Search of Allosteric Inhibitors and Associated Proteins of an AKT-like Kinase from Trypanosoma cruzi
Abstract
1. Introduction
2. Results
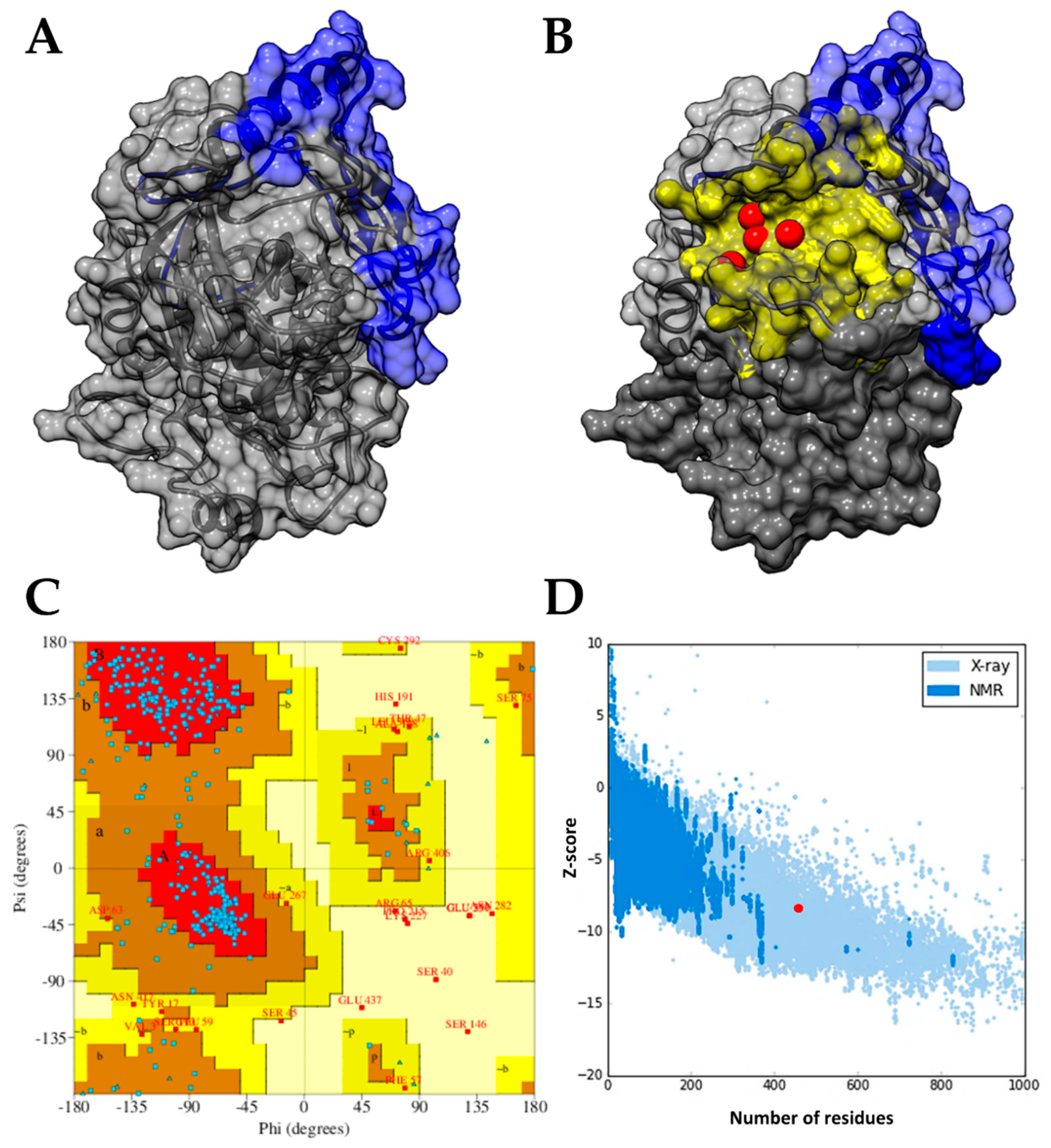
2.1. Modeling and Prediction of Pockets in the AKT-Like Protein of T. cruzi
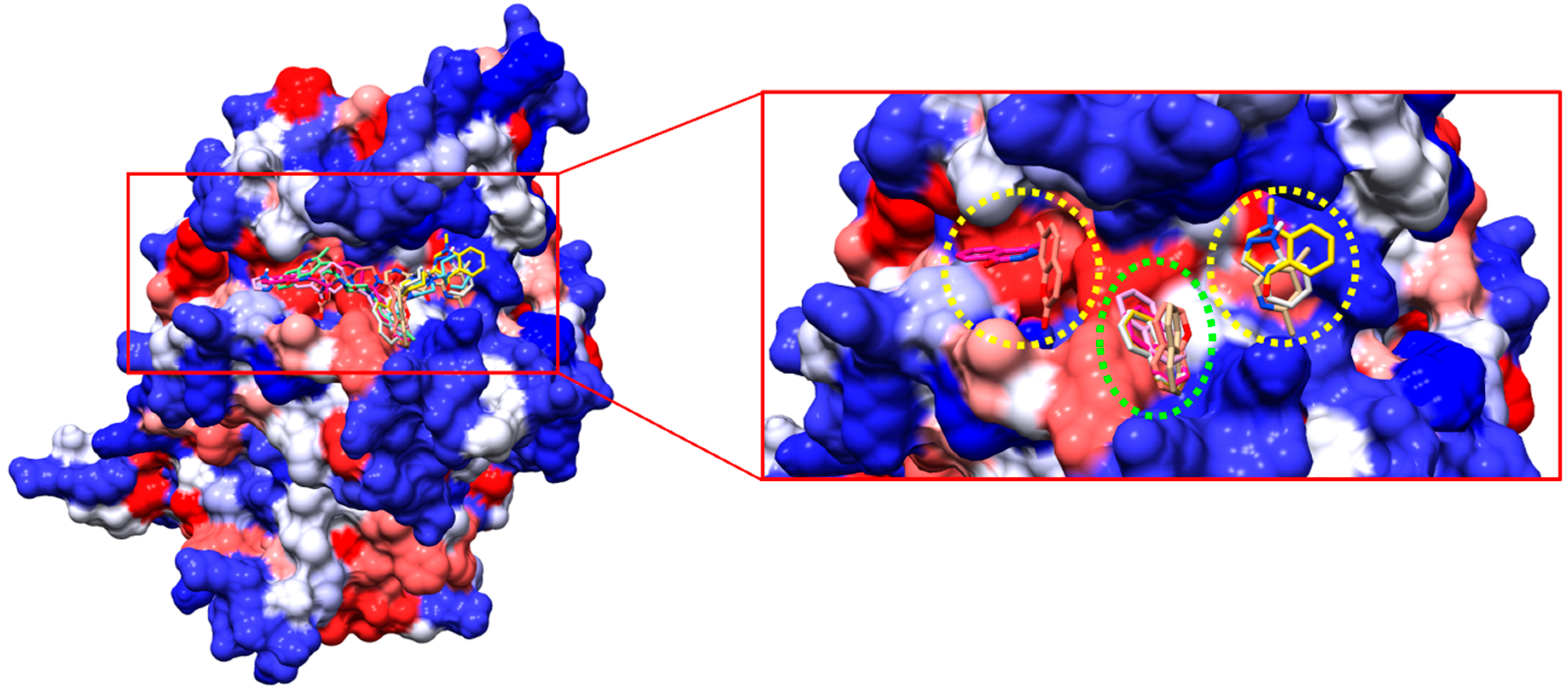
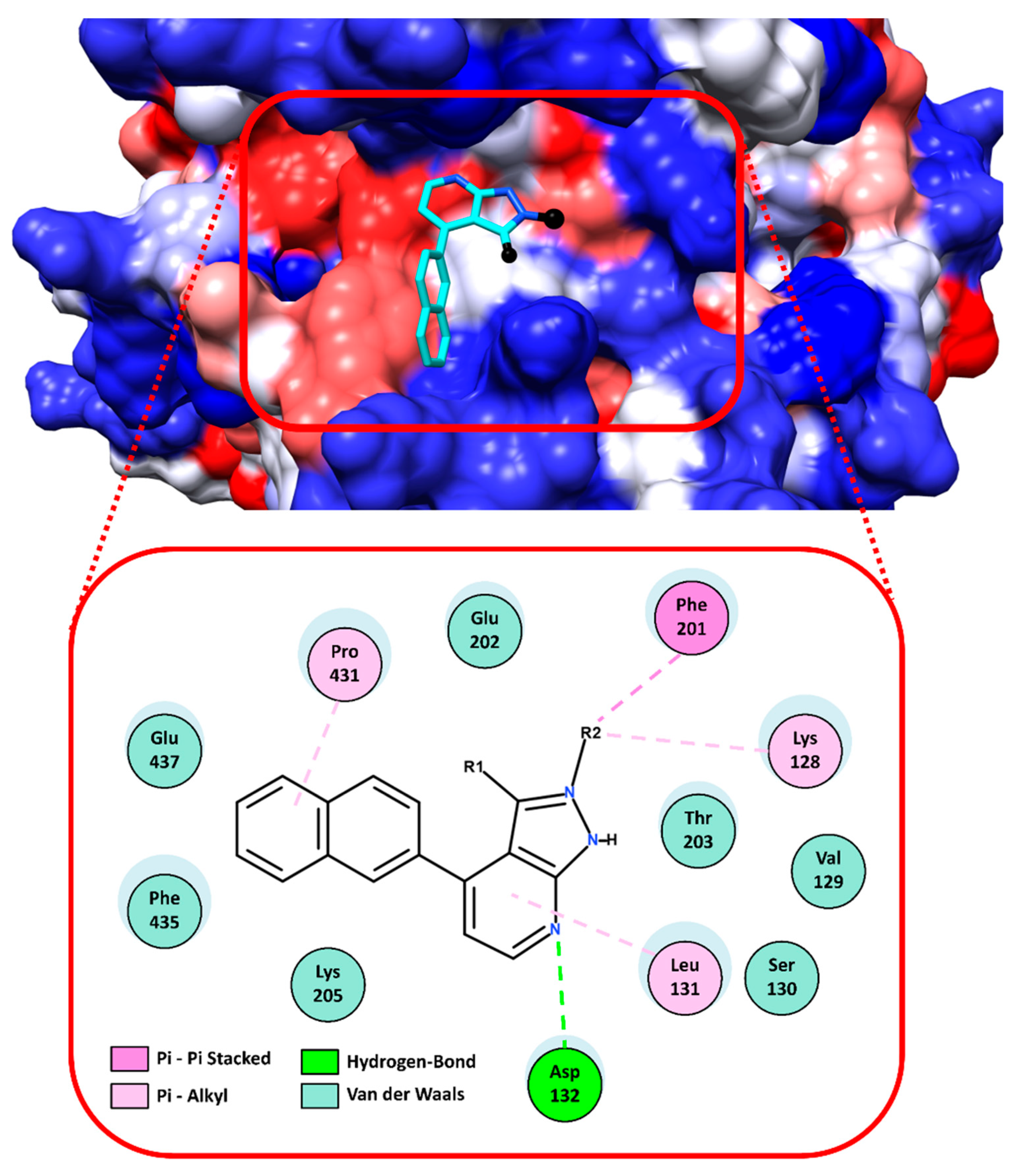
2.2. Virtual Screening and Ligand Property Predictions
2.3. Results of the In Vitro Assay
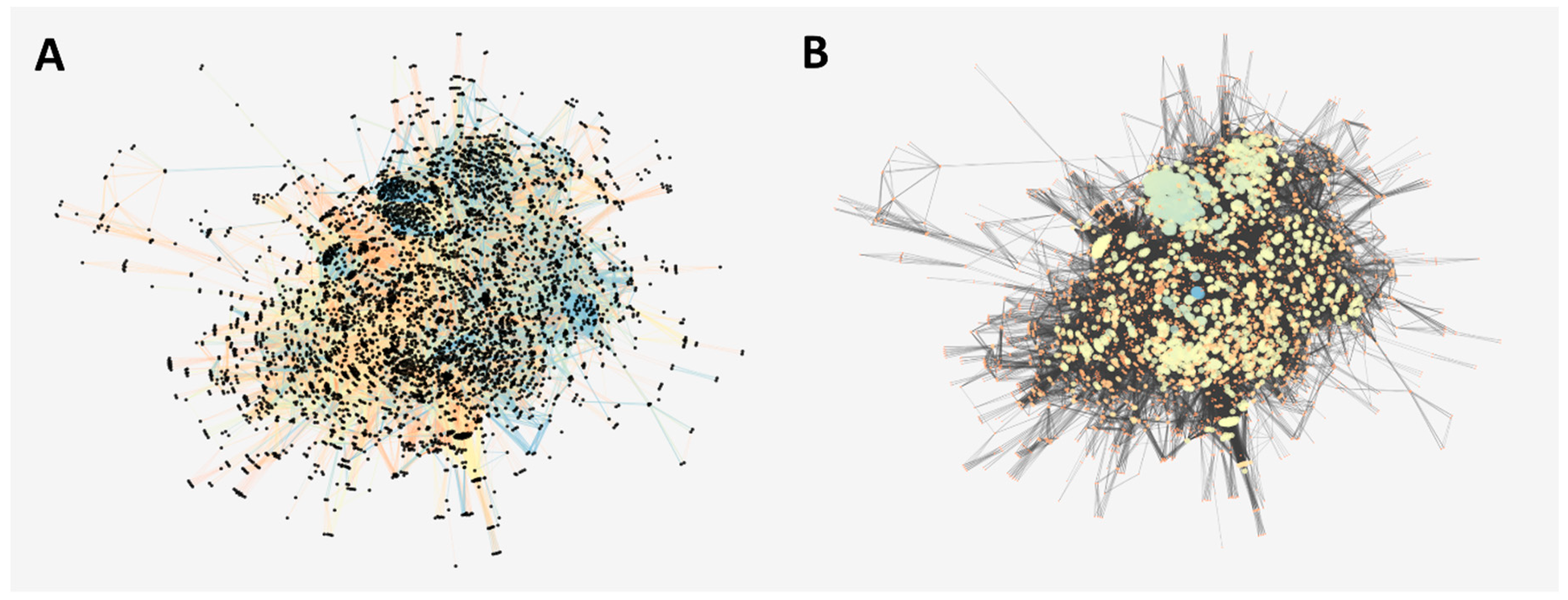
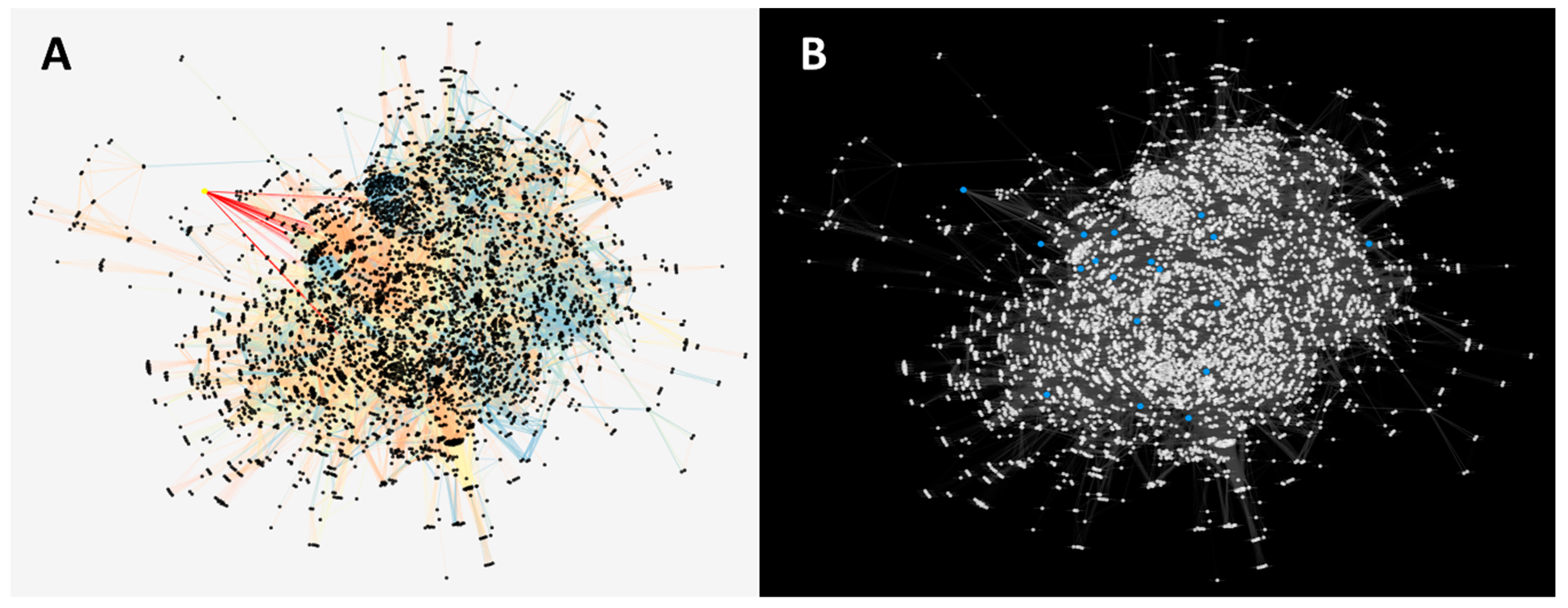
2.4. Role of the AKT-Like Protein in a Reconstructed Protein–Protein Interaction Network of T. cruzi
2.5. Identification of Proteins Potentially Involved in the AKT-Like Pathway from the Parasite
3. Discussion
4. Materials and Methods
4.1. Structural Modeling of the AKT-Like Protein of T. cruzi
4.2. Virtual Screening and Molecular Docking Simulations
4.3. In Silico Prediction of Toxicological Risks
4.4. In Vitro Assay of the Antitrypanosomal Activity of the UBMC Compounds
4.5. In Vitro Assay of Cytotoxicity of UBMC Compounds
4.6. Reconstruction of the Protein–Protein Interaction Network of T. cruzi
4.7. Detection of Orthologs between T. cruzi and Those Reported in the PI3/AKT/mTOR Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PKB | Protein kinase B |
| RAC | Related to A- and C-kinases |
| PH | Pleckstrin homology domain |
| PI2P | Phosphatidylinositol bisphosphate |
| PI3P | Phosphatidylinositol trisphosphate |
| TcAKT-like | AKT-like protein of Trypanosoma cruzi |
| OD | Optical density |
References
- Kim, J.H.; Ryu, W.; Shim, H.; Park, H. Development of new and selective Trypanosoma cruzi trans-sialidase inhibitors from sulfonamide chalcones and their derivatives. Chembiochem 2009, 10, 2475–2479. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Liu, X.; Stinson, M.; Rivera, I.; Groessl, T.; Tuntland, T.; Yeh, V.; Wen, B.; Molteni, V.; Glynne, R.; et al. Antitrypanosomal treatment with benznidazole is superior to posaconazole regimens in mouse models of chagas disease. Antimicrob. Agents Chemother. 2015, 59, 6385–6394. [Google Scholar] [CrossRef] [PubMed]
