Lipopolysaccharide-Preconditioned Periodontal Ligament Stem Cells Induce M1 Polarization of Macrophages through Extracellular Vesicles
Abstract
1. Introduction
2. Results
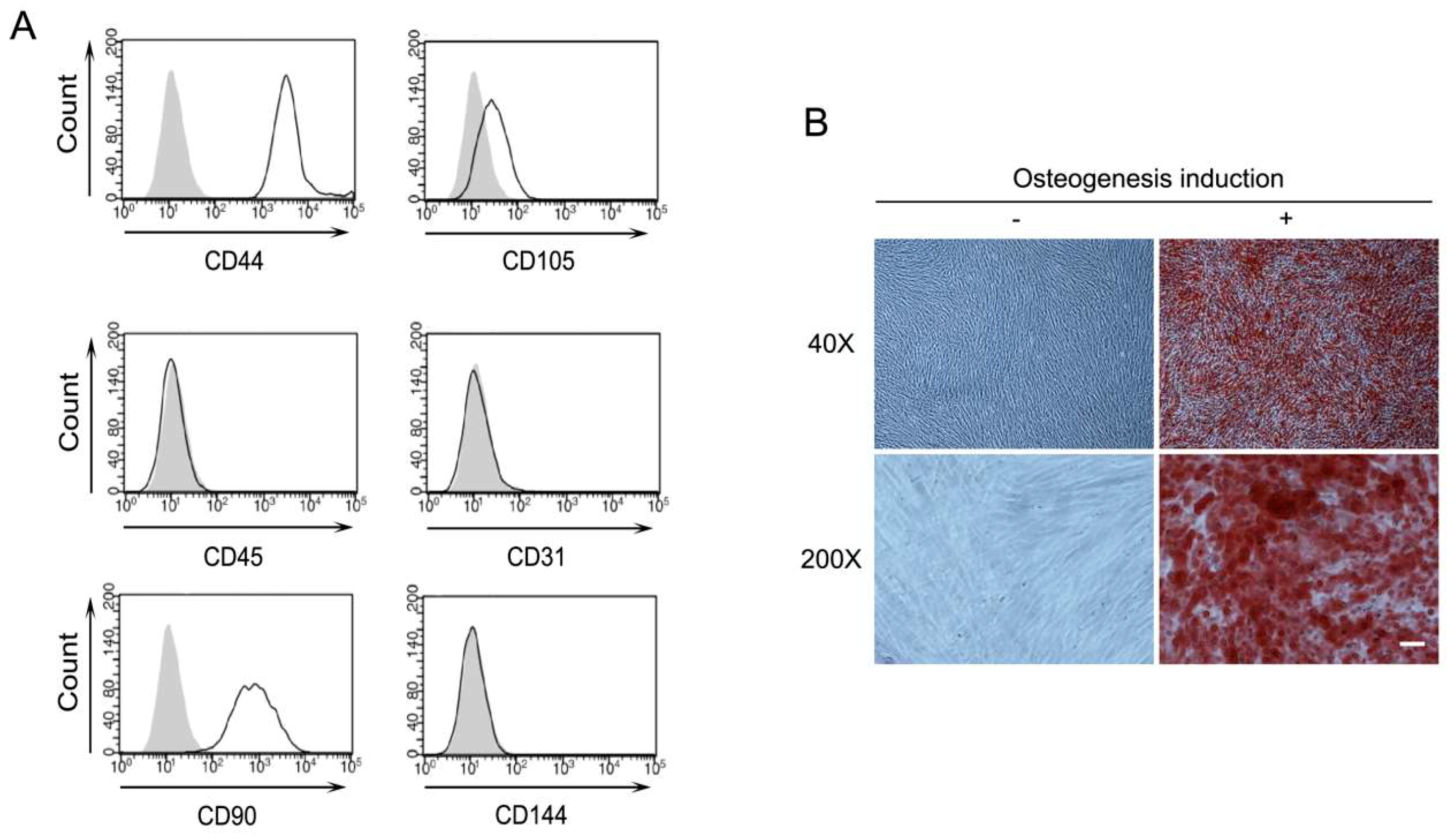
2.1. Characterization of Human Periodontal Ligament Stem Cells (PDLSCs)
