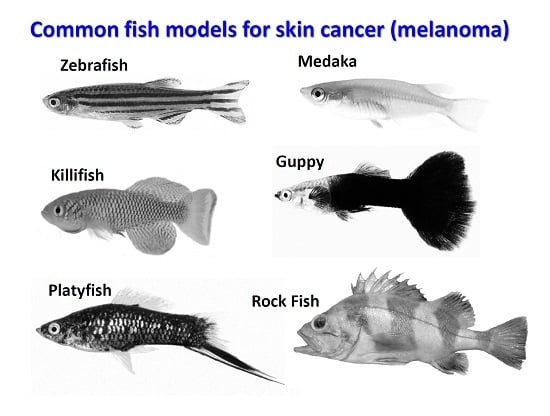
The Power of Fish Models to Elucidate Skin Cancer Pathogenesis and Impact the Discovery of New Therapeutic Opportunities
Abstract
1. Introduction
2. Teleost Models for Melanoma Studies
3. Zebrafish
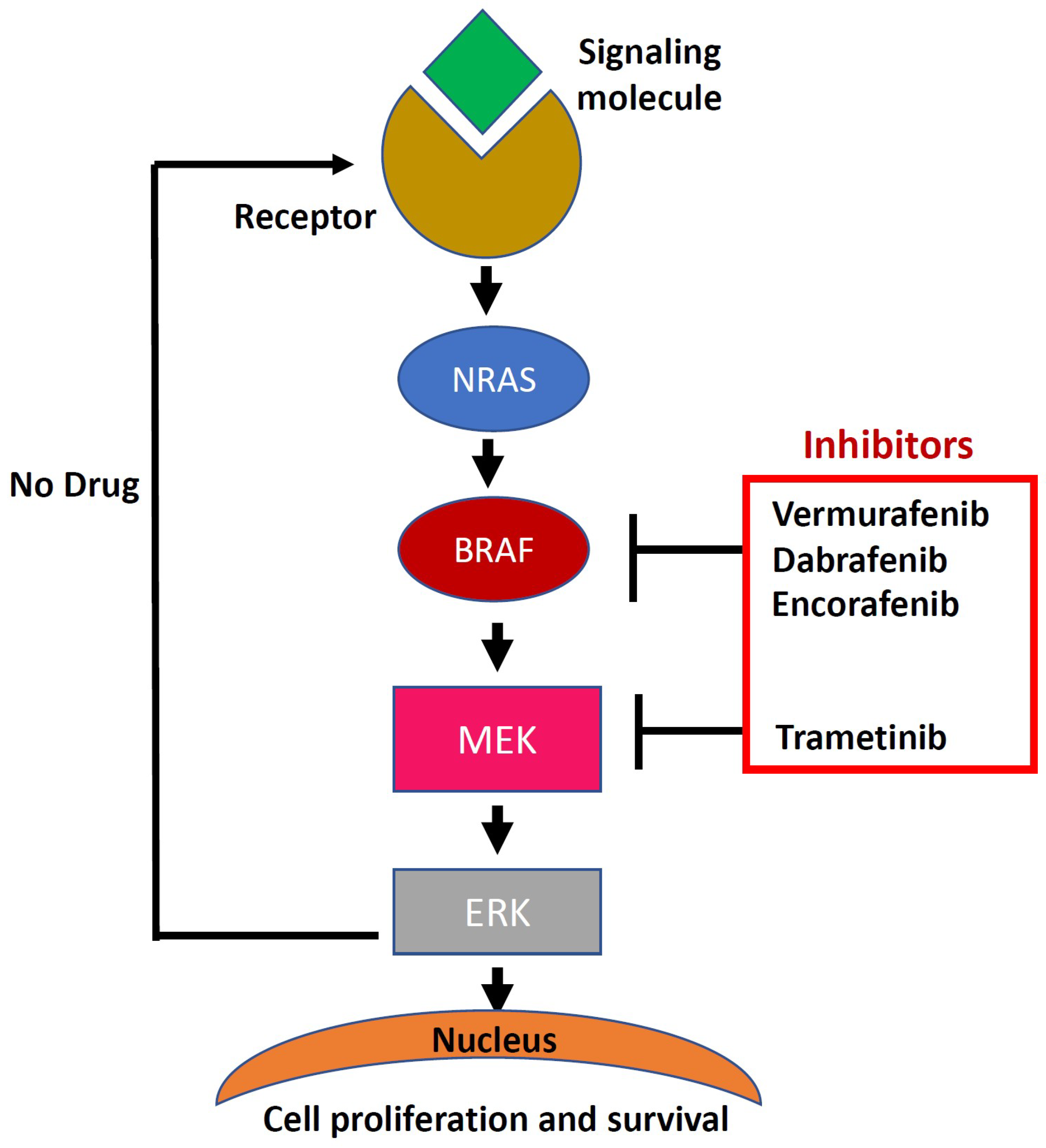
4. Downstream Signaling in BRAF/Mitogen-activated protein (MAP) Kinase Pathway
5. Non-Melanoma Skin Cancer Pathogenesis in Zebrafish Model
6. Platyfish (Xiphophorus Sp.)
7. Medaka (Oryzias latipes)
8. Annual Fish or Killifish
9. Rock Fish (Sebastes Sp.)
10. Guppy (Poecilia reticulata)
11. Limitations of Animal Models in Skin Cancer Research
11.1. Laboratory Mice
11.2. Fruitfly (Drosophila Melanogaster) as a Model for Skin Cancer Research
11.3. Teleost
12. Cutting Edge Methods to Boost Skin Cancer Studies in Fish
13. Small Molecule and Drug Screening in Zebrafish
14. Research is Changing in Advanced Melanoma (Stage III) Treatment
15. Injections of Modified Herpes Virus Kills Cancer Cells
16. Summarizing Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- von Mässenhausen, A.; Sanders, C.; Brägelmann, J.; Konantz, M.; Queisser, A.; Vogel, W.; Kristiansen, G.; Duensing, S.; Schröck, A.; Bootz, F. Targeting ddr2 in head and neck squamous cell carcinoma with dasatinib. Int. J. Cancer 2016, 139, 2359–2369. [Google Scholar] [CrossRef]
- Key Statistics for Melanoma Skin Cancer; American Cancer Society: Atlanta, GA, USA, 2018.
