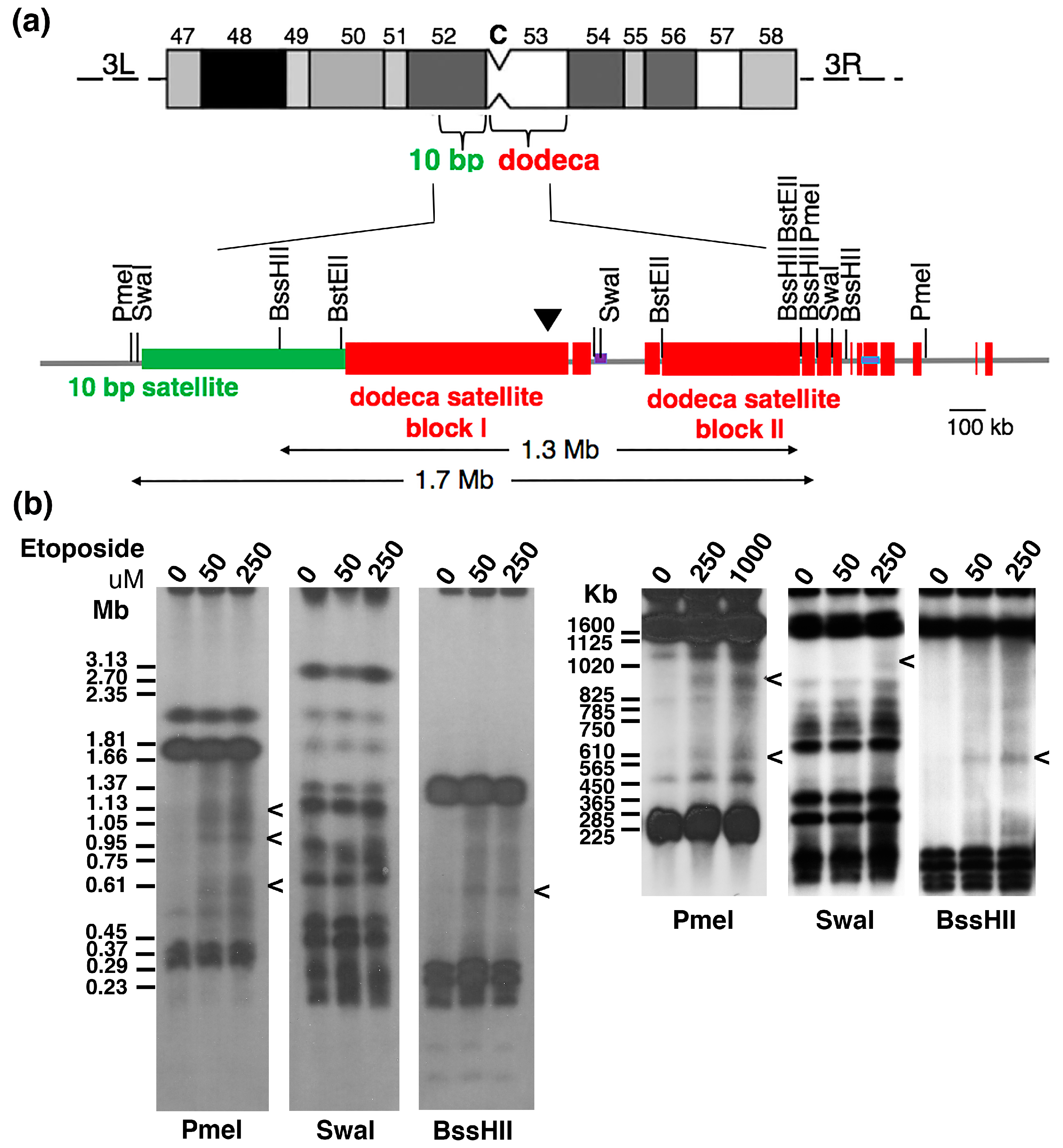
2.2. Topo 2 Cleavage within Drosophila Centromere 3
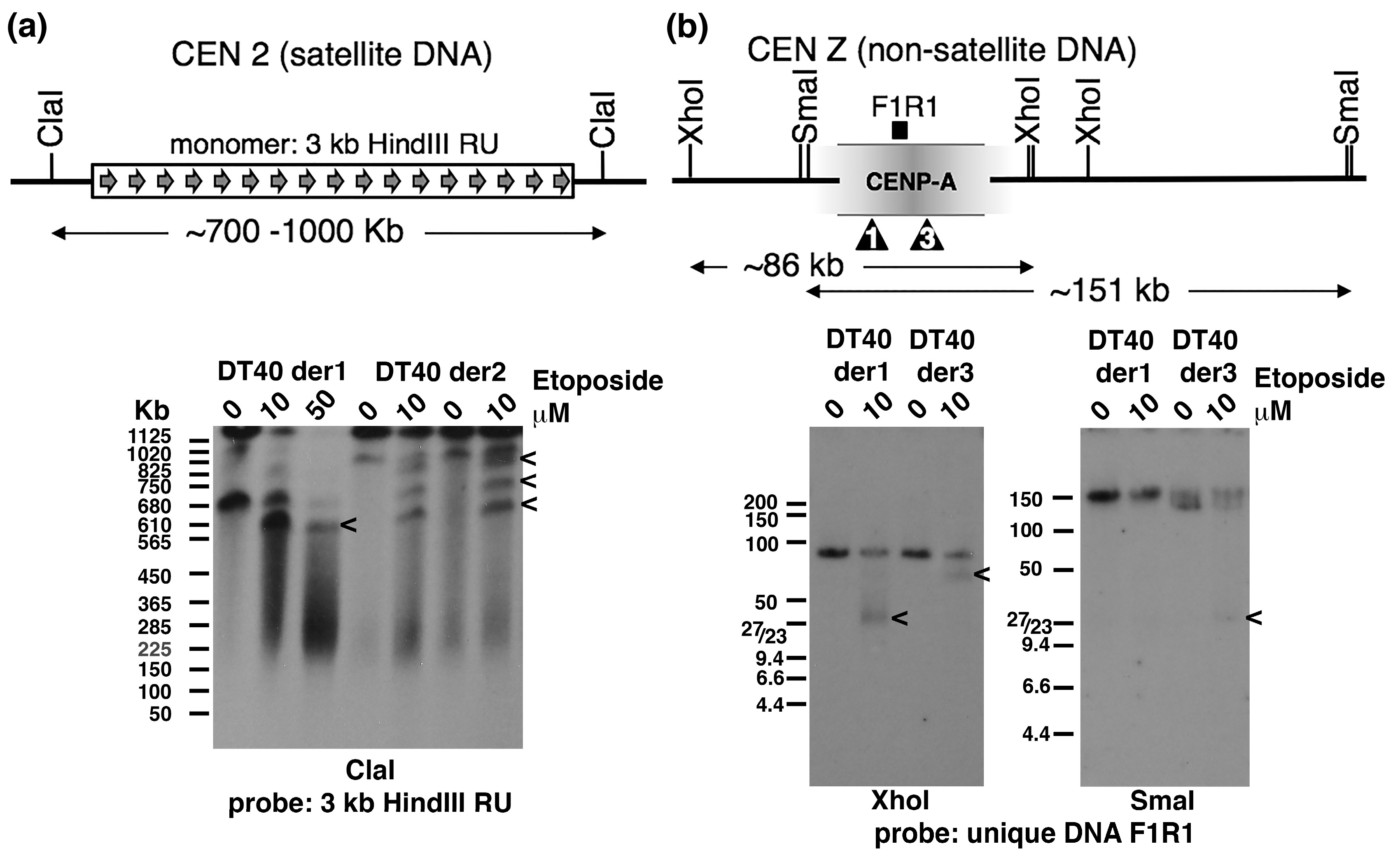
2.3. Topo 2 Cleavage within Satellite and Non-Satellite DNA-Based Centromeres in Chicken DT40 Cells
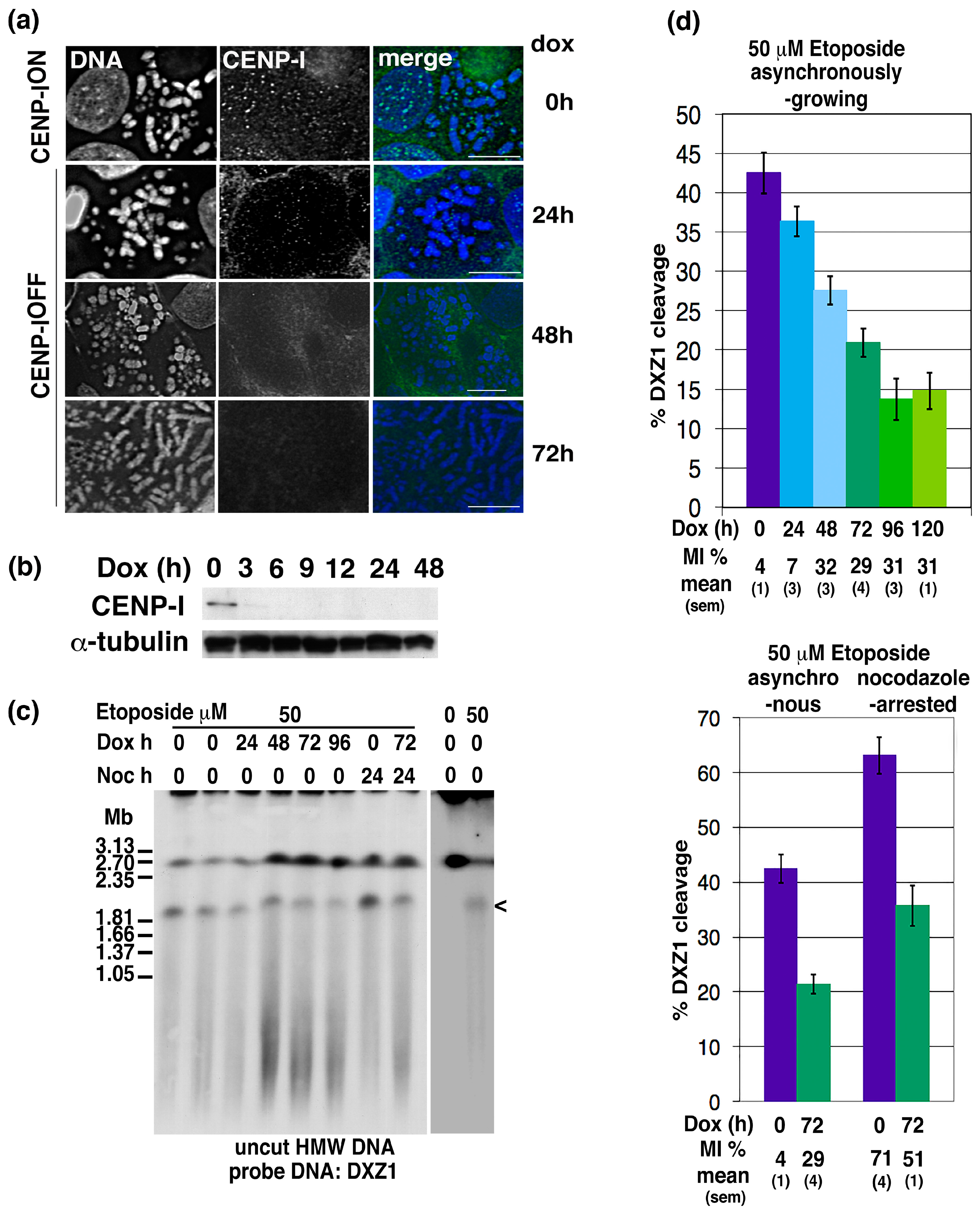
2.4. CENP-I Depletion Reduces Centromeric Site-Specific Topo 2 Cleavage
3. Discussion
4. Materials and Methods
4.1. S. pombe Strains
4.2. Insect and Vertebrate Cell Culture
4.3. Topoisomerase 2 Poisons
4.4. Molecular Biology
4.4.1. Probe DNAs
4.4.2. Gel Electrophoresis and Southern Blotting
4.5. Centromeric Topo 2-Cleavage Assay in DT40 Cells
4.6. Indirect Immunofluorescence and Microscopy
4.7. Western Blot Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Topo 2 | Topoisomerase 2 |
| CEN | Centromere |
| CCAN | Constitutive centromere-associated network |
| DSB | Double strand break |
| DMSO | Dimethylsulphoxide |
| PAGE | Polyacrylamide gel electrophoresis |
| PFGE | Pulsed-field gel electrophoresis |
| SDS | Sodium dodecyl sulphate |
References
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Rattner, J.B.; Hendzel, M.J.; Furbee, C.S.; Muller, M.T.; Bazett-Jones, D.P. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 1996, 134, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.A.; Come, M.G.; Hudson, J.R.; Mo, Y.Y.; Beck, W.T.; Gorbsky, G.J. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 2002, 158, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.O.; Larsen, M.K.; Barthelmes, H.U.; Hock, R.; Andersen, C.L.; Kjeldsen, E.; Knudsen, B.R.; Westergaard, O.; Boege, F.; Mielke, C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002, 157, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Wandall, A.; Kjeldsen, E.; Mielke, C.; Koch, J. Active, but not inactive, human centromeres display topoisomerase II activity in vivo. Chromosome Res. 2002, 10, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.C.; Farr, C.J. Topoisomerase II: Untangling its contribution at the centromere. Chromosome Res. 2004, 12, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.M.; Fournier, R.E.; Oshimura, M.; Regnier, V.; Farr, C.J. Topoisomerase II cleavage activity within the human D11Z1 and DXZ1 alpha-satellite arrays. Chromosome Res. 2005, 13, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Linka, R.M.; Porter, A.C.; Volkov, A.; Mielke, C.; Boege, F.; Christensen, M.O. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007, 35, 3810–3822. [Google Scholar] [CrossRef] [PubMed]
