Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses
Abstract
1. Introduction
2. Results
2.1. Gene Arrays
2.2. Gene Set Enrichment Analysis (GSEA)
2.3. Cytoscape/ClueGO
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Spaceflight Conditions and Ground Based Controls.
4.3. RNA Extraction
4.4. Gene Arrays
4.5. Data Analysis and Statistics
4.6. Gene Ontology (GO) Enrichment Analysis
4.7. Cytoscape Analysis Using ClueGO
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.M.; Yoshida, M.C.; Candelario, T.L.; Hughes-Fulford, M. Spaceflight and simulated microgravity cause a significant reduction of key gene expression in early T-cell activation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R480–R488. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Pecaut, M.J.; Jonscher, K.R. Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver. Int. J. Mol. Sci. 2017, 18, 2062. [Google Scholar] [CrossRef] [PubMed]
- Pecaut, M.J.; Mao, X.W.; Bellinger, D.L.; Jonscher, K.R.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Mohney, R.P.; Gridley, D.S. Is spaceflight-induced immune dysfunction linked to systemic changes in metabolism? PLoS ONE 2017, 12, e0174174. [Google Scholar] [CrossRef] [PubMed]
- Baqai, F.P.; Gridley, D.S.; Slater, J.M.; Luo-Owen, X.; Stodieck, L.S.; Ferguson, V.; Chapes, S.K.; Pecaut, M.J. Effects of spaceflight on innate immune function and antioxidant gene expression. J. Appl. Physiol. 2009, 106, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Nishimura, M.; Kuwahara, Y.; Ueda, N.; Naitoh, S.; Kume, M.; Yamamoto, Y.; Fujita, J.; Funae, Y.; Fukumoto, M. Analysis of gene and protein expression of cytochrome P450 and stress-associated molecules in rat liver after spaceflight. Pathol. Int. 2008, 58, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.A.; Arterburn, L.M.; Miller, A.P.; Cowger, N.L.; Hartley, S.M.; Andrews, A.; Silber, P.M.; Li, A.P. Maintenance of liver functions in rat hepatocytes cultured as spheroids in a rotating wall vessel. In Vitro Cell. Dev. Biol. Anim. 2003, 39, 13–20. [Google Scholar] [CrossRef]
- Hammond, T.G.; Benes, E.; O’Reilly, K.C.; Wolf, D.A.; Linnehan, R.M.; Taher, A.; Kaysen, J.H.; Allen, P.L.; Goodwin, T.J. Mechanical culture conditions effect gene expression: Gravity-induced changes on the space shuttle. Physiol. Genom. 2000, 3, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.G.; Allen, P.L.; Birdsall, H.H. Gene pathways analysis of the effects of suspension culture on primary human renal proximal tubular cells. Microgravity Sci. Technol. 2018. [Google Scholar] [CrossRef]