- Thompson, J.F.; Scolyer, R.A.; Kefford, R.F. Cutaneous melanoma. Lancet 2005, 365, 687–701. [Google Scholar] [CrossRef]
- Maddodi, N.; Setaluri, V. Role of uv in cutaneous melanoma. Photochem. Photobiol. 2008, 84, 528–536. [Google Scholar] [CrossRef]
- Gilchrest, B.A.; Eller, M.S.; Geller, A.C.; Yaar, M. The pathogenesis of melanoma induced by ultraviolet radiation. N. Engl. J. Med. 1999, 340, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Hodis, E.; Heffernan, T.P.; Deribe, Y.L.; Lawrence, M.S.; Protopopov, A.; Ivanova, E.; Watson, I.R.; Nickerson, E.; Ghosh, P. Melanoma genome sequencing reveals frequent prex2 mutations. Nature 2012, 485, 502. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Paus, R. Bomirski melanomas-a versatile and powerful model for pigment cell and melanoma research. Int. J. Oncol. 1993, 2, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Shen, Y.; Maurus, K.; Walter, R.; Tomlinson, C.; Wilson, R.K.; Postlethwait, J.; Warren, W.C. Whole body melanoma transcriptome response in medaka. PLoS ONE 2015, 10, e0143057. [Google Scholar] [CrossRef]
- Plonka, P.M.; Slominski, A.T.; Pajak, S.; Urbanska, K. Transplantable melanomas in gerbils (meriones unguiculatus). Ii: Melanogenesis. Exp. Dermatol. 2003, 12, 356–364. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I. Taking human cancer genes to the fish: A transgenic model of melanoma in zebrafish. Zebrafish 2005, 1, 363–368. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498. [Google Scholar] [CrossRef]
- Schartl, M. Beyond the zebrafish: Diverse fish species for modeling human disease. Dis. Models Mech. 2014, 7, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.H.; Woods, I.G.; Ngo-Hazelett, P.; Yan, Y.L.; Kelly, P.D.; Chu, F.; Huang, H.; Hill-Force, A.; Talbot, W.S. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000, 10, 1890–1902. [Google Scholar] [CrossRef]
- Anderson, J.L.; Mulligan, T.S.; Shen, M.-C.; Wang, H.; Scahill, C.M.; Tan, F.J.; Du, S.J.; Busch-Nentwich, E.M.; Farber, S.A. Mrna processing in mutant zebrafish lines generated by chemical and crispr-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 2017, 13, e1007105. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Nikaido, M.; Hagino-Yamagishi, K.; Okada, N. Distinct functions of two olfactory marker protein genes derived from teleost-specific whole genome duplication. BMC Evol. Biol. 2015, 15, 245. [Google Scholar] [CrossRef] [PubMed]
- Schartl, A.; Malitschek, B.; Kazianis, S.; Borowsky, R.; Schartl, M. Spontaneous melanoma formation in nonhybrid xiphophorus. Cancer Res. 1995, 55, 159–165. [Google Scholar] [PubMed]
- Patton, E.E.; Mitchell, D.L.; Nairn, R.S. Genetic and environmental melanoma models in fish. Pigm. Cell Melanoma Res. 2010, 23, 314–337. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, J.P.; Dadras, S.S.; Silva, K.A.; Kennedy, V.E.; Murray, S.A.; Denegre, J.M.; Schofield, P.N.; King, L.E., Jr.; Wiles, M.V.; Pratt, C.H. Excavating the genome: Large-scale mutagenesis screening for the discovery of new mouse models. J. Investig. Dermatol. Symp. Proc. 2015, 17, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Prykhozhij, S.V.; Steele, S.L.; Razaghi, B.; Berman, J.N. A rapid and effective method for screening, sequencing and reporter verification of engineered frameshift mutations in zebrafish. Dis. Model Mech. 2017, 10, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Lenz, S.; Karsten, P.; Schulz, J.B.; Voigt, A. Drosophila as a screening tool to study human neurodegenerative diseases. J. Neurochem. 2013, 127, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Dovey, M.; White, R.M.; Zon, L.I. Oncogenic nras cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 2009, 6, 397–404. [Google Scholar] [CrossRef]
- Yen, J.; White, R.M.; Wedge, D.C.; Van Loo, P.; de Ridder, J.; Capper, A.; Richardson, J.; Jones, D.; Raine, K.; Watson, I.R. The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biol. 2013, 14, R113. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Houvras, Y.; White, R.M.; Zon, L.I. Melanoma biology and the promise of zebrafish. Zebrafish 2008, 5, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, S. Conditional Control of Mitf Reveals Cellular Subpopulations Essential for Melanoma Survival and Recurrence in New Zebrafish Models. 2018. Available online: https://www.era.lib.ed.ac.uk/handle/1842/29645 (accessed on 6 December 2018).
- Chen, L.; Ren, X.; Liang, F.; Li, S.; Zhong, H.; Lin, S. Characterization of two novel small molecules targeting melanocyte development in zebrafish embryogenesis. Pigm. Cell Melanoma Res. 2012, 25, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, A.; Gavriouchkina, D.; Zorman, J.; Chong-Morrison, V.; Napolitani, G.; Cerundolo, V.; Sauka-Spengler, T. Generation of a double binary transgenic zebrafish model to study myeloid gene regulation in response to oncogene activation in melanocytes. Dis. Models Mech. 2018. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, C.; Jones, M.; Walker, P.; Kamarashev, J.; Kelly, A.; Hurlstone, A.F. Dissecting the roles of raf-and pi3k-signalling pathways in melanoma formation and progression in a zebrafish model. Dis. Models Mech. 2009. [Google Scholar] [CrossRef] [PubMed]
- Anelli, V.; Zettler, N.; Mione, M. Insights from genetic models of melanoma in fish. Curr. Pathobiol. Rep. 2014, 2, 85–92. [Google Scholar] [CrossRef]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef]
- Chen, C.-H.; Lin, D.-S.; Cheng, C.-W.; Lin, C.-J.; Lo, Y.-K.; Yen, C.-C.; Lee, A.Y.-L.; Hsiao, C.-D. Cdc6 cooperates with c-myc to promote genome instability and epithelial to mesenchymal transition (emt) in zebrafish. Oncotarget 2014, 5, 6300. [Google Scholar] [CrossRef]
- Yin, A.; Campbell, N.R.; Jones, L.W.; White, R.M. Zebrafish as a model to investigate the effects of exercise in cancer. bioRxiv 2018, 279232. [Google Scholar] [CrossRef]