- Westhorpe, F.G.; Straight, A.F. The centromere: Epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 2014, 7, a015818. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Kschonsak, M.; Haering, C.H. Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. Bioessays 2015, 37, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Baxter, J. “Breaking up is hard to do”: The formation and resolution of sister chromatid intertwines. J. Mol. Biol. 2015, 427, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Nielsen, C.F.; Yao, Q.; Hickson, I.D. The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev. 2014, 26, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Yoshida, M.M.; Sridharan, V.; Kumagai, A.; Dunphy, W.G.; Dasso, M.; Azuma, Y. SUMOylation of the C-terminal domain of DNA topoisomerase IIalpha regulates the centromeric localization of Claspin. Cell Cycle 2015, 14, 2777–2784. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, H.; Johansson, M.; Keifenheim, D.; Mukherjee, S.; Chacon, J.M.; Bachant, J.; Gardner, M.K.; Clarke, D.J. A noncatalytic function of the topoisomerase II CTD in Aurora B recruitment to inner centromeres during mitosis. J. Cell Biol. 2016, 213, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.M.; Ting, L.; Gygi, S.P.; Azuma, Y. SUMOylation of DNA topoisomerase IIalpha regulates histone H3 kinase Haspin and H3 phosphorylation in mitosis. J. Cell Biol. 2016, 213, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.M.; Azuma, Y. Mechanisms behind Topoisomerase II SUMOylation in chromosome segregation. Cell Cycle 2016, 15, 3151–3152. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.J.; Azuma, Y. Non-Catalytic Roles of the Topoisomerase IIalpha C-Terminal Domain. Int. J. Mol. Sci. 2017, 18, 2438. [Google Scholar] [CrossRef] [PubMed]
- Deweese, J.E.; Osheroff, N. The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 2009, 37, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Kas, E.; Laemmli, U.K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992, 11, 705–716. [Google Scholar] [PubMed]
- Govoni, M.; Neri, S.; Labella, T.; Sylvester, J.E.; Novello, F.; Pession, A. Topoisomerase-II-mediated DNA cleavage within the human ribosomal genes. Biochem. Biophys. Res. Commun. 1995, 213, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Borgnetto, M.E.; Zunino, F.; Tinelli, S.; Kas, E.; Capranico, G. Drug-specific sites of topoisomerase II DNA cleavage in Drosophila chromatin: Heterogeneous localization and reversibility. Cancer Res. 1996, 56, 1855–1862. [Google Scholar] [PubMed]