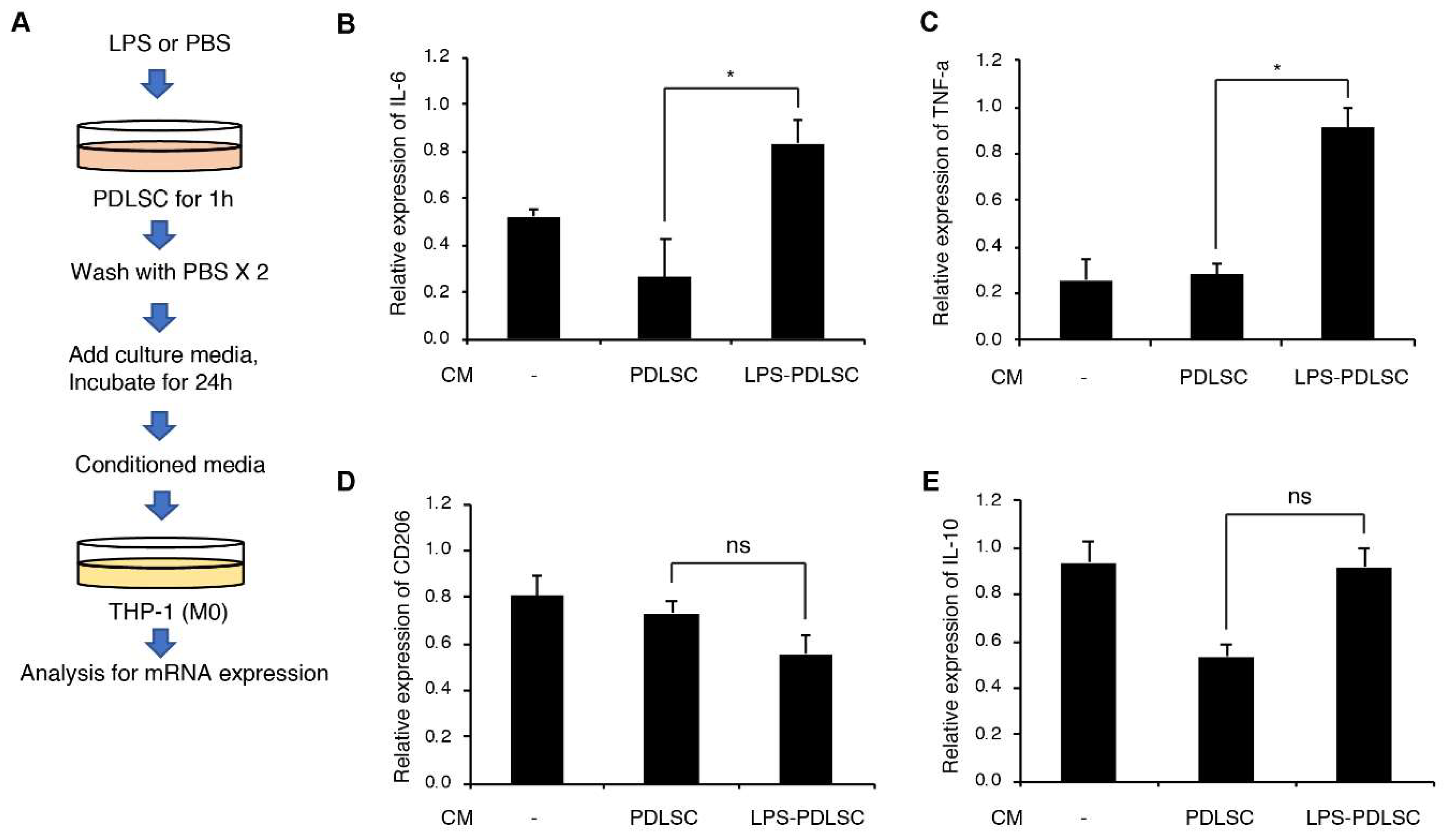
2.2. Effect of the Supernatant from PDLSCs on Macrophage Polarization
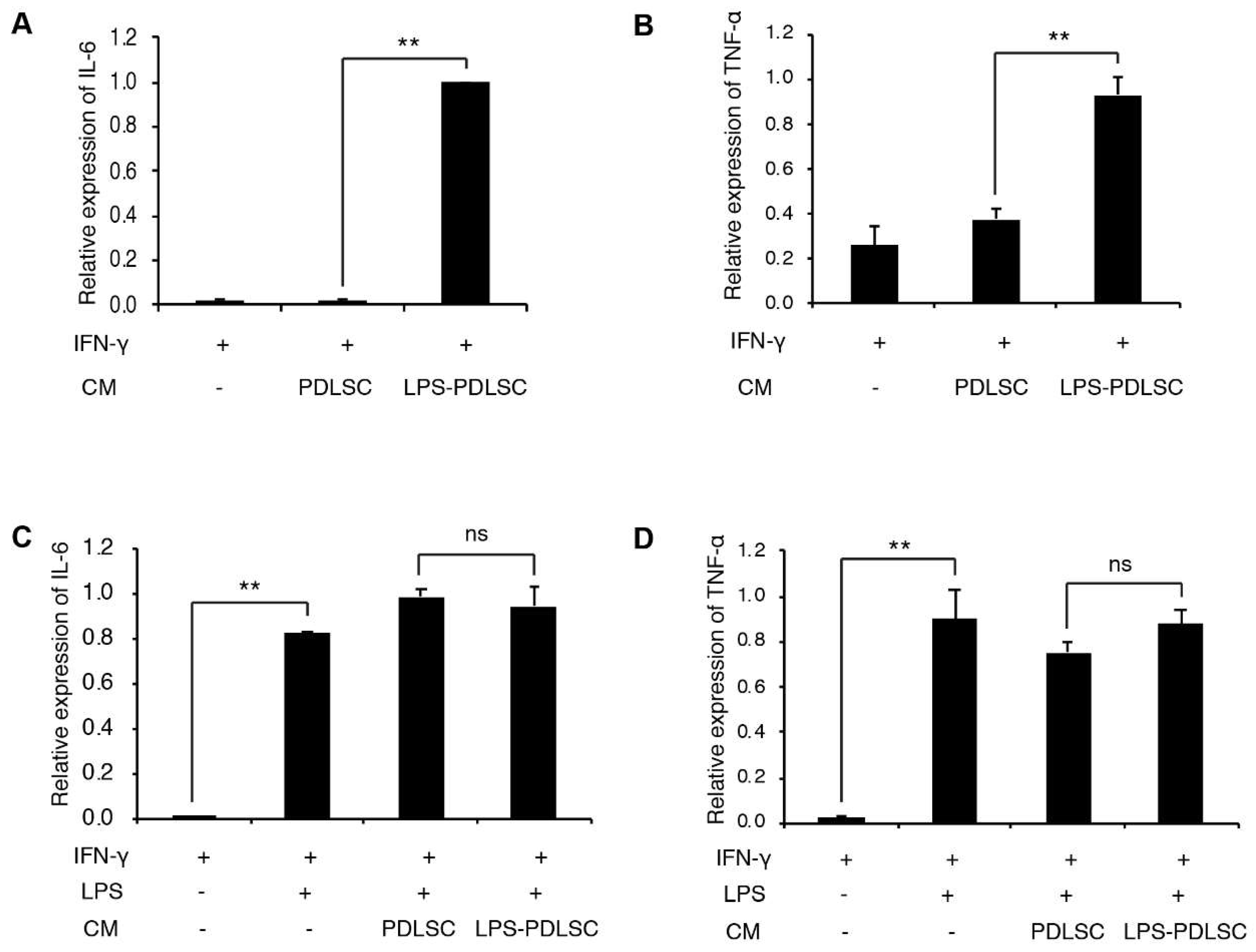