- Nislow, C.; Lee, A.Y.; Allen, P.L.; Giaever, G.; Smith, A.; Gebbia, M.; Stodieck, L.S.; Hammond, J.S.; Birdsall, H.H.; Hammond, T.G. Genes required for survival in microgravity revealed by genome-wide yeast deletion collections cultured during spaceflight. BioMed Res. Int. 2015, 2015, 976458. [Google Scholar] [CrossRef] [PubMed]
- Moestrup, S.K.; Kozyraki, R.; Kristiansen, M.; Kaysen, J.H.; Rasmussen, H.H.; Brault, D.; Pontillon, F.; Goda, F.O.; Christensen, E.I.; Hammond, T.G.; et al. The intrinsic factor-vitamin B12 receptor and target of teratogenic antibodies is a megalin-binding peripheral membrane protein with homology to developmental proteins. J. Biol. Chem. 1998, 273, 5235–5242. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
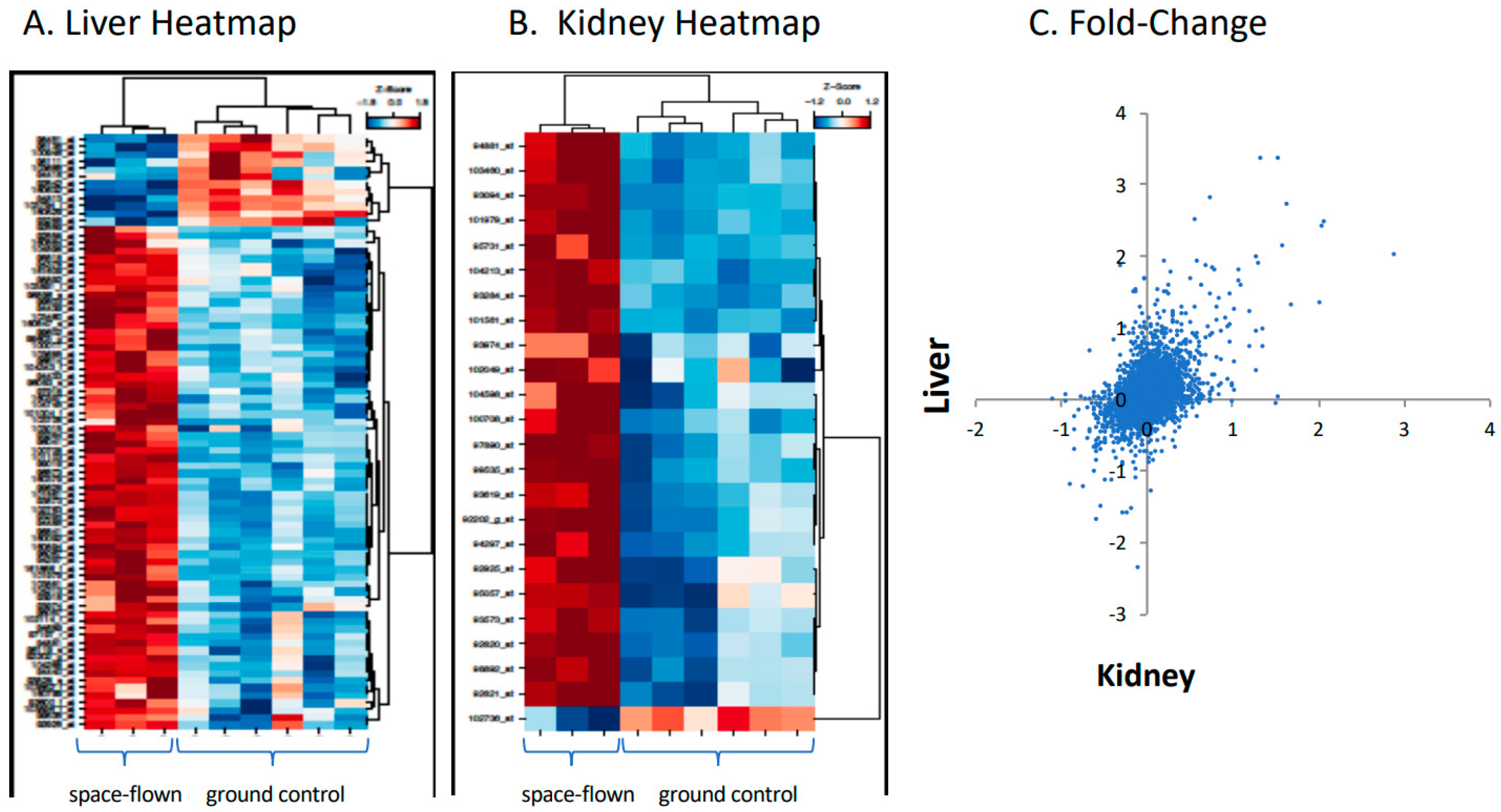
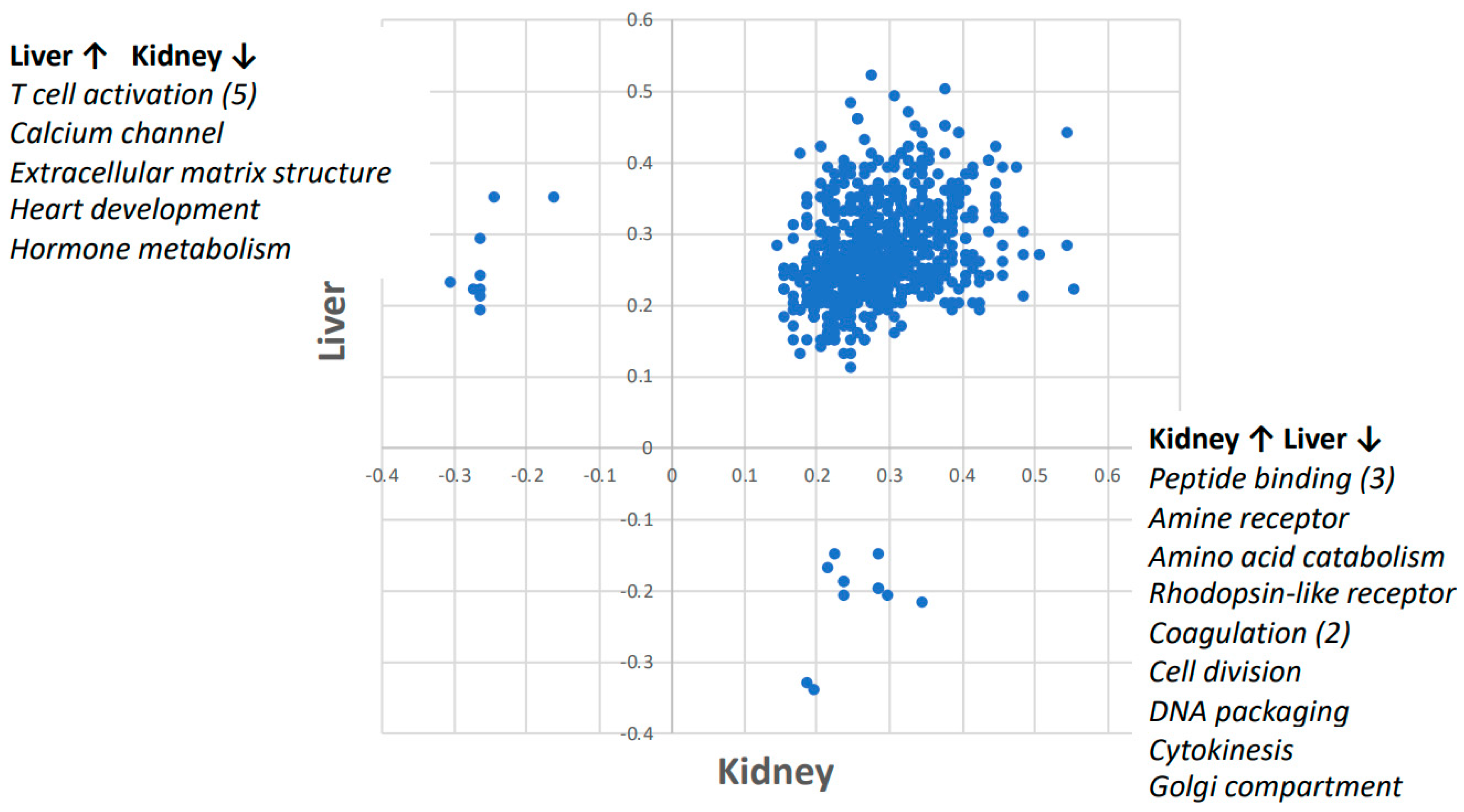
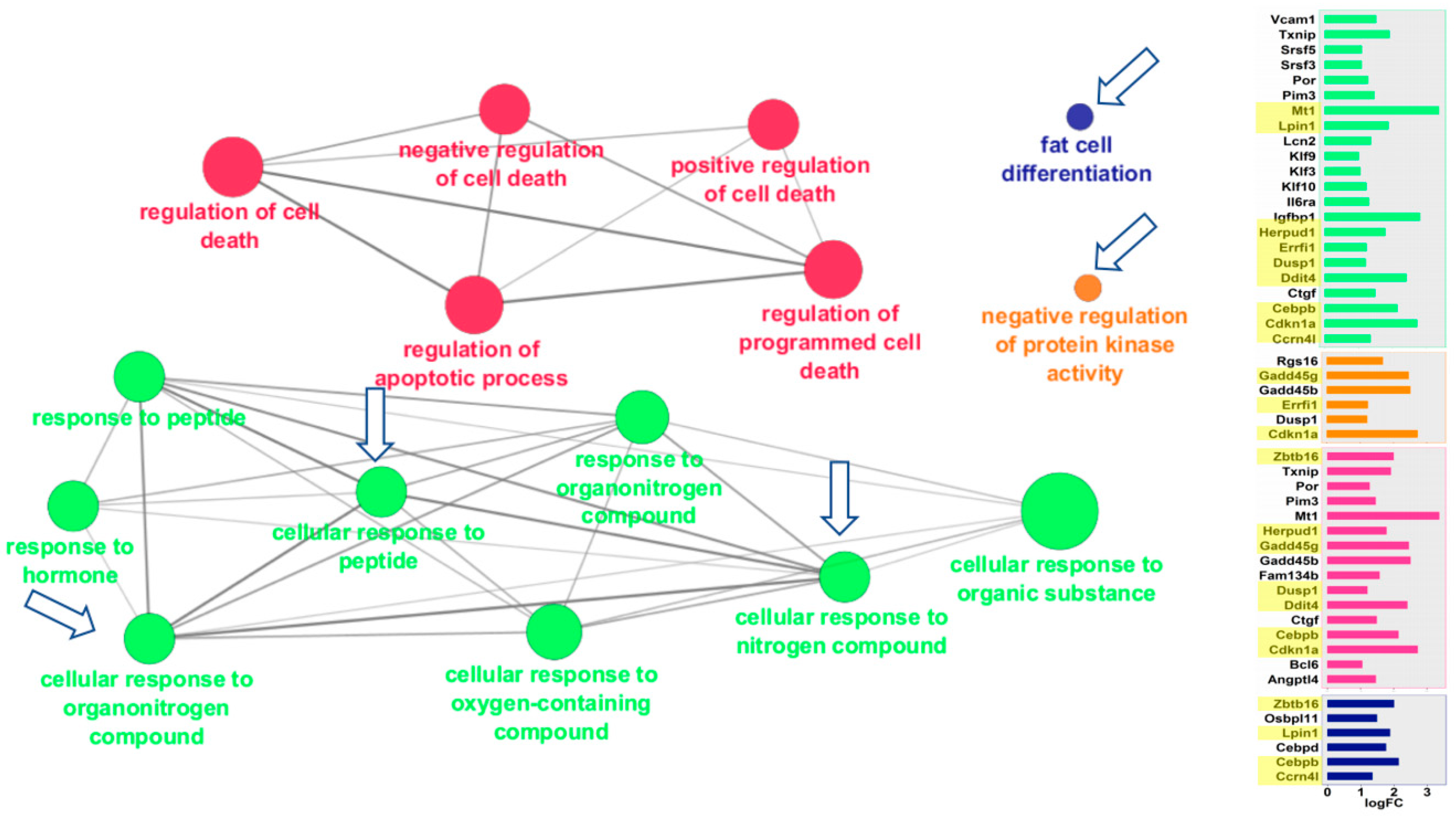
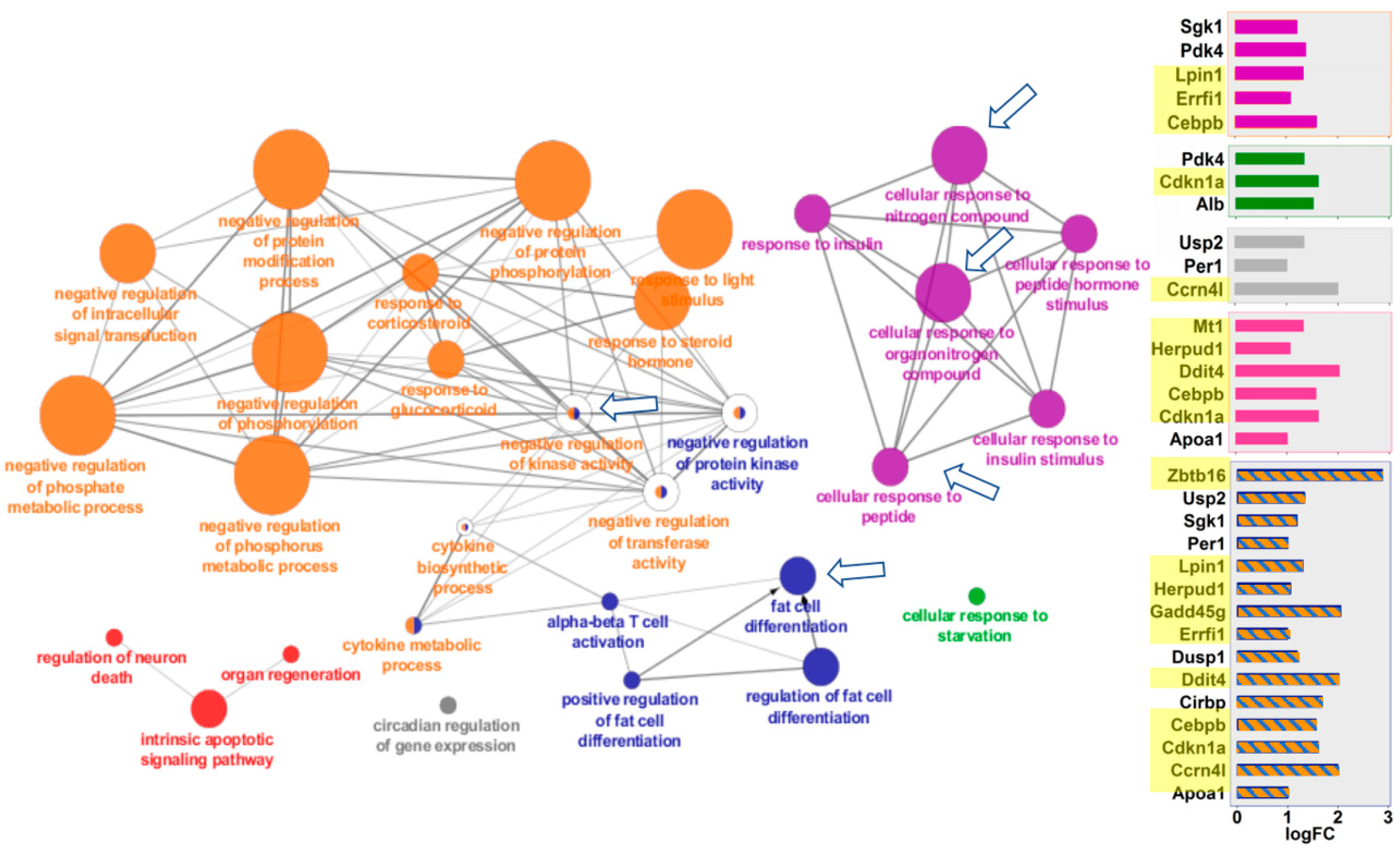
| Gene Set | Kidney Gene # | Liver Gene # | Kidney ES | Liver ES | Kidney NES | Liver NES | Kidney FWERp | Liver FWERp | Kidney Np | Liver Np | Kidney FDRq | Liver FDRq | Kidney Rank | Liver Rank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T cell activation | 29 | 29 | −0.26 | 0.19 | −1.37 | 0.67 | 0.338 | 1 | 0 | 0.933 | 0.186 | 1 | 6874 | 6586 |