- Perez, A.N.; Oehlers, L.; Heater, S.J.; Booth, R.E.; Walter, R.B.; David, W.M. Proteomic analyses of the xiphophorus gordon–kosswig melanoma model. Comp. Biochem. Physiol. Part C 2012, 155, 81–88. [Google Scholar] [CrossRef]
- Herrera, M.; Jagadeeswaran, P. Annual fish as a genetic model for aging. J. Gerontol. Ser. A 2004, 59, B101–B107. [Google Scholar] [CrossRef]
- Tanaka, M.; Kinoshita, M.; Kobayashi, D.; Nagahama, Y. Establishment of medaka (oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: A useful model to monitor germ cells in a live vertebrate. Proc. Natl. Acad. Sci. USA 2001, 98, 2544–2549. [Google Scholar] [CrossRef] [PubMed]
- Sandel, M.W.; Aguilar, A.; Buonaccorsi, V.P.; Herstein, J.; Evgrafov, O. Complete mitochondrial genome sequences of five rockfishes (perciformes: Sebastes). Mitochondrial DNA Part B 2018, 3, 825–826. [Google Scholar] [CrossRef]
- Künstner, A.; Hoffmann, M.; Fraser, B.A.; Kottler, V.A.; Sharma, E.; Weigel, D.; Dreyer, C. The genome of the trinidadian guppy, poecilia reticulata, and variation in the guanapo population. PLoS ONE 2016, 11, e0169087. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.R.; Steffen, L.S.; Howell, M.H.; Pusack, T.J.; Lawrence, C.; Kunkel, L.M. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta 2007, 1772, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Rennekamp, A.J.; Peterson, R.T. 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 2015, 24, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S.; Slack, B.E.; Uchiyama, J.; Zhdanova, I.V. Zebrafish as a genetic model in biological and behavioral gerontology: Where development meets aging in vertebrates—A mini-review. Gerontology 2009, 55, 430–441. [Google Scholar] [CrossRef]
- White, R.M.; Zon, L.I. Melanocytes in development, regeneration, and cancer. Cell Stem Cell 2008, 3, 242–252. [Google Scholar] [CrossRef]
- Kaufman, C.K.; Mosimann, C.; Fan, Z.P.; Yang, S.; Thomas, A.J.; Ablain, J.; Tan, J.L.; Fogley, R.D.; van Rooijen, E.; Hagedorn, E.J. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 2016, 351, aad2197. [Google Scholar] [CrossRef]
- Topczewska, J.M.; Postovit, L.-M.; Margaryan, N.V.; Anthony, S.; Hess, A.R.; Wheaton, W.W.; Nickoloff, B.J.; Topczewski, J.; Hendrix, M.J. Embryonic and tumorigenic pathways converge via nodal signaling: Role in melanoma aggressiveness. Nat. Med. 2006, 12, 925. [Google Scholar] [CrossRef]
- Krauss, J.; Geiger-Rudolph, S.; Koch, I.; Nüsslein-Volhard, C.; Irion, U. A dominant mutation in tyrp1a leads to melanophore death in zebrafish. Pigm. Cell Melanoma Res. 2014, 27, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Storer, N.Y.; Zon, L.I. Zebrafish models of p53 functions. Cold Spring Harbor Perspect. Biol. 2010, 2, a001123. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Capper, A.; Zeng, Z.; Mathers, M.E.; Richardson, J.; Paranthaman, K.; Jackson, I.J.; Patton, E.E. A conditional zebrafish mitf mutation reveals mitf levels are critical for melanoma promotion vs. Regression in vivo. J. Investig. Dermatol. 2014, 134, 133–140. [Google Scholar] [CrossRef] [PubMed]
- White, R.M.; Zon, L.I. Abstract Lb-335: Dhodh Regulates Transcriptional Elongation during Melanoma Evolution: Insights from Zebrafish Screening; AACR: Philadelphia, PA, USA, 2011. [Google Scholar]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L. Frequency of tert promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Puzanov, I.; Kim, K.B.; Ribas, A.; McArthur, G.A.; Sosman, J.A.; O’dwyer, P.J.; Lee, R.J.; Grippo, J.F.; Nolop, K. Inhibition of mutated, activated braf in metastatic melanoma. N. Engl. J. Med. 2010, 363, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Gabellini, C.; Gómez, E.; de Oliveira, S.; Del Bufalo, D.; Mulero, V. Bcl-Xl Protein Overexpression Enhances Tumor Progression of Human Melanoma Cells in Zebrafish Xenograft Model: Involvement of cxcl8 Chemokine; AACR: Philadelphia, PA, USA, 2014. [Google Scholar]
- Joung, J.K.; Sander, J.D. Talens: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Suster, M.L.; Kikuta, H.; Urasaki, A.; Asakawa, K.; Kawakami, K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 2009, 561, 41–63. [Google Scholar]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W. Mutations of the braf gene in human cancer. Nature 2002, 417, 949. [Google Scholar] [CrossRef]
- Busca, R.; Ballotti, R. Cyclic amp a key messenger in the regulation of skin pigmentation. Pigm. Cell Melanoma Res. 2000, 13, 60–69. [Google Scholar] [CrossRef]
- Goel, V.K.; Lazar, A.J.; Warneke, C.L.; Redston, M.S.; Haluska, F.G. Examination of mutations in braf, nras, and pten in primary cutaneous melanoma. J. Investig. Dermatol. 2006, 126, 154–160. [Google Scholar] [CrossRef]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J. Integrative genomic analyses identify mitf as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117. [Google Scholar] [CrossRef] [PubMed]
- McGill, G.G.; Horstmann, M.; Widlund, H.R.; Du, J.; Motyckova, G.; Nishimura, E.K.; Lin, Y.-L.; Ramaswamy, S.; Avery, W.; Ding, H.-F. Bcl2 regulation by the melanocyte master regulator mitf modulates lineage survival and melanoma cell viability. Cell 2002, 109, 707–718. [Google Scholar] [CrossRef]