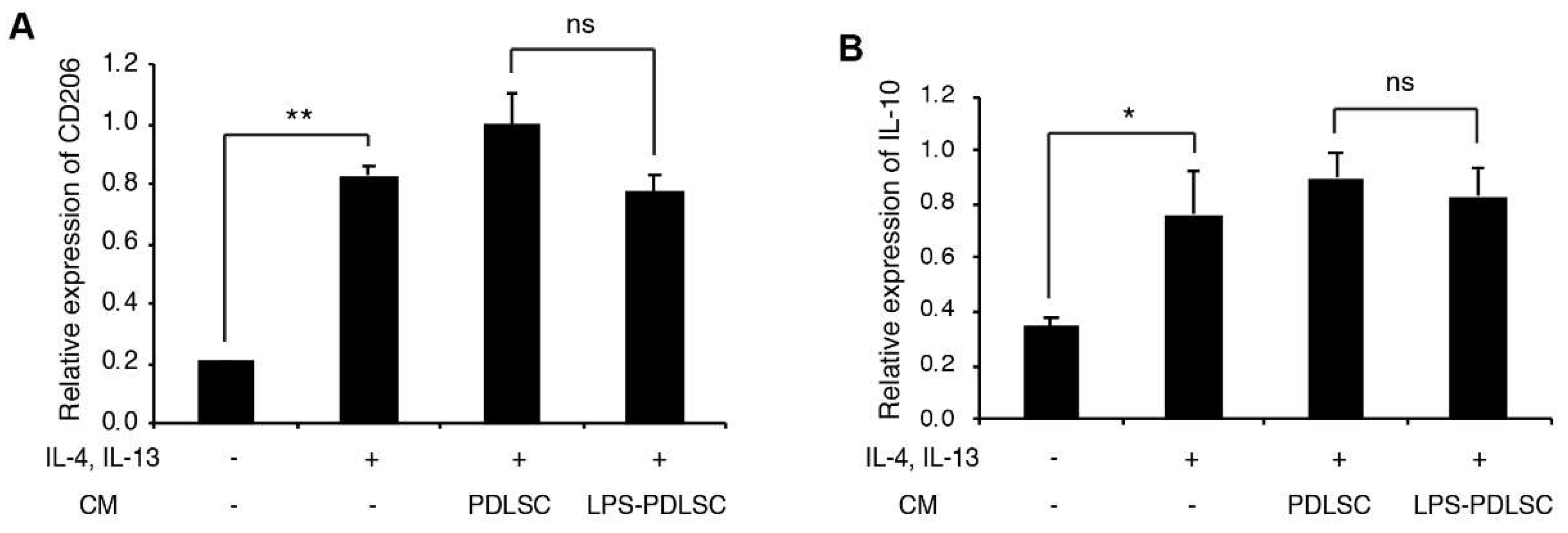
2.3. LPS-Preconditioned PDLSCs Accentuate the M1 Polarization of IFN-γ Treated Macrophages