- Borgnetto, M.E.; Tinelli, S.; Carminati, L.; Capranico, G. Genomic sites of topoisomerase II activity determined by comparing DNA breakage enhanced by three distinct poisons. J. Mol. Biol. 1999, 285, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Floridia, G.; Zatterale, A.; Zuffardi, O.; Tyler-Smith, C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000, 1, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.M.; Critcher, R.; Ebersole, T.A.; Valdivia, M.M.; Earnshaw, W.C.; Fukagawa, T.; Farr, C.J. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002, 21, 5269–5280. [Google Scholar] [CrossRef] [PubMed]
- Obado, S.O.; Bot, C.; Nilsson, D.; Andersson, B.; Kelly, J.M. Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol. 2007, 8, R37. [Google Scholar] [CrossRef] [PubMed]
- Obado, S.O.; Bot, C.; Echeverry, M.C.; Bayona, J.C.; Alvarez, V.E.; Taylor, M.C.; Kelly, J.M. Centromere-associated topoisomerase activity in bloodstream form Trypanosoma brucei. Nucleic Acids Res. 2011, 39, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Echeverry, M.C.; Bot, C.; Obado, S.O.; Taylor, M.C.; Kelly, J.M. Centromere-associated repeat arrays on Trypanosoma brucei chromosomes are much more extensive than predicted. BMC Genom. 2012, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Obado, S.O.; Taylor, M.C.; Wilkinson, S.R.; Bromley, E.V.; Kelly, J.M. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16-kb GC-rich transcriptional “strand-switch” domain as a major feature. Genome Res. 2005, 15, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; McRobert, L.; Baker, D.A. Evidence on the chromosomal location of centromeric DNA in Plasmodium falciparum from etoposide-mediated topoisomerase-II cleavage. Proc. Natl. Acad. Sci. USA 2006, 103, 6706–6711. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, S.; Khan, S.M.; Kaneko, I.; Christodoulou, Z.; Newbold, C.; Yuda, M.; Janse, C.J.; Waters, A.P. Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe 2010, 7, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.F.; Francia, M.E.; Gissot, M.; Croken, M.M.; Kim, K.; Striepen, B. Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc. Natl. Acad. Sci. USA 2011, 108, 3767–3772. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, A.L.; Allshire, R.C. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 2004, 12, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Byl, J.A.; Cline, S.D.; Utsugi, T.; Kobunai, T.; Yamada, Y.; Osheroff, N. DNA topoisomerase II as the target for the anticancer drug TOP-53: Mechanistic basis for drug action. Biochemistry 2001, 40, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.; Nitiss, J.L. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot Cell 2004, 3, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Duca, M.; Guianvarc’h, D.; Meresse, P.; Bertounesque, E.; Dauzonne, D.; Kraus-Berthier, L.; Thirot, S.; Leonce, S.; Pierre, A.; Pfeiffer, B.; et al. Synthesis and biological study of a new series of 4′-demethylepipodophyllotoxin derivatives. J. Med. Chem. 2005, 48, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Guianvarc’h, D.; Duca, M.; Boukarim, C.; Kraus-Berthier, L.; Leonce, S.; Pierre, A.; Pfeiffer, B.; Renard, P.; Arimondo, P.B.; Monneret, C.; et al. Synthesis and biological activity of sulfonamide derivatives of epipodophyllotoxin. J. Med. Chem. 2004, 47, 2365–2374. [Google Scholar]
- Matsumoto, T.; Fukui, K.; Niwa, O.; Sugawara, N.; Szostak, J.W.; Yanagida, M. Identification of healed terminal DNA fragments in linear minichromosomes of Schizosaccharomyces pombe. Mol. Cell. Biol. 1987, 7, 4424–4430. [Google Scholar] [CrossRef] [PubMed]
- Niwa, O.; Matsumoto, T.; Chikashige, Y.; Yanagida, M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989, 8, 3045–3052. [Google Scholar] [PubMed]
- Murakami, S.; Yanagida, M.; Niwa, O. A large circular minichromosome of Schizosaccharomyces pombe requires a high dose of type II DNA topoisomerase for its stabilization. Mol. Gen. Genet. 1995, 246, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, C.; Clarke, L. The chromatin structure of centromeres from fission yeast: Differentiation of the central core that correlates with function. J. Cell Biol. 1991, 112, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Murakami, S.; Chikashige, Y.; Funabiki, H.; Niwa, O.; Yanagida, M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 1992, 3, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Garavis, M.; Mendez-Lago, M.; Gabelica, V.; Whitehead, S.L.; Gonzalez, C.; Villasante, A. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015, 5, 13307. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Abad, J.P.; Villasante, A.; Gonzalez, C. The Drosophila melanogaster dodecasatellite sequence is closely linked to the centromere and can form connections between sister chromatids during mitosis. J. Cell Sci. 1993, 105 Pt 1, 41–50. [Google Scholar] [PubMed]