- Gabelli, S.B.; McLellan, J.S.; Montalvetti, A.; Oldfield, E.; Docampo, R.; Amzel, L.M. Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design. Proteins 2005, 62, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Fayard, E.; Tintignac, L.A.; Baudry, A.; Hemmings, B.A. Protein Kinase B/Akt at a Glance. J. Cell Sci. 2005, 118, 5675–5678. [Google Scholar] [CrossRef] [PubMed]
- Bachmaier, S.; Boshart, M. Kinetoplastid AGC kinases. In Protein Phosphorylation in Parasites: Novel Targets for Antiparasitic Intervention; Willey: Hoboken, NJ, USA, 2013; pp. 99–122. [Google Scholar]
- Mabuchi, S.; Kuroda, H.; Takahashi, R.; Sasano, T. The PI3K/AKT/MTOR pathway as a therapeutic target in ovarian cancer. Gynecol. Oncol. 2015, 137, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K.M.; Anderson, N.G. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 2002, 14, 381–395. [Google Scholar] [CrossRef]
- Prêtre, V.; Wicki, A. Inhibition of Akt and other AGC kinases: A target for clinical cancer therapy? Semin. Cancer Biol. 2018, 48, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Leroux, A.E.; Schulze, J.O.; Biondi, R.M. AGC Kinases, Mechanisms of regulation and innovative drug development. Semin. Cancer Biol. 2018, 48, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tirado-Duarte, D.; Marín-Villa, M.; Ochoa, R.; Blandón-Fuentes, G.; Soares, M.J.; Robledo, S.M.; Varela-Miranda, R.E. The Akt-like kinase of Leishmania panamensis: As a new molecular target for drug discovery. Acta Trop. 2018, 177, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Varela, R.E.M.; Ochoa, R.; Muskus, C.E.; Muro, A.; Mollinedo, F. Identification