- Haq, R.; Yokoyama, S.; Hawryluk, E.B.; Jönsson, G.B.; Frederick, D.T.; McHenry, K.; Porter, D.; Tran, T.-N.; Love, K.T.; Langer, R. Bcl2a1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to braf inhibition. Proc. Natl. Acad. Sci. USA 2013, 110, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Dhomen, N.; Marais, R. Braf signaling and targeted therapies in melanoma. Hematol. Oncol. Clin. North Am. 2009, 23, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M. Improved survival with vemurafenib in melanoma with braf v600e mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Menzies, A.M.; Nagrial, A.M.; Haydu, L.E.; Hamilton, A.L.; Mann, G.J.; Hughes, T.M.; Thompson, J.F.; Scolyer, R.A.; Kefford, R.F. Prognostic and clinicopathologic associations of oncogenic braf in metastatic melanoma. J. Clin. Oncol. 2011, 29, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Daniotti, M.; Oggionni, M.; Ranzani, T.; Vallacchi, V.; Campi, V.; Di Stasi, D.; Della Torre, G.; Perrone, F.; Luoni, C.; Suardi, S. Braf alterations are associated with complex mutational profiles in malignant melanoma. Oncogene 2004, 23, 5968. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, W.K.; Nevis, K.R.; Qu, P.; Ibrahim, J.G.; Zhou, T.; Zhou, Y.; Simpson, D.A.; Helms-Deaton, J.; Cordeiro-Stone, M.; Moore, D.T. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J. Investig. Dermatol. 2008, 128, 175–187. [Google Scholar] [CrossRef]
- Anchelin, M.; Alcaraz-Pérez, F.; Martínez, C.M.; Bernabé-García, M.; Mulero, V.; Cayuela, M.L. Premature aging in telomerase-deficient zebrafish. Dis. Models Mech. 2013, 6, 1101–1112. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef]
- Barut, B.A.; Zon, L.I. Realizing the potential of zebrafish as a model for human disease. Physiol. Genom. 2000, 2, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F. The histone methyltransferase setdb1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Houvras, Y.; Ceol, C.; Lin, W.; Jane-Valbuena, J.; Ferre, F.; Burke, C.; Bourque, C.; Turner, L.; Uong, A.; Garraway, L. Abstract# 2078: A Novel Screening Approach in Zebrafish Identifies Setdb1, a Histone Methyltransferase, as an Oncogene in Human Melanoma; AACR: Philadelphia, PA, USA, 2009. [Google Scholar]
- Casa, A.; Bourque, C.; Zack, Y.; Houvras, Y. Genetic and Epigenetic Analysis of Tumors in a Zebrafish Model of Melanoma; AACR: Philadelphia, PA, USA, 2012. [Google Scholar]
- Gabellini, C.; Gómez-Abenza, E.; Ibáñez-Molero, S.; Tupone, M.G.; Pérez-Oliva, A.B.; de Oliveira, S.; Del Bufalo, D.; Mulero, V. Interleukin 8 mediates bcl-xl-induced enhancement of human melanoma cell dissemination and angiogenesis in a zebrafish xenograft model. Int. J. Cancer 2018, 142, 584–596. [Google Scholar] [CrossRef] [PubMed]
- McNeill, M.S.; Paulsen, J.; Bonde, G.; Burnight, E.; Hsu, M.-Y.; Cornell, R.A. Cell death of melanophores in zebrafish trpm7 mutant embryos depends on melanin synthesis. J. Investig. Dermatol. 2007, 127, 2020–2030. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Maruyama, K.; Takenaka, H.; Furukawa, T.; Saga, T. A medaka model of cancer allowing direct observation of transplanted tumor cells in vivo at a cellular-level resolution. Proc. Natl. Acad. Sci. USA 2009, 106, 13832–13837. [Google Scholar] [CrossRef] [PubMed]
- Rawls, J.F.; Johnson, S.L. Zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration. Development 2000, 127, 3715–3724. [Google Scholar]
- Sarasamma, S.; Varikkodan, M.M.; Liang, S.-T.; Lin, Y.-C.; Wang, W.-P.; Hsiao, C.-D. Zebrafish: A premier vertebrate model for biomedical research in indian scenario. Zebrafish 2017, 14, 589–605. [Google Scholar] [CrossRef]
- Ju, B.; Spitsbergen, J.; Eden, C.J.; Taylor, M.R.; Chen, W.J.M.C. Co-activation of hedgehog and akt pathways promote tumorigenesis in zebrafish. Mol. Cancer 2009, 8, 40. [Google Scholar] [CrossRef]
- Gordon, M. The genetics of a viviparous top-minnow platypoecilus; the inheritance of two kinds of melanophores. Genetics 1927, 12, 253. [Google Scholar]
- HSussler, G. Uber melanombildung bei bastarden von xiphophorus helleri und platypoecilus maculatus var. Rubra. KIm. Wochschr 1928, 7, 1561–1562. [Google Scholar] [CrossRef]
- Kosswig, C. ber bastarde der teleostier platypoecilus und xiphophorus. Mol. Gen. Genet. MGG 1927, 44, 253. [Google Scholar] [CrossRef]
- Gordon, M. The melanoma cell as an incompletely differentiated pigment cell. Pigm. Cell Biol. 1959, 215–240. [Google Scholar]
- Vielkind, U. Genetic control of cell differentiation in platyfish-swordtail melanomas. J. Exp.Zool. Part A 1976, 196, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.A.; Bowser, P.R. Selection for a dominant oncogene and large male size as a risk factor for melanoma in the xiphophorus animal model. Mol. Ecol. 2010, 19, 3114–3123. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Walter, R.B.; Shen, Y.; Garcia, T.; Catchen, J.; Amores, A.; Braasch, I.; Chalopin, D.; Volff, J.-N.; Lesch, K.-P. The genome of the platyfish, xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat. Genet. 2013, 45, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, Y. Establishment of a cell line from the platyfish-swordtail hybrid melanoma. Cancer Res. 1981, 41, 679–680. [Google Scholar] [PubMed]
- Sobel, H.; Marquet, E.; Kallman, K.; Corley, G. Melanomas in platy/swordtail hybrids. Pathol. Fish. 1975, 945–981. [Google Scholar]
- Riehl, R.; Schartl, M.; Kollinger, G. Comparative studies on the ultrastructure of malignant melanoma in fish and human by freeze-etching and transmission electron microscopy. J. Cancer Res. Clin. Oncol. 1984, 107, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Conti, I.; Woodhead, A.D.; Harshbarger, J.C.; Kazianis, S.; Setlow, R.B.; Nairn, R.S.; Walter, R.B. A proposed classification scheme for xiphophorus melanomas based on histopathologic analyses. Mar. Biotechnol. 2001, 3, S100–S106. [Google Scholar]
- Wakamatsu, Y.; Oikawa, A.; Obika, M.; Hirobe, T.; Ozato, K. Fish hereditary melanoma cell lines of different degrees of cell differentiation. Dev. Growth Differ. 1984, 26, 503–513. [Google Scholar] [CrossRef]
- Meierjohann, S. Oxidative stress in melanocyte senescence and melanoma transformation. Eur. J. Cell Biol. 2014, 93, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.R.; Berwick, M.; Ley, R.D.; Walter, R.B.; Setlow, R.B.; Timmins, G.S. Uv causation of melanoma in xiphophorus is dominated by melanin photosensitized oxidant production. Proc. Natl. Acad. Sci. USA 2006, 103, 4111–4115. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.; Paniker, L.; Sanchez, G.; Trono, D.; Nairn, R. The etiology of sunlight-induced melanoma in xiphophorus hybrid fish. Mol. Carcinog. 2007, 46, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Sakakibara, K.; Kurumado, K.; Shimada, H.; Yamaguchi, K. Morphologic and microspectrophotometric studies on spontaneous melanomas in xiphophorus helleri. J. Natl. Cancer Inst. 1975, 54, 1373–1378. [Google Scholar] [CrossRef] [PubMed]