- Losada, A.; Abad, J.P.; Agudo, M.; Villasante, A. Long-range analysis of the centromeric region of Drosophila melanogaster chromosome 3. Chromosome Res. 2000, 8, 651–653. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010, 20, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.H.; Hara, M.; Ariyoshi, M.; Arimura, Y.; Fujita, R.; Kurumizaka, H.; Fukagawa, T. Association of M18BP1/KNL2 with CENP-A Nucleosome Is Essential for Centromere Formation in Non-mammalian Vertebrates. Dev. Cell 2017, 42, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Fukagawa, T. Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN). Prog. Mol. Subcell. Biol. 2017, 56, 29–57. [Google Scholar] [PubMed]
- Nishihashi, A.; Haraguchi, T.; Hiraoka, Y.; Ikemura, T.; Regnier, V.; Dodson, H.; Earnshaw, W.C.; Fukagawa, T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2002, 2, 463–476. [Google Scholar] [CrossRef]
- Spence, J.M.; Phua, H.H.; Mills, W.; Carpenter, A.J.; Porter, A.C.; Farr, C.J. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 2007, 120, 3952–3964. [Google Scholar] [CrossRef] [PubMed]
- Vagnarelli, P.; Hudson, D.F.; Ribeiro, S.A.; Trinkle-Mulcahy, L.; Spence, J.M.; Lai, F.; Farr, C.J.; Lamond, A.I.; Earnshaw, W.C. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 2006, 8, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Heck, M.M.; Hittelman, W.N.; Earnshaw, W.C. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA 1988, 85, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.A.; Gatlin, J.C.; Dong, Y.; Joglekar, A.; Cameron, L.; Hudson, D.F.; Farr, C.J.; McEwen, B.F.; Salmon, E.D.; Earnshaw, W.C.; et al. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell 2009, 20, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.A.; Vagnarelli, P.; Earnshaw, W.C. DNA content of a functioning chicken kinetochore. Chromosome Res. 2014, 22, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Kagawa, N.; Toyoda, A.; Fujiyama, A.; Misu, S.; Monma, N.; Makino, F.; Ikeo, K.; Fukagawa, T. Constitutive centromere-associated network controls centromere drift in vertebrate cells. J. Cell Biol. 2017, 216, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Saitoh, S.; Yanagida, M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999, 13, 1664–1677. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Akiyoshi, B. Evolutionary Lessons from Species with Unique Kinetochores. Prog. Mol. Subcell. Biol. 2017, 56, 111–138. [Google Scholar] [PubMed]