of a RAC/AKT-like gene in Leishmania parasites as a putative therapeutic target in leishmaniasis. Parasit. Vectors 2017, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Davoli-Ferreira, M.; Medina, T.S.; Sesti-Costa, R.; Silva, G.K.; Lopes, C.D.; Cardozo, L.E.; Gava, F.N.; Lyroni, K.; Dias, F.C.; et al. Canonical PI3Kγ signaling in myeloid cells restricts Trypanosoma cruzi infection and dampens chagasic myocarditis. Nat. Commun. 2018, 9, 1513. [Google Scholar] [CrossRef] [PubMed]
- Márquez, J.D.R.; Ana, Y.; Baigorrí, R.E.; Stempi, C.C.; Cerban, F.M. Mammalian target of rapamycin inhibition in Trypanosoma cruzi-infected macrophages leads to an intracellular profile that is detrimental for infection. Front. Immunol. 2018, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Pascuccelli, V.; Labriola, C.; Téllez-Iñón, M.T.; Parodi, A.J. Molecular and biochemical characterization of a protein kinase B from Trypanosoma cruzi. Mol. Biochem. Parasitol. 1999, 102, 21–33. [Google Scholar] [CrossRef]
- Scheffzek, K.; Welti, S. Pleckstrin homology (PH) like domains—Versatile modules in protein-protein interaction platforms. FEBS Lett. 2012, 586, 2662–2673. [Google Scholar] [CrossRef] [PubMed]
- Ahad, A.M.; Zuohe, S.; Du-Cuny, L.; Moses, S.A.; Zhou, L.L.; Zhang, S.; Powis, G.; Meuillet, E.J.; Mash, E.A. Development of sulfonamide AKT PH domain inhibitors. Bioorganic Med. Chem. 2011, 19, 2046–2054. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, U.; Tomlinson, S.M.; Fonner, J.M.; Mock, S.A.; Watowich, S.J. Identification of a novel inhibitor of dengue virus protease through use of a virtual screening drug discovery web portal. J. Chem. Inf. Model. 2014, 54, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2013, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Lin, B.; Schroeder, M.; Huang, B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics 2011, 27, 2083–2088. [Google Scholar] [CrossRef] [PubMed]
