Tissue-Specific Down-Regulation of the Long Non-Coding RNAs PCAT18 and LINC01133 in Gastric Cancer Development
Abstract
1. Introduction
2. Results
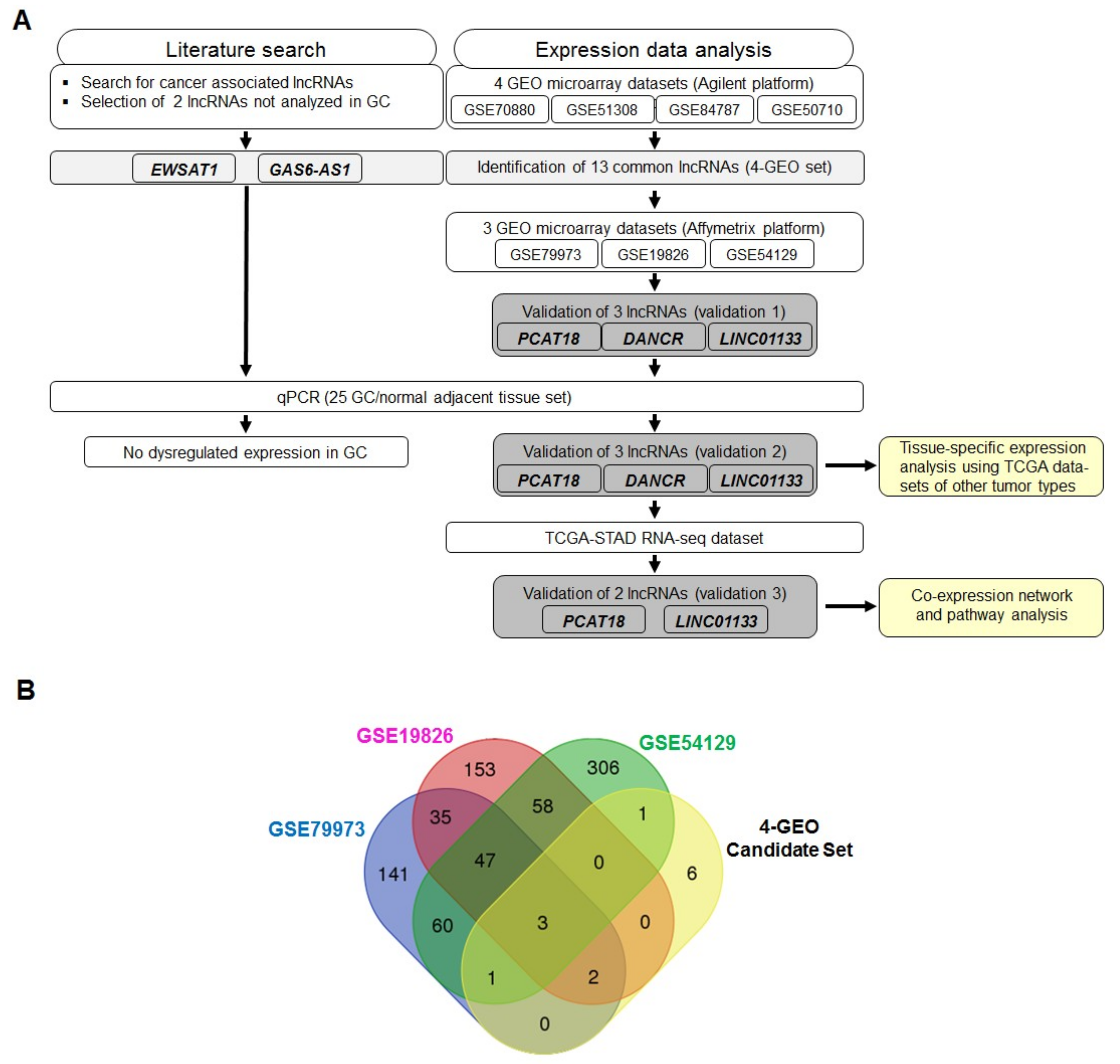
2.1. Identification of LncRNA Candidates Associated with Gastric Cancer Using Gene Expression Omnibus (GEO) Datasets and Literature Data
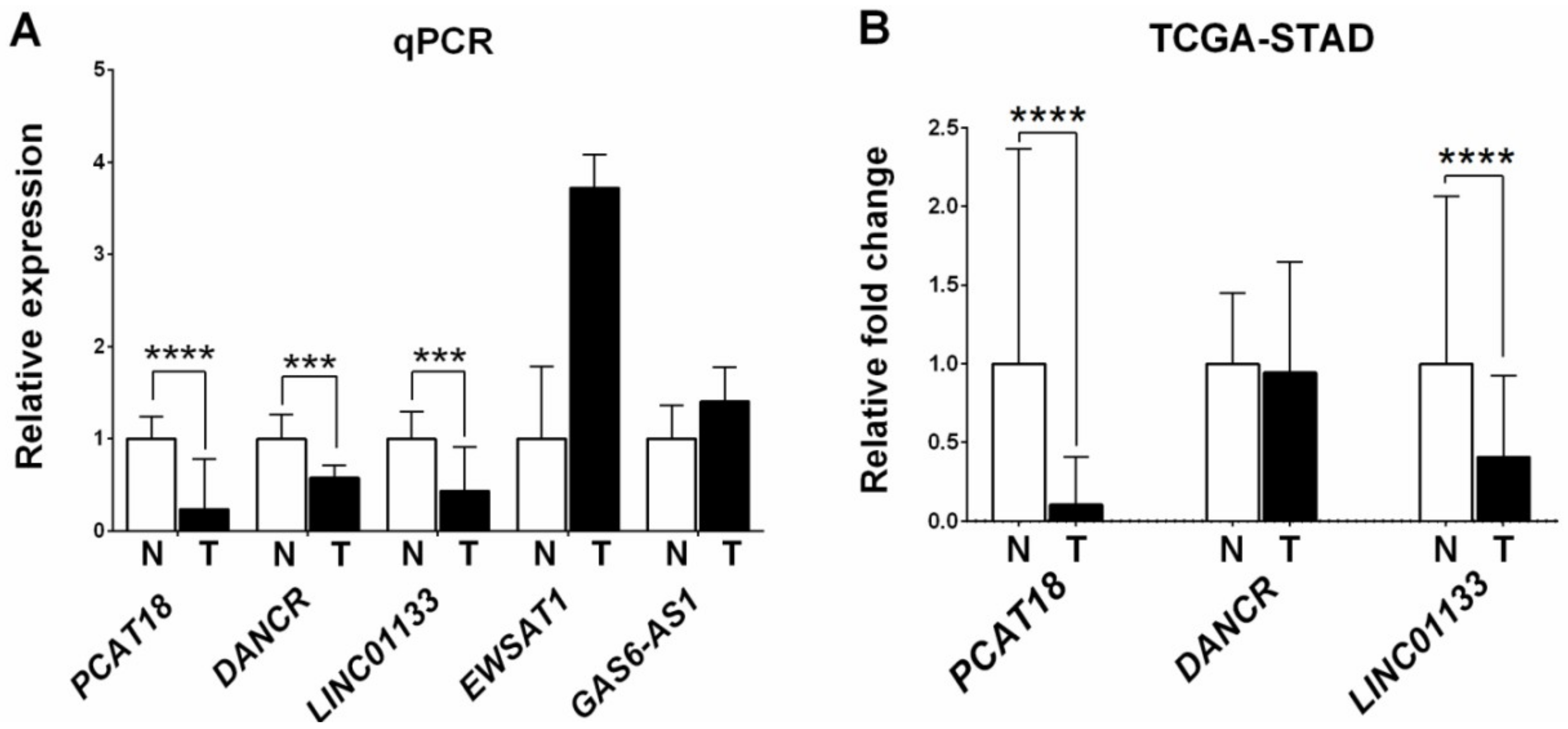
2.2. LncRNA Expression Levels in the 25 GC/Normal Tissue Sample Set and Associations with Clinical, Histopathological, and Epidemiological Parameters
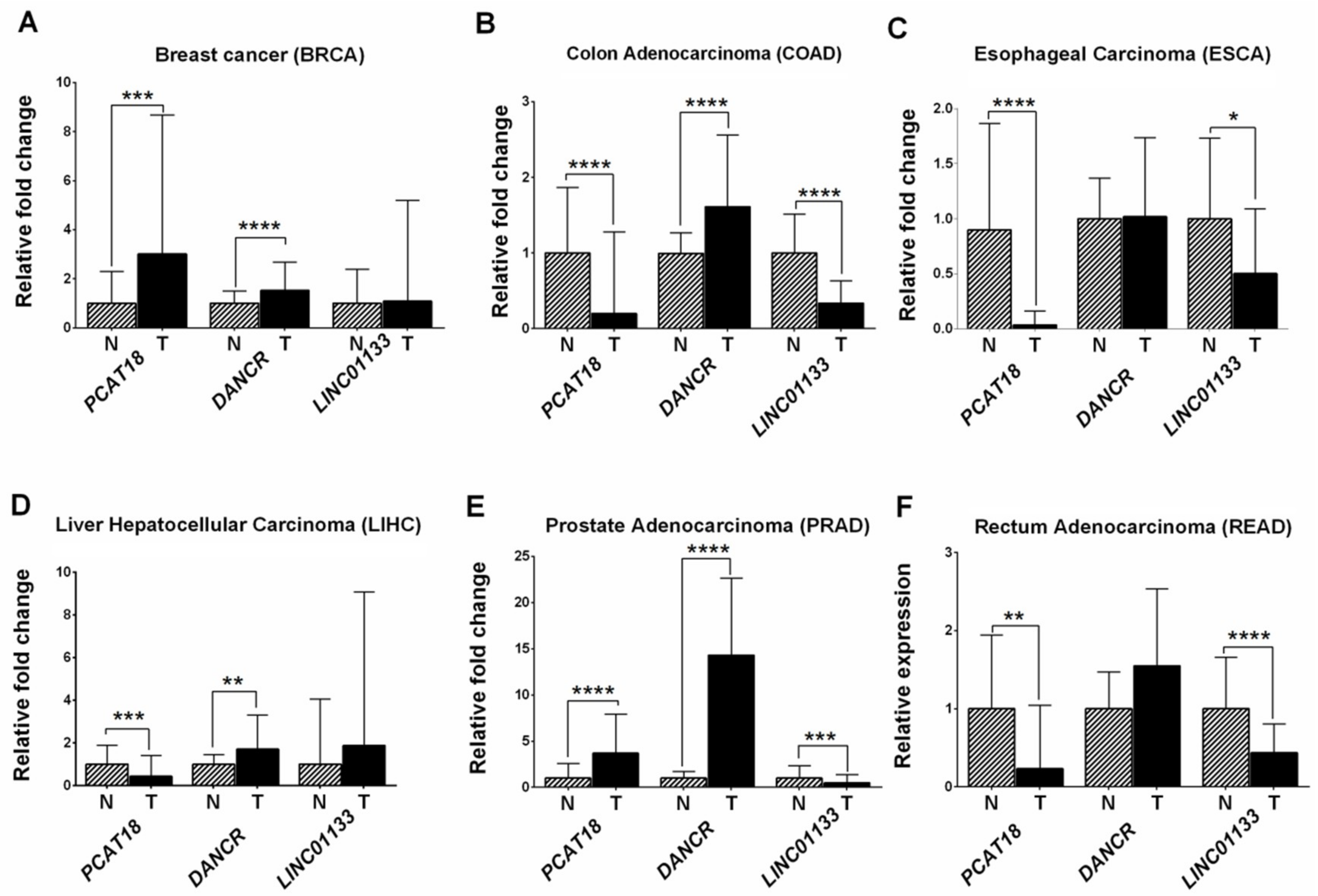
2.3. Analysis of LncRNA Expression Levels Using RNA-Sequencing Data from The Cancer Genome Atlas- Stomach Adenocarcinoma (TCGA-STAD)
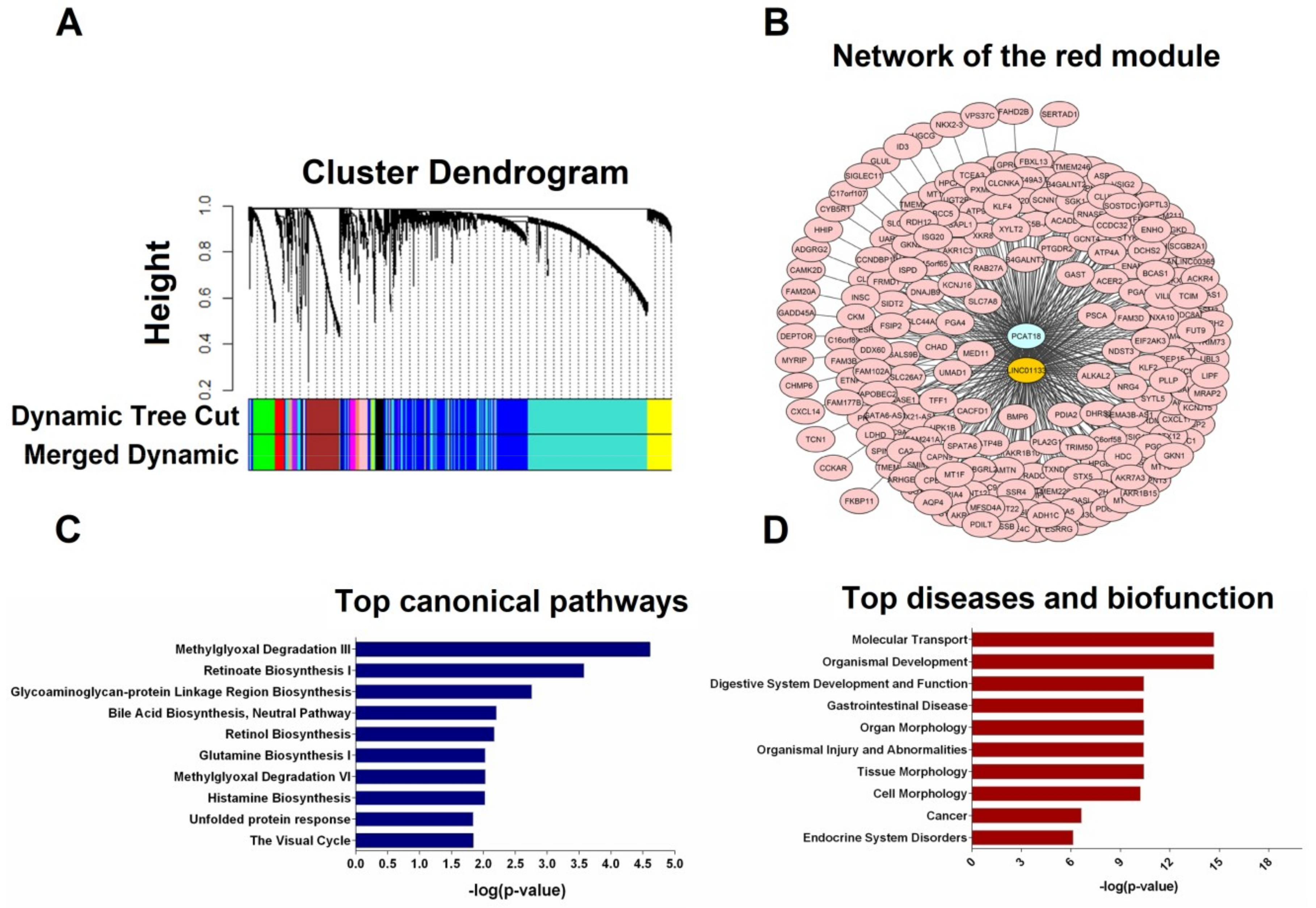
2.4. Weighted Gene Correlation Network Analysis and Functional Annotation of Co-Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Data Extraction from the GEO Database and Literature Review
4.2. Patient Samples
4.3. RNA Extraction and cDNA Synthesis
4.4. Real-Time PCR
4.5. Data Extraction from TCGA and Data Analyses
4.6. Weighted Gene Correlation Network Analysis
4.7. Functional Annotation of the Co-Expressed Genes in the Module
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval
Abbreviations
| GC | Gastric Cancer |
| GEO | Gene Expression Omnibus |
| TCGA | The Cancer Genome Atlas |
| lncRNAs | Long Non-Coding RNAs |
| WGCNA | Weighted Gene Correlation Network Analysis |
| RNA-seq | RNA-Sequencing |
| STAD | Stomach Adenocarcinoma |
| TMM | Trimmed Mean of M-Values |
| IPA | Ingenuity Pathway Analysis |
| SD | Standard Deviation |