- Akiyoshi, B.; Gull, K. Discovery of unconventional kinetochores in kinetoplastids. Cell 2014, 156, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- D’Archivio, S.; Wickstead, B. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J. Cell Biol. 2017, 216, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Koch, J. Neocentromeres and alpha satellite: A proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 2000, 9, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Abad, J.P.; Carmena, M.; Baars, S.; Saunders, R.D.; Glover, D.M.; Ludena, P.; Sentis, C.; Tyler-Smith, C.; Villasante, A. Dodeca satellite: A conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1992, 89, 4663–4667. [Google Scholar] [CrossRef] [PubMed]
- Kasinathan, S.; Henikoff, S. Non-B-form DNA is enriched at centromeres. Mol. Biol. Evol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pognan, F.; Paoletti, C. Does cruciform DNA provide a recognition signal for DNA-topoisomerase II? Biochimie 1992, 74, 1019–1023. [Google Scholar] [CrossRef]
- West, K.L.; Austin, C.A. Human DNA topoisomerase IIbeta binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999, 27, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Jonstrup, A.T.; Thomsen, T.; Wang, Y.; Knudsen, B.R.; Koch, J.; Andersen, A.H. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIalpha. Nucleic Acids Res. 2008, 36, 6165–6174. [Google Scholar] [CrossRef] [PubMed]
- Garavis, M.; Escaja, N.; Gabelica, V.; Villasante, A.; Gonzalez, C. Centromeric Alpha-Satellite DNA Adopts Dimeric i-Motif Structures Capped by AT Hoogsteen Base Pairs. Chemistry 2015, 21, 9816–9824. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Nishino, T.; Mayanagi, K.; Horikoshi, N.; Osakabe, A.; Tachiwana, H.; Hori, T.; Kurumizaka, H.; Fukagawa, T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 2014, 42, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Kim, B.W.; Earle, E.; Choo, K.H.; Wong, L.H. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA 2012, 109, 1979–1984. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007, 17, 1146–1160. [Google Scholar] [CrossRef] [PubMed]
- Quenet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife 2014, 3, e03254. [Google Scholar] [CrossRef] [PubMed]
- Perea-Resa, C.; Blower, M.D. Satellite Transcripts Locally Promote Centromere Formation. Dev. Cell 2017, 42, 201–202. [Google Scholar] [CrossRef] [PubMed]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Dev. Cell 2017, 42, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.S.; Stralfors, A.; Castillo, A.G.; Durand-Dubief, M.; Ekwall, K.; Allshire, R.C. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011, 286, 23600–23607. [Google Scholar] [CrossRef] [PubMed]
- Catania, S.; Pidoux, A.L.; Allshire, R.C. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet. 2015, 11, e1004986. [Google Scholar] [CrossRef] [PubMed]