- Luchtan, M. TcruziDB: An integrated Trypanosoma cruzi genome resource. Nucleic Acids Res. 2004, 32, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Balaña-Fouce, R.; Álvarez-Velilla, R.; Fernández-Prada, C.; García-Estrada, C.; Reguera, R.M. Trypanosomatids topoisomerase re-visited. new structural findings and role in drug discovery. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Leija, C.; Rijo-ferreira, F.; Chen, J.; Cestari, I.; Tu, B.P.; Phillips, M.A. GMP synthase is essential for viability and infectivity of Trypanosoma brucei despite a redundant purine salvage pathway. Mol. Microbiol. 2016, 97, 1006–1020. [Google Scholar] [CrossRef] [PubMed]
- Senkovich, O.; Schormann, N.; Chattopadhyay, D. Structures of Dihydrofolate Reductase-Thymidylate Synthase of Trypanosoma cruzi in the Folate-Free State and in Complex with Two Antifolate Drugs, Trimetrexate and Methotrexate. Acta Crystallogr. Sect. D Biol. 2009, 65, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Wilkowsky, S.E.; Barbieri, M.A.; Stahl, P.; Isola, E.L.D. Trypanosoma Cruzi: Phosphatidylinositol 3-kinase and protein kinase b activation is associated with parasite invasion. Exp. Cell Res. 2001, 264, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Chuenkova, M.V.; Furnari, F.B.; Cavenee, W.K.; Pereira, M.A. Trypanosoma cruzi trans-sialidase: A potent and specific survival factor for human schwann cells by means of phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 9936–9941. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dai, D.; Chen, B.; Tang, H.; Xie, X.; Wei, W. Efficacy of PI3K/AKT/MTOR pathway inhibitors for the treatment of advanced solid cancers: A literature-based meta-analysis of 46 randomised control trials. PLoS ONE 2018, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, D.; Powis, G.; Mash, E.A.; George, B.; Gokhale, V.M.; Zhang, S.; Shakalya, K.; Du-Cuny, L.; Berggren, M.; Ali, M.A.; et al. Discovery of a novel class of AKT pleckstrin homology (PH) domain inhibitors. Mol. Cancer Ther. 2008, 7, 2621–2632. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.; Dangate, M.; Vetro, M.; Donvito, G.; Gabrielli, L.; Amigoni, L.; Cassinelli, G.; Lanzi, C.; Ceriani, M.; de Gioia, L.; et al. Synthetic sulfoglycolipids targeting the serine–threonine protein kinase Akt. Bioorganic Med. Chem. 2016, 24, 3396–3405. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Oshimura, M.; Ito, H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis 2004, 9, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Franke, T.F. Intracellular signaling by Akt: Bound to be specific. Sci. Signal. 2008, 1, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Logan-Klumpler, F.J.; de Silva, N.; Boehme, U.; Rogers, M.B.; Velarde, G.; McQuillan, J.A.; Carver, T.; Aslett, M.; Olsen, C.; Subramanian, S.; et al. GeneDB-an annotation database for pathogens. Nucleic Acids Res. 2012, 40, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Albà, M.M. Zinc-finger domains in metazoans: Evolution gone wild. Genome Biol. 2017, 18, 168. [Google Scholar] [CrossRef] [PubMed]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Salvador, R.R.S.; Bello, M.L.; Barreto, I.R.L.; Vera, M.A.F.; Muri, E.M.F.; de Albuquerque, S.; Dias, L.R.S. New carbohydrazide derivatives of 1H-Pyrazolo[3,4-b]Pyridine and trypanocidal activity. An. Acad. Bras. Cienc. 2016, 88, 2341–2348. [Google Scholar] [CrossRef] [PubMed]