| PCAT18 | Prostate Cancer Associated Transcript 18 |
| LINC01133 | Long Intergenic Non-Coding RNA 1133 |
| DANCR | Differentiation Antagonizing Non-Protein Coding RNA |
| GTEx | Genotype-Tissue Expression Database |
| NSCLC | Non-Small Cell Lung Cancer |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bria, E.; de Manzoni, G.; Beghelli, S.; Tomezzoli, A.; Barbi, S.; di Gregorio, C.; Scardoni, M.; Amato, E.; Frizziero, M.; Sperduti, I.; et al. A clinical-biological risk stratification model for resected gastric cancer: Prognostic impact of Her2, Fhit, and APC expression status. Ann. Oncol. 2013, 24, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Satolli, M.A.; Buffoni, L.; Spadi, R.; Roato, I. Gastric cancer: The times they are a-changin. World J. Gastrointest. Oncol. 2015, 7, 303–316. [Google Scholar] [CrossRef]
- Shimizu, T.; Marusawa, H.; Matsumoto, Y.; Inuzuka, T.; Ikeda, A.; Fujii, Y.; Minamiguchi, S.; Miyamoto, S.; Kou, T.; Sakai, Y.; et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 2014, 147, 407-17.e3. [Google Scholar] [CrossRef] [PubMed]
- Nagini, S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J. Gastrointest. Oncol. 2012, 4, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Gigek, C.O.; Chen, E.S.; Calcagno, D.Q.; Wisnieski, F.; Burbano, R.R.; Smith, M.A. Epigenetic mechanisms in gastric cancer. Epigenomics 2012, 4, 279–294. [Google Scholar] [CrossRef]
- Bartonicek, N.; Maag, J.L.V.; Dinger, M.E. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol. Cancer 2016, 15, 43. [Google Scholar] [CrossRef]