| Positive regulation of T cell activation | 18 | 18 | −0.26 | 0.29 | −1.09 | 0.94 | 0.882 | 1 | 0.281 | 0.559 | 0.393 | 0.972 | 6874 | 4085 |
| Regulation of Lymphocyte activation | 26 | 26 | −0.26 | 0.21 | −1.44 | 0.75 | 0.208 | 1 | 0 | 0.869 | 0.163 | 1 | 6874 | 6695 |
| Positive regulation of Lymphocyte activation | 20 | 20 | −0.26 | 0.22 | −1.25 | 0.74 | 0.574 | 1 | 0.091 | 0.854 | 0.19 | 1 | 6874 | 4085 |
| Regulation of T cell activation | 21 | 21 | −0.26 | 0.24 | −1.26 | 0.8 | 0.535 | 1 | 0.071 | 0.786 | 0.206 | 0.997 | 6874 | 4085 |
| Calcium channel activity | 20 | 20 | −0.27 | 0.22 | −1.36 | 0.73 | 0.341 | 1 | 0.111 | 0.857 | 0.141 | 1 | 5840 | 2858 |
| Extracellular matrix structural constituent | 15 | 15 | −0.24 | 0.35 | −1.03 | 1.09 | 0.946 | 1 | 0.526 | 0.343 | 0.453 | 0.873 | 6985 | 4443 |
| Heart development | 24 | 24 | −0.3 | 0.23 | −1.49 | 0.78 | 0.173 | 1 | 0 | 0.828 | 0.272 | 1 | 6497 | 4327 |
| Hormone metabolic process | 16 | 16 | −0.16 | 0.35 | −0.65 | 1.1 | 0.999 | 1 | 0.919 | 0.336 | 0.97 | 0.835 | 7751 | 4932 |