- Rosic, S.; Kohler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Bowers, S.; Lipinszki, Z.; Palladino, J.; Trusiak, S.; Bettini, E.; Rosin, L.; Przewloka, M.R.; Glover, D.M.; O’Neill, R.J.; et al. Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Dev. Cell 2015, 34, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Martinez, L.A.; Gimenez-Abian, J.F.; Clarke, D.J. Chromosome cohesion—Rings, knots, orcs and fellowship. J. Cell Sci. 2008, 121, 2107–2114. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, S.G.; Baker, N.M.; Chapados, B.R.; Soutoglou, E.; Wang, J.Y.; Berns, M.W.; Cleveland, D.W. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl. Acad. Sci. USA 2009, 106, 15762–15767. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Davenport, J.W.; Urtishak, K.A.; Carillo, M.L.; Gosai, S.J.; Kolaris, C.P.; Byl, J.A.W.; Rappaport, E.F.; Osheroff, N.; Gregory, B.D.; et al. Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci. Genome Res. 2017, 27, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Jaco, I.; Canela, A.; Vera, E.; Blasco, M.A. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 2008, 181, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S. Near the edge of a chromosome’s “black hole”. Trends Genet. 2002, 18, 165–167. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [PubMed]
- Allshire, R.C.; Nimmo, E.R.; Ekwall, K.; Javerzat, J.P.; Cranston, G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995, 9, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Dieken, E.S.; Epner, E.M.; Fiering, S.; Fournier, R.E.; Groudine, M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat. Genet. 1996, 12, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Mills, W.; Critcher, R.; Lee, C.; Farr, C.J. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum. Mol. Genet. 1999, 8, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Regnier, V.; Vagnarelli, P.; Fukagawa, T.; Zerjal, T.; Burns, E.; Trouche, D.; Earnshaw, W.; Brown, W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 2005, 25, 3967–3981. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Mikami, Y.; Nishihashi, A.; Regnier, V.; Haraguchi, T.; Hiraoka, Y.; Sugata, N.; Todokoro, K.; Brown, W.; Ikemura, T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001, 20, 4603–4617. [Google Scholar] [CrossRef] [PubMed]
- Dieken, E.S.; Fournier, R.E.K. Homologous Modification of Human Chromosomal Genes in Chicken B-Cell X Human Microcell Hybrids. Methods 1996, 9, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russel, R.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Press: New York, NY, USA, 2001. [Google Scholar]