- Sun, W.; Yang, Y.; Xu, C.; Guo, J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017, 216–217, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.L.; Beck, A.H.; Edris, B.; Sweeney, R.T.; Zhu, S.X.; Li, R.; Montgomery, K.; Varma, S.; Gilks, T.; Guo, X.; et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012, 13, R75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.; Feng, L.; Li, F.; Sun, Z.; Wu, T.; Shi, X.; Li, J.; Li, X. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget 2016, 7, 64148–64167. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 2018, 28, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.Y.; Pan, H.F.; Leng, R.X.; Ye, D.Q. Long noncoding RNAs: Novel insights into gastric cancer. Cancer Lett. 2015, 356 (2 Pt B), 357–366. [Google Scholar] [CrossRef]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012, 279, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Shiroki, T.; Nakagawa, T.; Yokoyama, M.; Tamai, K.; Yamanami, H.; Fujiya, T.; Sato, I.; Yamaguchi, K.; Tanaka, N.; et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE 2013, 8, e77070. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Zhang, Y.; Zhou, F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed. Pharmacother. 2015, 72, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Xia, R.; Jin, F.; Xu, T.; Liu, Z.; De, W.; Liu, X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014, 35, 1065–1073. [Google Scholar] [CrossRef]
- Sun, M.; Jin, F.Y.; Xia, R.; Kong, R.; Li, J.H.; Xu, T.P.; Liu, Y.W.; Zhang, E.B.; Liu, X.H.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, X.; Li, G.; Shi, Y.; Li, L. Long noncoding RNAs as potential biomarkers in gastric cancer: Opportunities and challenges. Cancer Lett. 2016, 371, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Crea, F.; Watahiki, A.; Quagliata, L.; Xue, H.; Pikor, L.; Parolia, A.; Wang, Y.; Lin, D.; Lam, W.L.; Farrar, W.L.; et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget 2014, 5, 764–774. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Zhang, J.H.; Li, A.Y.; Wei, N. Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. European Rev. Med. Pharmacol. Sci. 2017, 21, 2103–2107. [Google Scholar]
- Zhang, J.; Zhu, N.; Chen, X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015, 36, 7465–7471. [Google Scholar] [CrossRef]
- Zang, C.; Nie, F.Q.; Wang, Q.; Sun, M.; Li, W.; He, J.; Zhang, M.; Lu, K.H. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget 2016, 7, 11696–11707. [Google Scholar] [CrossRef]
- Zeng, H.F.; Qiu, H.Y.; Feng, F.B. Long Noncoding RNA LINC01133 sponges miR-422a to aggravate the tumorigenesis of human osteosarcoma. Oncol. Res. 2017, 26, 335–343. [Google Scholar] [CrossRef]
- Yang, X.Z.; Cheng, T.T.; He, Q.J.; Lei, Z.Y.; Chi, J.; Tang, Z.; Liao, Q.X.; Zhang, H.; Zeng, L.S.; Cui, S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef]
- Ma, X.; Wang, X.; Yang, C.; Wang, Z.; Han, B.; Wu, L.; Zhuang, L. DANCR acts as a diagnostic biomarker and promotes tumor growth and metastasis in hepatocellular carcinoma. Anticancer Res. 2016, 36, 6389–6398. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.X.; Wang, J.; Yang, F.; Tao, Q.F.; Zhang, J.; Wang, L.L.; Yang, Y.; Liu, H.; Wang, Z.G.; Xu, Q.G.; et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology (Baltimore, Md.) 2016, 63, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, M.; Liang, L.; Li, J.; Chen, Y.