- Ojo, K.K.; Reid, M.C.; Siddaramaiah, L.K.; Müller, J.; Winzer, P.; Zhang, Z.; Keyloun, K.R.; Vidadala, R.S.R.; Merritt, E.A.; Hol, W.G.J.; et al. Neospora caninum calcium-dependent protein kinase 1 is an effective drug target for neosporosis therapy. PLoS ONE 2014, 9, e92929. [Google Scholar] [CrossRef] [PubMed]
- Rutaganira, F.U.; Barks, J.; Dhason, M.S.; Wang, Q.; Lopez, M.S.; Long, S.; Radke, J.B.; Jones, N.G.; Maddirala, A.R.; Janetka, J.W.; et al. Inhibition of calcium dependent protein kinase 1 (CDPK1) by pyrazolopyrimidine analogs decreases establishment and reoccurrence of central nervous system disease by Toxoplasma gondii. J. Med. Chem. 2017, 60, 9976–9989. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.A.; Libra, M.; Tsatsakis, A.M. Akt Inhibitors in cancer treatment: The long journey from drug discovery to clinical use. Int. J. Oncol. 2016, 48, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.-A.; Riccardi, L.; Minniti, E.; Borgogno, M.; Arencibia, J.M.; Greco, M.L.; Minarini, A.; Sissi, C.; de Vivo, M. Pharmacophore hybridization to discover novel topoisomerase II poisons with promising antiproliferative activity. J. Med. Chem. 2017, 61, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The current case of quinolones: Synthetic approaches and antibacterial activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Roa, C.; Cossio-Pérez, R.; Molina, D.; Patiño, J.; Cardona, N. In silico study of moxifloxacin derivatives with possible antibacterial activity against a resistant form of DNA gyrase from Porphyromonas gingivalis. Arch. Oral Biol. 2018, 95, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Gafter-Gvili, A.; Fraser, A.; Leibovici, L. The anti-cancer effects of quinolone antibiotics? Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Dalhoff, A. Antiviral, antifungal, and antiparasitic activities of fluoroquinolones optimized for treatment of bacterial infections: A puzzling paradox or a logical consequence of their mode of action? Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Martins-Duarte, E.S.; Dubar, F.; Lawton, P.; Da Silva, C.F.; Soeiro, M.D.N.C.; de Souza, W.; Biot, C.; Vommaro, R.C. Ciprofloxacin derivatives affect parasite cell division and increase the survival of mice infected with Toxoplasma gondii. PLoS ONE 2015, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Borrel, A.; Regad, L.; Xhaard, H.; Petitjean, M.; Camproux, A.C. PockDrug: A model for predicting pocket druggability that overcomes pocket estimation uncertainties. J. Chem. Inf. Model. 2015, 55, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. Datawarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ai, H.; Chen, W.; Yin, Z.; Hu, H.; Zhu, J.; Zhao, J.; Zhao, Q.; Liu, H. CarcinoPred-EL: Novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Finney, D.J. Statistical Method in Biological Assay; Charles Griffin and Company: London, UK, 1952. [Google Scholar]
- Pastrana Restrepo, M.; Galeano Jaramillo, E.; Martínez Martínez, A.; Robledo Restrepo, S. Synthesis and trypanocide activity of chloro-l-tyrosine and bromo-l-tyrosine derivatives. Med. Chem. Res. 2018, 27, 2454. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Hermjakob, H.; Montecchi-Pilazzi, L.; Lewington, C.; Mudali, S.; Kerrien, S.; Orchard, S.; Vingron, M.; Roechert, B.; Roepstorff, P.; Valencia, A.; et al. IntAct: An open source molecular interaction database. Nucleic Acids Res. 2004, 32, D452–D455. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
| Compound | Predicted Scores (kcal/mol) | Predicted Toxicity | |
|---|---|---|---|
| AutoDock Vina | SwissDock | ||
| UBMC-1 | −10.6 | -8.61 | Positive |
| UBMC-2 | −10.3 | −8.33 | Positive |
| UBMC-3 | −10.0 | −7.41 | Positive |
| UBMC-4 | −9.9 | −8.41 | Positive |
| UBMC-5 | −9.8 | −7.9 | Negative |
| UBMC-6 | −9.8 | −7.63 | Negative |