- Setlow, R.B.; Woodhead, A.D.; Grist, E. Animal model for ultraviolet radiation-induced melanoma: Platyfish-swordtail hybrid. Proc. Natl. Acad. Sci. USA 1989, 86, 8922–8926. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, M.; Schwab, M.; Anders, F. Lactate dehydrogenase isozymes in xiphophorin fish melanoma conditioned by the locussd. Cell. Mol. Life Sci. 1975, 31, 296–298. [Google Scholar] [CrossRef]
- Meierjohann, S.; Schartl, M. From mendelian to molecular genetics: The xiphophorus melanoma model. Trends Genet. 2006, 22, 654–661. [Google Scholar] [CrossRef]
- Adam, D.; Maueler, W.; Schartl, M. Transcriptional activation of the melanoma inducing xmrk oncogene in xiphophorus. Oncogene 1991, 6, 73–80. [Google Scholar]
- Kazianis, S.; Gutbrod, H.; Nairn, R.S.; McEntire, B.B.; Coletta, L.D.; Walter, R.B.; Borowsky, R.L.; Woodhead, A.D.; Setlow, R.B.; Schartl, M. Localization of a cdkn2 gene in linkage group v of xiphophorus fishes defines it as a candidate for the diff tumor suppressor. Genes Chromosomes Cancer 1998, 22, 210–220. [Google Scholar] [CrossRef]
- Rahn, J.J.; Trono, D.; Gimenez-Conti, I.; Butler, A.P.; Nairn, R.S. Etiology of mnu-induced melanomas in xiphophorus hybrids. Comp. Biochem. Physiol. Part C 2009, 149, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, N.; Winkler, C.; Wellbrock, C.; Gomez, A.; Duschl, J.; Altschmied, J.; Schartl, M. Activation of the xmrk proto-oncogene of xiphophorus by overexpression and mutational alterations. Oncogene 1998, 16, 1681. [Google Scholar] [CrossRef] [PubMed]
- Kazianis, S.; Coletta, L.D.; Morizot, D.C.; Johnston, D.A.; Osterndorff, E.A.; Nairn, R.S. Overexpression of a fish cdkn2 gene in a hereditary melanoma model. Carcinogenesis 2000, 21, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Negishi, S.; Masada, M.; Wakamatsu, Y.; Ohoka, T.; Obika, M. Epinephrine-induced changes in the cyclic nucleotide content of fish melanoma cells. Gen. Comp. Endocrinol. 1982, 47, 88–93. [Google Scholar] [CrossRef]
- Wittbrodt, J.; Adam, D.; Malitschek, B.; Maueler, W.; Raulf, F.; Telling, A.; Robertson, M.; Schartl, M. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing tu locus in xiphophorus. Nature 1989, 341, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Schartl, A.; Dimitrijevic, N.; Schartl, M. Evolutionary origin and molecular biology of the melanoma-inducing oncogene of xiphophorus. Pigm. Cell Melanoma Res. 1994, 7, 428–432. [Google Scholar] [CrossRef]
- Froschauer, A.; Körting, C.; Bernhardt, W.; Nanda, I.; Schmid, M.; Schartl, M.; Volff, J.-N. Genomic plasticity and melanoma formation in the fish xiphophorus. Mar. Biotechnol. 2001, 3, S72–S80. [Google Scholar] [CrossRef] [PubMed]
- Gómez, A.; Wellbrock, C.; Schartl, M. Constitutive activation and specific signaling of the xmrk receptor in xiphophorus melanoma. Mar. Biotechnol. 2002, 4, 208–217. [Google Scholar]
- Baudler, M.; Duschl, J.; Winkler, C.; Schartl, M.; Altschmied, J. Activation of transcription of the melanoma inducing xmrk oncogene by a gc box element. J. Biol. Chem. 1997, 272, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Delfgaauw, J.; Duschl, J.; Wellbrock, C.; Froschauer, C.; Schartl, M.; Altschmied, J. Mitf-m plays an essential role in transcriptional activation and signal transduction in xiphophorus melanoma. Gene 2003, 320, 117–126. [Google Scholar] [CrossRef]
- Woolcock, B.W.; Schmidt, B.M.; Kallman, K.D.; Vielkind, J.R. Differences in transcription and promoters of xmrk-1 and xmrk-2 genes suggest a role for xmrk-2 in pigment pattern development in the platyfish, xiphophorus maculatus. Cell Growth Differ. 1994, 5, 575–583. [Google Scholar] [PubMed]
- Ahuja, M.; Schwab, M.; Anders, F. Linkage between a regulatory locus for melanoma cell differentiation and an esterase locus in xiphophorus. J. Hered. 1980, 71, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Winnemoeller, D.; Wellbrock, C.; Schartl, M. Activating mutations in the extracellular domain of the melanoma inducing receptor xmrk are tumorigenic in vivo. Int. J. Cancer 2005, 117, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Morcinek, J.C.; Weisser, C.; Geissinger, E.; Schartl, M.; Wellbrock, C. Activation of stat5 triggers proliferation and contributes to anti-apoptotic signalling mediated by the oncogenic xmrk kinase. Oncogene 2002, 21, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Baudler, M.; Schartl, M.; Altschmied, J. Specific activation of a stat family member in xiphophorus melanoma cells. Exp. Cell Res. 1999, 249, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Meierjohann, S.; Wende, E.; Kraiss, A.; Wellbrock, C.; Schartl, M. The oncogenic epidermal growth factor receptor variant xiphophorus melanoma receptor kinase induces motility in melanocytes by modulation of focal adhesions. Cancer Res. 2006, 66, 3145–3152. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, C.; Lammers, R.; Ullrich, A.; Schartl, M. Association between the melanoma-inducing receptor tyrosine kinase xmrk and src family tyrosine kinases in xiphophorus. Oncogene 1995, 10, 2135–2143. [Google Scholar]
- Wellbrock, C.; Fischer, P.; Schartl, M. Pi3-kinase is involved in mitogenic signaling by the oncogenic receptor tyrosine kinase xiphophorus melanoma receptor kinase in fish melanoma. Exp. Cell Res. 1999, 251, 340–349. [Google Scholar] [CrossRef]
- Chen, H.C.; Pan, I.J.; Tu, W.J.; Lin, W.H.; Hong, C.C.; Brittelli, M.R. Neoplastic response in japanese medaka and channel catfish exposed to n-methyl-n’-nitro-n-nitrosoguanidine. Toxicol. Pathol. 1996, 24, 696–706. [Google Scholar] [CrossRef]