-X. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int. J.Clin. Exp. Pathol. 2015, 8, 11480–11484. [Google Scholar] [PubMed]
- Jia, J.; Li, F.; Tang, X.-S.; Xu, S.; Gao, Y.; Shi, Q.; Guo, W.; Wang, X.; He, D.; Guo, P. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget 2016, 7, 37868–37881. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wang, X.; Xie, X.; Liao, Y.; Liu, N.; Liu, J.; Miao, N.; Shen, J.; Peng, T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017, 405, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.P.; Qiu, J.H.; Zhang, D.B.; Yu, C.G. Long non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancer. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Hung, C.L.; Wang, L.Y.; Yu, Y.L.; Chen, H.W.; Srivastava, S.; Petrovics, G.; Kung, H.J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 18697–18702. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Yang, M.; Huang, C.Z. Mitogen-activated protein kinase signaling pathway and invasion and metastasis of gastric cancer. World J. Gastroenterol. 2015, 21, 11673–11679. [Google Scholar] [CrossRef]
- Joo, M.K.; Park, J.J.; Chun, H.J. Impact of homeobox genes in gastrointestinal cancer. World J. Gastroenterol. 2016, 22, 8247–8256. [Google Scholar] [CrossRef]
- Huh, W.J.; Esen, E.; Geahlen, J.H.; Bredemeyer, A.J.; Lee, A.H.; Shi, G.; Konieczny, S.F.; Glimcher, L.H.; Mills, J.C. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 2010, 139, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Flockhart, R.J.; Webster, D.E.; Qu, K.; Mascarenhas, N.; Kovalski, J.; Kretz, M.; Khavari, P.A. BRAF(V600E) remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012, 22, 1006–1014. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Liu, Z.Q.; Jiang, B.; Lu, X.Y.; Ning, X.F.; Yuan, C.T.; Wang, A.L. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem. Biophys. Res. Commun. 2015, 465, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Tang, X.; Tian, F.; Shi, F.; Liu, F.; Gao, G. AnnoLnc: A web server for systematically annotating novel human lncRNAs. BMC Genom. 2016, 17, 931. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The gene expression omnibus database. Methods Mol. Biol. (Clifton, N.J.) 2016, 1418, 93–110. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery Rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
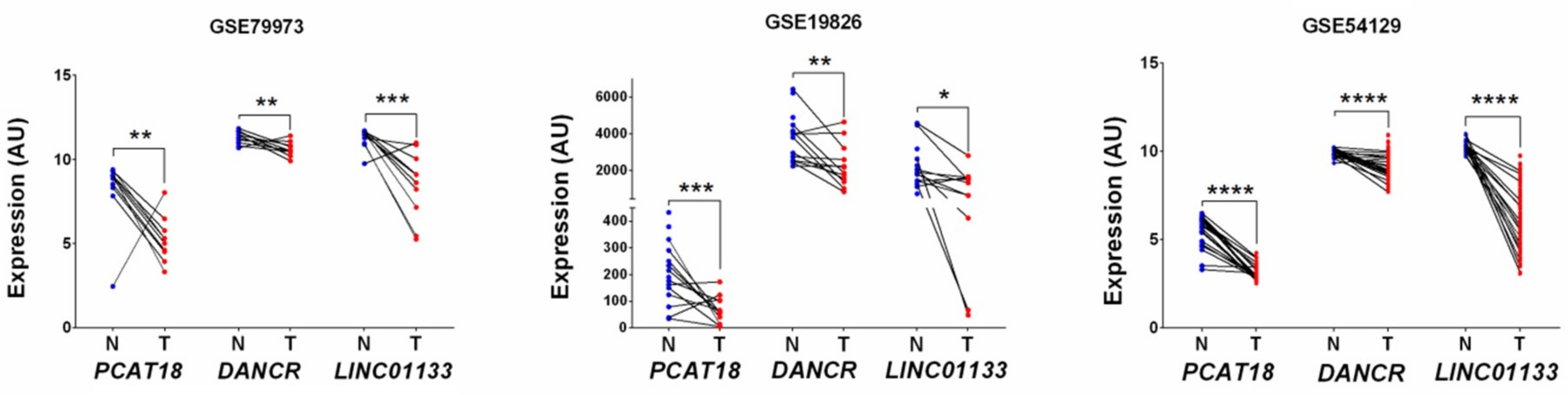
| Gene ID * | Gene Symbol | Gene Name | Log2FC | p Value | Regulatory Status |
|---|---|---|---|---|---|
| 728606 | PCAT18 | Prostate cancer associated transcript 18 | −2.62 | 1.58 × 10−4 | Down |
| 57291 | DANCR | Differentiation antagonizing non-protein coding RNA | −0.86 | 1.37 × 10−3 | Down |
| 283738 | NTRK3-AS1 | Neurotrophic receptor tyrosine kinase 3 antisense RNA 1 | 1.24 | 2.0 × 10−4 | Up |
| 101928223 | LINC01612 | Long intergenic non-protein coding RNA 1612 | −1.50 | 2.4 × 10−4 | Down |
| 404201 | WDFY3-AS2 | WD repeat and FYVE domain containing 3 antisense RNA 2 | −0.77 | 8.85 × 10−4 | Down |
| 100505875 | LINC01088 | Long intergenic non-protein coding RNA 1088 | −2.69 | 4.44 × 10−9 | Down |