| Gene Set | Kidney Gene # | Liver Gene # | Kidney ES | Liver ES | Kidney NES | Liver NES | Kidney FWERp | Liver FWERp | Kidney Np | Liver Np | Kidney FDRq | Liver FDRq | Kidney Rank | Liver Rank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amine receptor activity | 23 | 23 | 0.24 | −0.21 | 0.79 | −1.01 | 1 | 0.983 | 0.8 | 0.55 | 1 | 0.964 | 2860 | 7304 |
| Amino acid catabolic process | 19 | 19 | 0.22 | −0.17 | 0.68 | −0.79 | 1 | 1 | 0.917 | 0.964 | 1 | 0.836 | 3122 | 6652 |
| Blood coagulation | 35 | 35 | 0.24 | −0.19 | 0.82 | −1.1 | 1 | 0.94 | 0.81 | 0.4 | 1 | 0.833 | 6444 | 6951 |
| Coagulation | 35 | 35 | 0.24 | −0.19 | 0.82 | −0.98 | 1 | 0.991 | 0.79 | 0.75 | 1 | 0.684 | 6444 | 6951 |
| Cell division | 18 | 18 | 0.19 | −0.33 | 0.59 | −1.3 | 1 | 0.548 | 0.968 | 0.158 | 0.998 | 0.326 | 4143 | 6233 |
| DNA packaging | 23 | 23 | 0.3 | −0.21 | 0.97 | −0.98 | 1 | 0.991 | 0.532 | 0.577 | 1 | 0.782 | 5378 | 7345 |
| Cytokinesis | 16 | 16 | 0.2 | −0.34 | 0.6 | −1.46 | 1 | 0.229 | 0.964 | 0.02 | 1 | 0.159 | 448 | 6088 |
| ER Golgi intermediate compartment | 16 | 16 | 0.35 | −0.22 | 1.06 | −0.94 | 1 | 0.994 | 0.39 | 0.566 | 0.923 | 0.712 | 5924 | 7226 |
| Neuropeptide binding | 17 | 17 | 0.29 | −0.2 | 0.89 | −0.93 | 1 | 0.994 | 0.67 | 0.679 | 1 | 0.648 | 4808 | 6250 |