- Hyodo-Taguchi, Y.; Matsudaira, H. Induction of transplantable melanoma by treatment with n-methyl-n’-nitro-n-nitrosoguanidine in an inbred strain of the teleost oryzias latipes. J. Natl. Cancer Inst. 1984, 73, 1219–1227. [Google Scholar]
- Hyodo-taguchi, Y.; Matsudaira, H. Higher susceptibility to n-methyl-n’-nitro-n-nitrosoguanidine-induced tumorigenesis in an interstrain hybrid of the fish, oryzias latipes (medaka). Jpn. J. Cancer Res. GANN 1987, 78, 487–493. [Google Scholar] [PubMed]
- Chen, S.; Hong, Y.; Scherer, S.J.; Schartl, M. Lack of ultraviolet-light inducibility of the medakafish (oryzias latipes) tumor suppressor gene p53. Gene 2001, 264, 197–203. [Google Scholar] [CrossRef]
- Armstrong, T.N.; Reimschuessel, R.; Bradley, B.P. DNA damage, histologial changes and DNA repair in larval japanese medaka (oryzias latipes) exposed to ultraviolet-b radiation. Aquat. Toxicol. 2002, 58, 1–14. [Google Scholar] [CrossRef]
- Kneitz, S.; Mishra, R.R.; Chalopin, D.; Postlethwait, J.; Warren, W.C.; Walter, R.B.; Schartl, M. Germ cell and tumor associated pirnas in the medaka and xiphophorus melanoma models. BMC Genom. 2016, 17, 357. [Google Scholar] [CrossRef] [PubMed]
- Béjar, J.; Hong, Y.; Schartl, M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development 2003, 130, 6545–6553. [Google Scholar] [CrossRef]
- Schartl, M.; Wilde, B.; Laisney, J.A.; Taniguchi, Y.; Takeda, S.; Meierjohann, S. A mutated egfr is sufficient to induce malignant melanoma with genetic background-dependent histopathologies. J. Investig. Dermatol. 2010, 130, 249–258. [Google Scholar] [CrossRef]
- Menescal, L.A. In Vivo Characterization of Genetic Factors Involved in Xmrk Driven Melanoma Formation in Medaka (Oryzias Latipes): A Closer Look at Braf, Stat5 and C-myc. Doctoral Dissertation, Universität Würzburg, Würzburg, Germany, 2012. [Google Scholar]
- Matsuzaki, Y.; Hosokai, H.; Mizuguchi, Y.; Shimizu, A.; Saya, H. Establishment of Transgenic Medaka as a Stable Tumor Model for In Vivo Screening of Anticancer Drugs; AACR: Philadelphia, PA, USA, 2013. [Google Scholar]
- Kelsh, R.N.; Inoue, C.; Momoi, A.; Kondoh, H.; Furutani-Seiki, M.; Ozato, K.; Wakamatsu, Y. The tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 2004, 121, 841–859. [Google Scholar] [CrossRef]
- Mishra, R.R.; Kneitz, S.; Schartl, M. Comparative analysis of melanoma deregulated mirnas in the medaka and xiphophorus pigment cell cancer models. Comp. Biochem. Physiol. Part C 2014, 163, 64–76. [Google Scholar] [CrossRef]
- Meller, C.L.; Meller, R.; Simon, R.P.; Culpepper, K.M.; Podrabsky, J.E. Cell cycle arrest associated with anoxia-induced quiescence, anoxic preconditioning, and embryonic diapause in embryos of the annual killifish austrofundulus limnaeus. J. Comp. Physiol. B 2012, 182, 909–920. [Google Scholar] [CrossRef]
- Matias, J. The stage-dependent resistance of the chorion to external chemical damage and its relationship to embryonic diapause in the annual fish, nothobranchius guentheri. Experientia 1984, 40, 753–754. [Google Scholar] [CrossRef]
- Podrabsky, J.E.; Carpenter, J.F.; Hand, S.C. Survival of water stress in annual fish embryos: Dehydration avoidance and egg envelope amyloid fibers. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2001, 280, R123–R131. [Google Scholar] [CrossRef] [PubMed]
- Hers, I.; Vincent, E.E.; Tavaré, J.M. Akt signalling in health and disease. Cell. Signal. 2011, 23, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Mangel, M.; Kindsvater, H.K.; Bonsall, M.B. Evolutionary analysis of life span, competition, and adaptive radiation, motivated by the pacific rockfishes (sebastes). Evolution 2007, 61, 1208–1224. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.R.; Kimbrell, C.; Robertson, L.; Clifford, K.; Lynn, E.; Vetter, R. Multiple paternity and maintenance of genetic diversity in the live-bearing rockfishes sebastes spp. Mar. Ecol. Prog. Ser. 2008, 357, 245–253. [Google Scholar] [CrossRef]
- Sivasundar, A.; Palumbi, S. Parallel amino acid replacements in the rhodopsins of the rockfishes (sebastes spp.) associated with shifts in habitat depth. J. Evol. Biol. 2010, 23, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Darias, M.J.; Andree, K.B.; Boglino, A.; Fernández, I.; Estévez, A.; Gisbert, E. Coordinated regulation of chromatophore differentiation and melanogenesis during the ontogeny of skin pigmentation of solea senegalensis (kaup, 1858). PLoS ONE 2013, 8, e63005. [Google Scholar] [CrossRef] [PubMed]
- Okihiro, M.S.; Whipple, J.A.; Groff, J.M.; Hinton, D.E. Chromatophoromas and chromatophore hyperplasia in pacific rockfish (sebastes spp.). Cancer Res. 1993, 53, 1761–1769. [Google Scholar] [PubMed]
- Hoffmann, M.; Tripathi, N.; Henz, S.R.; Lindholm, A.K.; Weigel, D.; Breden, F.; Dreyer, C. Opsin gene duplication and diversification in the guppy, a model for sexual selection. Proc. R. Soc. London B 2007, 274, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kodric-Brown, A. Female preference and sexual selection for male coloration in the guppy (poecilia reticulata). Behav. Ecol. Sociobiol. 1985, 17, 199–205. [Google Scholar] [CrossRef]
- Grether, G.F.; Hudon, J.; Endler, J.A. Carotenoid scarcity, synthetic pteridine pigments and the evolution of sexual coloration in guppies (poecilia reticulata). Proc. R. Soc. London B 2001, 268, 1245–1253. [Google Scholar] [CrossRef]
- Kottler, V.A.; Koch, I.; Flötenmeyer, M.; Hashimoto, H.; Weigel, D.; Dreyer, C. Multiple pigment cell types contribute to the black, blue, and orange ornaments of male guppies (poecilia reticulata). PLoS ONE 2014, 9, e85647. [Google Scholar] [CrossRef] [PubMed]
- Walinski, H. Studying Gene Function: Creating Knockout Mice. 2004. Available online: https://www.scq.ubc.ca/studying-gene-function-creating-knockout-mice/ (accessed on 6 December 2018).