| 102723487 | LINC01497 | Long intergenic non-protein coding RNA 1497 | −1.42 | 1.82 × 10−7 | Down |
| 100505633 | LINC01133 | Long intergenic non-protein coding RNA 1133 | −1.42 | 9.32 × 10−3 | Down |
| 348761 | SPATA3-AS1 | Spermatogenesis associated 3 antisense RNA 1 | −1.96 | 2.02 × 10−9 | Down |
| 401172 | LINC02145 | Long intergenic non-protein coding RNA 2145 | −1.95 | 3.79 × 10−9 | Down |
| 100507099 | FRY-AS1 | FRY microtubule binding protein antisense RNA 1 | −1.30 | 0.02946 | Down |
| 286190 | LACTB2-AS1 | Lactamase beta 2 antisense RNA 1 | −1.30 | 0.02946 | Down |
| 101927171 | LINC01564 | Long intergenic non-protein coding RNA 1564 | −3.30 | 0.03868 | Down |
| Parameter | Cases n (%) | p Value | ||
|---|---|---|---|---|
| PCAT18 | DANCR | LINC01133 | ||
| Age (years) | ||||
| <60 | 12 (48) | 0.623 | 0.965 | 0.504 |
| ≥60 | 13 (52) | |||
| Gender | ||||
| Male | 21 (84) | 0.856 | 0.006 | 0.002 |
| Female | 4 (16) | |||
| Site of primary tumor | ||||
| Gastric cardia | 4 (16) | 0.758 | 0.359 | 0.536 |
| Antrum | 5 (20) | |||
| Stomach | 16 (64) | |||
| Tumor size (cm) | ||||
| <5 | 11 (44) | 0.597 | 0.809 | 0.666 |
| ≥5 | 12 (48) | |||
| Unknown | 2 (8) | |||
| Histological grade | ||||
| 1, 2 | 17 (68) | 0.316 | 0.879 | 0.982 |
| 3, 4 | 8 (32) | |||
| Lymph node status | ||||
| N0 | 19 (76) | 0.404 | 0.831 | 0.771 |
| ≥N1 | 6 (24) | |||
| Vascular invasion | ||||
| Yes | 19 (76) | 0.404 | 0.831 | 0.771 |
| No | 6 (24) | |||
| Perineural invasion | ||||
| Yes | 16 (64) | 0.966 | 0.843 | 0.938 |
| No | 9 (36) | |||
| Serosal invasion | ||||