| UBMC-7 | −9.7 | −7.07 | Positive |
| UBMC-8 | −9.3 | −7.27 | Negative |
| Benznidazole | −6.2 | −6.48 | Positive |
| Compound | IC50 (µM) a | LC50 (µM) b | SI c |
|---|---|---|---|
| UBMC-6 | 14.25 ± 1 | 34.11 ± 3.14 | 2.39 |
| UBMC-8 | 18.26 ± 1.30 | 55.2 ± 2.25 | 3.02 |
| UBMC-7 | 19.44 ± 0.35 | 62.3 ± 5.63 | 3.2 |
| UBMC-1 | 34.92 ± 3.87 | 61.66 ± 456 | 1.76 |
| UBMC-3 | 37.53 ± 2.65 | 197.9 ± 7.84 | 5.27 |
| UBMC-2 | 62.44 ± 4.62 | 44.5 ± 2.88 | 0.71 |
| UBMC-5 | 69.82 ± 9.23 | 43.25 ± 7.53 | 0.61 |
| UBMC-4 | 72.13 ± 8.58 | 70.88 ± 6.33 | 0.98 |
| Benznidazole | 16.61 ± 4 | >200 (µg/mL) | 12.04 |
| Doxorrubicin | NA d | 3.91 ± 0.40 | NC e |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa, R.; Rocha-Roa, C.; Marín-Villa, M.; Robledo, S.M.; Varela-M, R.E. Search of Allosteric Inhibitors and Associated Proteins of an AKT-like Kinase from Trypanosoma cruzi. Int. J. Mol. Sci. 2018, 19, 3951. https://doi.org/10.3390/ijms19123951
Ochoa R, Rocha-Roa C, Marín-Villa M, Robledo SM, Varela-M RE. Search of Allosteric Inhibitors and Associated Proteins of an AKT-like Kinase from Trypanosoma cruzi. International Journal of Molecular Sciences. 2018; 19(12):3951. https://doi.org/10.3390/ijms19123951
Chicago/Turabian StyleOchoa, Rodrigo, Cristian Rocha-Roa, Marcel Marín-Villa, Sara M. Robledo, and Rubén E. Varela-M. 2018. "Search of Allosteric Inhibitors and Associated Proteins of an AKT-like Kinase from Trypanosoma cruzi" International Journal of Molecular Sciences 19, no. 12: 3951. https://doi.org/10.3390/ijms19123951
APA StyleOchoa, R., Rocha-Roa, C., Marín-Villa, M., Robledo, S. M., & Varela-M, R. E. (2018). Search of Allosteric Inhibitors and Associated Proteins of an AKT-like Kinase from Trypanosoma cruzi. International Journal of Molecular Sciences, 19(12), 3951. https://doi.org/10.3390/ijms19123951