- Schneider, L.S.; Mangialasche, F.; Andreasen, N.; Feldman, H.; Giacobini, E.; Jones, R.; Mantua, V.; Mecocci, P.; Pani, L.; Winblad, B. Clinical trials and late-stage drug development for alzheimer’s disease: An appraisal from 1984 to 2014. J. Intern. Med. 2014, 275, 251–283. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114. [Google Scholar] [PubMed]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Shay, T.; Jojic, V.; Zuk, O.; Rothamel, K.; Puyraimond-Zemmour, D.; Feng, T.; Wakamatsu, E.; Benoist, C.; Koller, D.; Regev, A. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. USA 2013, 110, 2946–2951. [Google Scholar] [CrossRef] [PubMed]
- Husain, Z.; Pathak, M.A.; Flotte, T.; Wick, M.M. Role of ultraviolet radiation in the induction of melanocytic tumors in hairless mice following 7, 12-dimethylbenz (a) anthracene application and ultraviolet irradiation. Cancer Res. 1991, 51, 4964–4970. [Google Scholar] [PubMed]
- Tu, S.; Johnson, S.L. Clonal analyses reveal roles of organ founding stem cells, melanocyte stem cells and melanoblasts in establishment, growth and regeneration of the adult zebrafish fin. Development 2010, 137, 3931–3939. [Google Scholar] [CrossRef]
- Dee, C.T.; Nagaraju, R.T.; Athanasiadis, E.I.; Gray, C.; del Ama, L.F.; Johnston, S.A.; Secombes, C.J.; Cvejic, A.; Hurlstone, A.F. Cd4-transgenic zebrafish reveal tissue-resident th2-and regulatory t cell–like populations and diverse mononuclear phagocytes. J. Immunol. 2016, 197, 3520–3530. [Google Scholar] [CrossRef]
- Cenci, G.; Ciapponi, L.; Gatti, M. The mechanism of telomere protection: A comparison between drosophila and humans. Chromosoma 2005, 114, 135–145. [Google Scholar] [CrossRef]
- Serrano, M.; Lee, H.-W.; Chin, L.; Cordon-Cardo, C.; Beach, D.; DePinho, R.A. Role of the ink4a locus in tumor suppression and cell mortality. Cell 1996, 85, 27–37. [Google Scholar] [CrossRef]
- VanBrocklin, M.W.; Robinson, J.P.; Lastwika, K.J.; Khoury, J.D.; Holmen, S.L. Targeted delivery of nrasq61r and cre-recombinase to post-natal melanocytes induces melanoma in ink4a/arflox/lox mice. Pigm. Cell Melanoma Res. 2010, 23, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.C.; Chinnasamy, N.; Morgan, R.A.; Varmus, H.E. Development of an avian leukosis-sarcoma virus subgroup a pseudotyped lentiviral vector. J. Virol. 2001, 75, 9339–9344. [Google Scholar] [CrossRef] [PubMed]
- Kamb, A.; Shattuck-Eidens, D.; Eeles, R.; Liu, Q.; Gruis, N.; Ding, W.; Hussey, C.; Tran, T.; Miki, Y.; Weaver-Feldhaus, J. Analysis of the p16 gene (cdkn2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994, 8, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Xie, X.; Walker, S.; White, D.T.; Mumm, J.S.; Cowell, J.K. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 2013, 13, 453. [Google Scholar] [CrossRef] [PubMed]
- Albertson, R.C.; Cresko, W.; Detrich, H.W., 3rd; Postlethwait, J.H. Evolutionary mutant models for human disease. Trends Genet. 2009, 25, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Ribas, L.; Valdivieso, A.; Diaz, N.; Piferrer, F. Response to “the importance of controlling genetic variation—Remarks on ‘appropriate rearing density in domesticated zebrafish to avoid masculinization: Links with the stress response’”. J. Exp. Biol. 2017, 220, 4079–4080. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas III, C.F. Zfn, talen, and crispr/cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Regan, S.N.; Xia, Y.; Oostrom, L.A.; Cowan, C.A.; Musunuru, K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing talens with crisprs. Cell Stem Cell 2013, 12, 393–394. [Google Scholar] [CrossRef]
- Zu, Y.; Tong, X.; Wang, Z.; Liu, D.; Pan, R.; Li, Z.; Hu, Y.; Luo, Z.; Huang, P.; Wu, Q. Talen-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 2013, 10, 329. [Google Scholar] [CrossRef]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.-R.J. Targeted gene disruption in somatic zebrafish cells using engineered talens. Nat. Biotechnol. 2011, 29, 697. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized talens. Nat. Biotechnol. 2011, 29, 699. [Google Scholar] [CrossRef] [PubMed]
- Perles, Z.; Moon, S.; Ta-Shma, A.; Yaacov, B.; Francescatto, L.; Edvardson, S.; Rein, A.J.; Elpeleg, O.; Katsanis, N. A human laterality disorder caused by a homozygous deleterious mutation in mmp21. J. Med. Genet. 2015, 52, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, L.; Xie, Y.; Wang, N.; Tang, R.; Zheng, W.; Jiang, X. Genome editing for cancer therapy: Delivery of cas9 protein/sgrna plasmid via a gold nanocluster/lipid core–shell nanocarrier. Adv. Sci. 2017, 4, 1700175. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Zager, J.S. Clinical development of dabrafenib in braf mutant melanoma and other malignancies. Expert Opin. Drug Metab. Toxicol. 2013, 9, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.J.; Joung, J.K. Efficient genome editing in zebrafish using a crispr-cas system. Nat. Biotechnol. 2013, 31, 227. [Google Scholar] [CrossRef] [PubMed]
- Manguso, R.T.; Pope, H.W.; Zimmer, M.D.; Brown, F.D.; Yates, K.B.; Miller, B.C.; Collins, N.B.; Bi, K.; LaFleur, M.W.; Juneja, V.R. In vivo crispr screening identifies ptpn2 as a cancer immunotherapy target. Nature 2017, 547, 413. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Pan, L.; Groen, R.W.; Baleydier, F.; Kentsis, A.; Marineau, J.; Grebliunaite, R.; Kozakewich, E.; Reed, C.; Pflumio, F. Phenothiazines induce pp2a-mediated apoptosis in t cell acute lymphoblastic leukemia. J. Clin. Investig. 2014, 124, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Oppedal, D.; Goldsmith, M.I. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish 2010, 7, 53–60. [Google Scholar] [CrossRef]