- Allshire, R.C. Introduction of large linear minichromosomes into Schizosaccharomyces pombe by an improved transformation procedure. Proc. Natl. Acad. Sci. USA 1990, 87, 4043–4047. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Vogelstein, B. “A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity”. Addendum. Anal. Biochem. 1984, 137, 266–267. [Google Scholar] [PubMed]
- Johnson, M.; Phua, H.H.; Bennett, S.C.; Spence, J.M.; Farr, C.J. Studying vertebrate topoisomerase 2 function using a conditional knockdown system in DT40 cells. Nucleic Acids Res. 2009, 37, e98. [Google Scholar] [CrossRef] [PubMed]
| Strain | Genotype |
|---|---|
| #521 | h− ade6-210 leu1-32 ura4-DS/E [Ch16 ade6-216 m23::ura4] |
| #2115 | h+ ade6-704 his3-D1 leu1-32 ura4-D18 [CM3112 sup3e] |
| S. pombe Locus | Forward Primer 5′-3′ | Reverse Primer 5′-3′ | Size (nt) |
|---|---|---|---|
| ura4 | CAGAGGAAGCCTTTTTGCCAG | CCCATCTCACCGACCAACTTAG | 618 |
| c1259.02 | CGTTCAATACGGAAACCTTACAG | GCAGAATCAGCAGAAGCAATGG | 1041 |
| c4B3.03 | CGTTAGTGCTGAAAAGGATTCGG | AGGTGATTGCGACATTGACCC | 1285 |
| ade6 | TCCTACTGCCATCAAAGCACTTG | GCAATAATCACACGCAACCCTC | 846 |
| imr1 | TTTTTTACGCCACAATGTCGC | CAAACAGATACTTAGCCTCAAG | 857 |
| cnt2-1 | TGCTATGTGGGTCTGTATTGCCTC | GGGGTTACCTTTTGCGATGG | 1482 |
| cnt2-2 | TGAAGAAGCATGGTTAGTCCTGG | CCTGAACGGAACAAAACTCCTG | 1058 |
| cnt3 | TCAGATTGTGGTCATAACAGTGC | TAGGTGAAGCGTAAGTGAGTGG | 1642 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mills, W.E.; Spence, J.M.; Fukagawa, T.; Farr, C.J. Site-Specific Cleavage by Topoisomerase 2: A Mark of the Core Centromere. Int. J. Mol. Sci. 2018, 19, 534. https://doi.org/10.3390/ijms19020534
Mills WE, Spence JM, Fukagawa T, Farr CJ. Site-Specific Cleavage by Topoisomerase 2: A Mark of the Core Centromere. International Journal of Molecular Sciences. 2018; 19(2):534. https://doi.org/10.3390/ijms19020534
Chicago/Turabian StyleMills, Walter E., Jennifer M. Spence, Tatsuo Fukagawa, and Christine J. Farr. 2018. "Site-Specific Cleavage by Topoisomerase 2: A Mark of the Core Centromere" International Journal of Molecular Sciences 19, no. 2: 534. https://doi.org/10.3390/ijms19020534
APA StyleMills, W. E., Spence, J. M., Fukagawa, T., & Farr, C. J. (2018). Site-Specific Cleavage by Topoisomerase 2: A Mark of the Core Centromere. International Journal of Molecular Sciences, 19(2), 534. https://doi.org/10.3390/ijms19020534