| Absent | 11 (44) | 0.243 | 0.913 | 0.732 |
| Present | 14 (56) | |||
| Clinical stage | ||||
| 1, 2 | 9 (36) | 0.223 | 0.374 | 0.221 |
| 3, 4 | 16 (64) | |||
| Family history of gastric cancer | ||||
| Yes | 9 (36) | 0.798 | 0.663 | 0.431 |
| No | 16 (64) | |||
| Smoking status | ||||
| Ever | 14 (56) | 0.056 | 0.005 | 0.057 |
| Never | 11 (44) | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foroughi, K.; Amini, M.; Atashi, A.; Mahmoodzadeh, H.; Hamann, U.; Manoochehri, M. Tissue-Specific Down-Regulation of the Long Non-Coding RNAs PCAT18 and LINC01133 in Gastric Cancer Development. Int. J. Mol. Sci. 2018, 19, 3881. https://doi.org/10.3390/ijms19123881
Foroughi K, Amini M, Atashi A, Mahmoodzadeh H, Hamann U, Manoochehri M. Tissue-Specific Down-Regulation of the Long Non-Coding RNAs PCAT18 and LINC01133 in Gastric Cancer Development. International Journal of Molecular Sciences. 2018; 19(12):3881. https://doi.org/10.3390/ijms19123881
Chicago/Turabian StyleForoughi, Kobra, Mohammad Amini, Amir Atashi, Habibollah Mahmoodzadeh, Ute Hamann, and Mehdi Manoochehri. 2018. "Tissue-Specific Down-Regulation of the Long Non-Coding RNAs PCAT18 and LINC01133 in Gastric Cancer Development" International Journal of Molecular Sciences 19, no. 12: 3881. https://doi.org/10.3390/ijms19123881
APA StyleForoughi, K., Amini, M., Atashi, A., Mahmoodzadeh, H., Hamann, U., & Manoochehri, M. (2018). Tissue-Specific Down-Regulation of the Long Non-Coding RNAs PCAT18 and LINC01133 in Gastric Cancer Development. International Journal of Molecular Sciences, 19(12), 3881. https://doi.org/10.3390/ijms19123881