- Namdaran, P.; Reinhart, K.E.; Owens, K.N.; Raible, D.W.; Rubel, E.W. Identification of modulators of hair cell regeneration in the zebrafish lateral line. J. Neurosci. 2012, 32, 3516–3528. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Hodi, F.S.; Fisher, D.E. From genes to drugs: Targeted strategies for melanoma. Nat. Rev. Cancer 2012, 12, 349. [Google Scholar] [CrossRef]
- Cheng, C.; Yang, H.-W.; Shang, J.-F.; Li, W.-W.; Sun, Q.-Z.; Chen, X.; Cao, Z.-X.; Yao, S.-H.; Yang, S.-Y. Identification of a small molecule that downregulates mitf expression and mediates antimelanoma activity in vitro. Melanoma Res. 2016, 26, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, M.; Feng, Y.; Jones, R.; Martin, P. Inflammation drives wound hyperpigmentation by recruiting pigment cells to sites of tissue damage. Dis. Models Mech. 2013, 6, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.E.; Mercer, C.A.; Carnevalli, L.S.; Park, J.; Andersen, J.B.; Conner, E.A.; Tanaka, K.; Matsutani, T.; Iwanami, A.; Aronow, B.J. Mtor inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci. Transl. Med. 2012, 3003923. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.L.; Fogley, R.D.; Flynn, R.A.; Ablain, J.; Yang, S.; Saint-André, V.; Fan, Z.P.; Do, B.T.; Laga, A.C.; Fujinaga, K. Stress from nucleotide depletion activates the transcriptional regulator hexim1 to suppress melanoma. Mol. Cell 2016, 62, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Lasithiotakis, K.G.; Sinnberg, T.W.; Schittek, B.; Flaherty, K.T.; Kulms, D.; Maczey, E.; Garbe, C.; Meier, F.E. Combined inhibition of mapk and mtor signaling inhibits growth, induces cell death, and abrogates invasive growth of melanoma cells. J. Investig. Dermatol. 2008, 128, 2013–2023. [Google Scholar] [CrossRef] [PubMed]
- Amaria, R.N.; Prieto, P.A.; Tetzlaff, M.T.; Reuben, A.; Andrews, M.C.; Ross, M.I.; Glitza, I.C.; Cormier, J.; Hwu, W.-J.; Tawbi, H.A. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: A. single-centre, open-label, randomised, phase 2 trial. Lancet Oncol. 2018, 19, 181–193. [Google Scholar] [CrossRef]
- Andtbacka, R.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.J. Toxicity and cytopathogenic properties toward human melanoma cells of activated cancer therapeutics in zebra fish. Integr. Cancer Ther. 2010, 9, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.C.; Hynes, R.O. Intravital imaging of metastasis in adult zebrafish. BMC Cancer 2017, 17, 660. [Google Scholar] [CrossRef]
- Stewart, A.M.; Nguyen, M.; Wong, K.; Poudel, M.K.; Kalueff, A.V. Developing zebrafish models of autism spectrum disorder (asd). Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2014, 50, 27–36. [Google Scholar] [CrossRef]
| Transgene | Key Features | References |
|---|---|---|
| mitfa : BRAF (V600E) | BRAF (V600E) expression leads to melanoma on a tp53 mutant background | [21,22] |
| mitfa : BRAF (V600E) | Zebrafish melanoma model used for functional genetic screens to identify new oncogenes | [23] |
| mitfa : BRAF (V600E) | Genetic modulation of mitf leads to melanoma regression | [24] |
| mitfa : BRAF (V600E) | Zebrafish melanoma model used to screen for novel anti-melanoma drugs | [25] |
| mitfa : NRAS (Q61K) | Transgenic expression of NRAS (Q61K) leads to melanomas | [26] |
| mitfa : HRAS (G12V) | HRAS acts through PI3K signaling to induce melanoma | [27] |
| kita : Gal4 x UAS : HRAS (G12V) | HRAS expression in kit-expressing cells lead to highly penetrant and invasive phenotypes | [28,29] |
| krt4: cmyc x krt4:cdc6 | Co-activation of c-myc and cdc6 in zebrafish skin to induce skin cancer formation | [30] |
| Model Organism | Disease Model | Possibility to Maintain in Lab | Genomic Resource | Transgenic Methods | References |
|---|---|---|---|---|---|
| Zebrafish (Danio rerio) | Many different types of cancer | Yes | Danio rerio (Tuebingen) | No | [11,31] |
| Platy fish (Xiphophorus maculatus) | Cancer, specifically melanoma | Yes | Three species completed | No | [32] |
| Annual fish (Cynolebias nigripinnis Nothobranchius rachovii) | Used as a model for aging | Yes | Two species completed | Yes | [33] |
| Medaka fish (Oryzias latipes) | Used as a model most notably in toxicology | Yes, short life span and is easy to breed | One species | Yes | [34] |
| Rockfish (Sebastes) | Complete mitochondrial genome sequences | Yes, easy to rear in the laboratory | Five species | No | [35] |
| Guppy (Poecilia reticulate) | Model systems for the study of evolution, ecology, behavior, tumor genetics and genomics | Yes | One species | No | [36] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarasamma, S.; Lai, Y.-H.; Liang, S.-T.; Liu, K.; Hsiao, C.-D. The Power of Fish Models to Elucidate Skin Cancer Pathogenesis and Impact the Discovery of New Therapeutic Opportunities. Int. J. Mol. Sci. 2018, 19, 3929. https://doi.org/10.3390/ijms19123929
Sarasamma S, Lai Y-H, Liang S-T, Liu K, Hsiao C-D. The Power of Fish Models to Elucidate Skin Cancer Pathogenesis and Impact the Discovery of New Therapeutic Opportunities. International Journal of Molecular Sciences. 2018; 19(12):3929. https://doi.org/10.3390/ijms19123929
Chicago/Turabian StyleSarasamma, Sreeja, Yu-Heng Lai, Sung-Tzu Liang, Kechun Liu, and Chung-Der Hsiao. 2018. "The Power of Fish Models to Elucidate Skin Cancer Pathogenesis and Impact the Discovery of New Therapeutic Opportunities" International Journal of Molecular Sciences 19, no. 12: 3929. https://doi.org/10.3390/ijms19123929
APA StyleSarasamma, S., Lai, Y.-H., Liang, S.-T., Liu, K., & Hsiao, C.-D. (2018). The Power of Fish Models to Elucidate Skin Cancer Pathogenesis and Impact the Discovery of New Therapeutic Opportunities. International Journal of Molecular Sciences, 19(12), 3929. https://doi.org/10.3390/ijms19123929