| Neuropeptide receptor Activity | 17 | 17 | 0.29 | −0.2 | 0.89 | −0.77 | 1 | 1 | 0.638 | 0.853 | 1 | 0.792 | 4808 | 6250 |
| Peptide receptor activity | 38 | 38 | 0.29 | −0.15 | 1.01 | −1 | 1 | 0.988 | 0.477 | 0.5 | 0.99 | 0.84 | 4920 | 6917 |
| Rhodopsin like receptor activity | 80 | 80 | 0.23 | −0.15 | 0.86 | 1 | 0 | 0.834 | 1 | 1 | 5403 | 7907 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammond, T.G.; Allen, P.L.; Birdsall, H.H. Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses. Int. J. Mol. Sci. 2018, 19, 4106. https://doi.org/10.3390/ijms19124106
Hammond TG, Allen PL, Birdsall HH. Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses. International Journal of Molecular Sciences. 2018; 19(12):4106. https://doi.org/10.3390/ijms19124106
Chicago/Turabian StyleHammond, Timothy G., Patricia L. Allen, and Holly H. Birdsall. 2018. "Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses" International Journal of Molecular Sciences 19, no. 12: 4106. https://doi.org/10.3390/ijms19124106
APA StyleHammond, T. G., Allen, P. L., & Birdsall, H. H. (2018). Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses. International Journal of Molecular Sciences, 19(12), 4106. https://doi.org/10.3390/ijms19124106