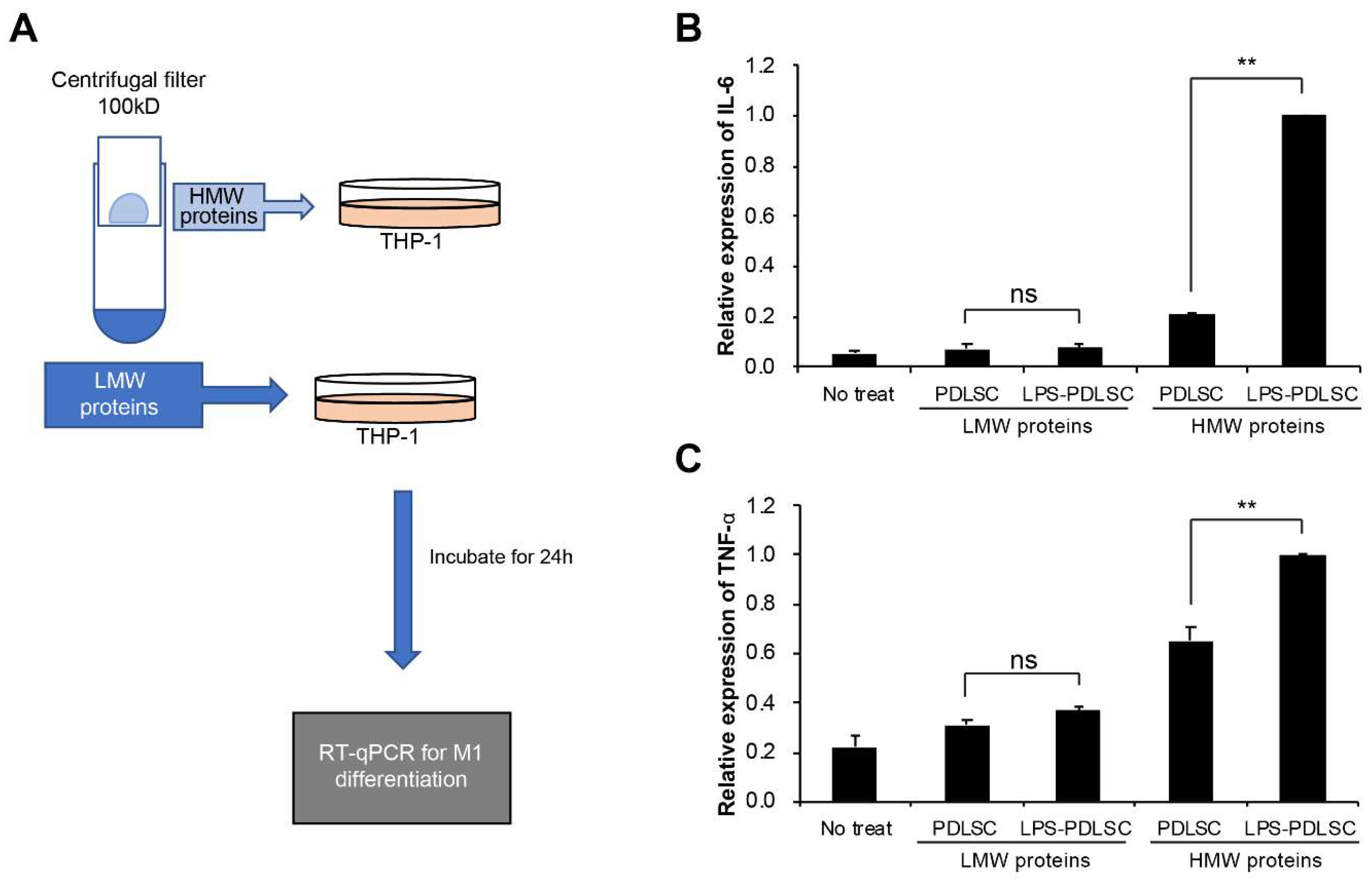
2.4. M1 Polarization Was Not Mediated by Free (Soluble) Cytokines from PDLSCs
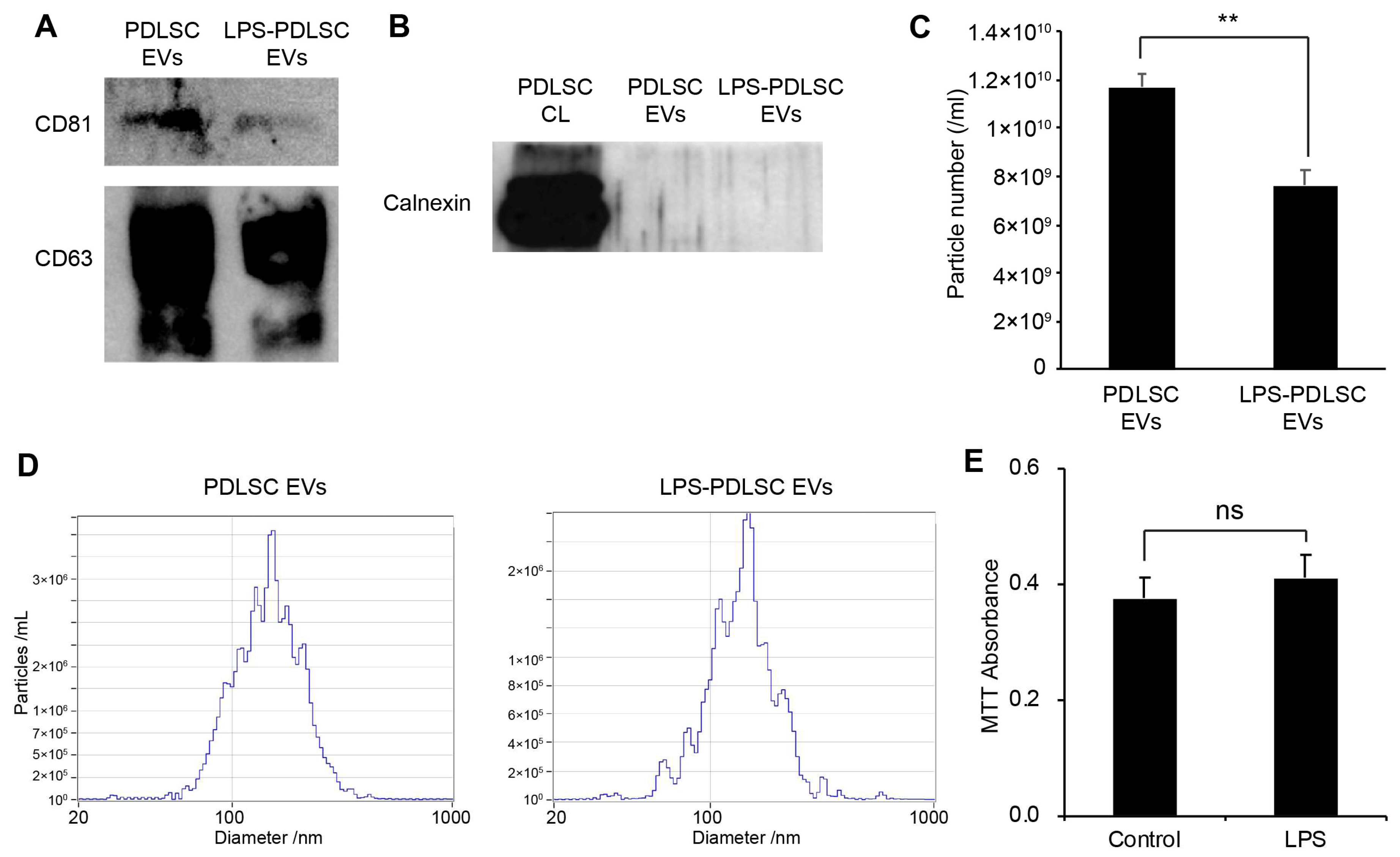
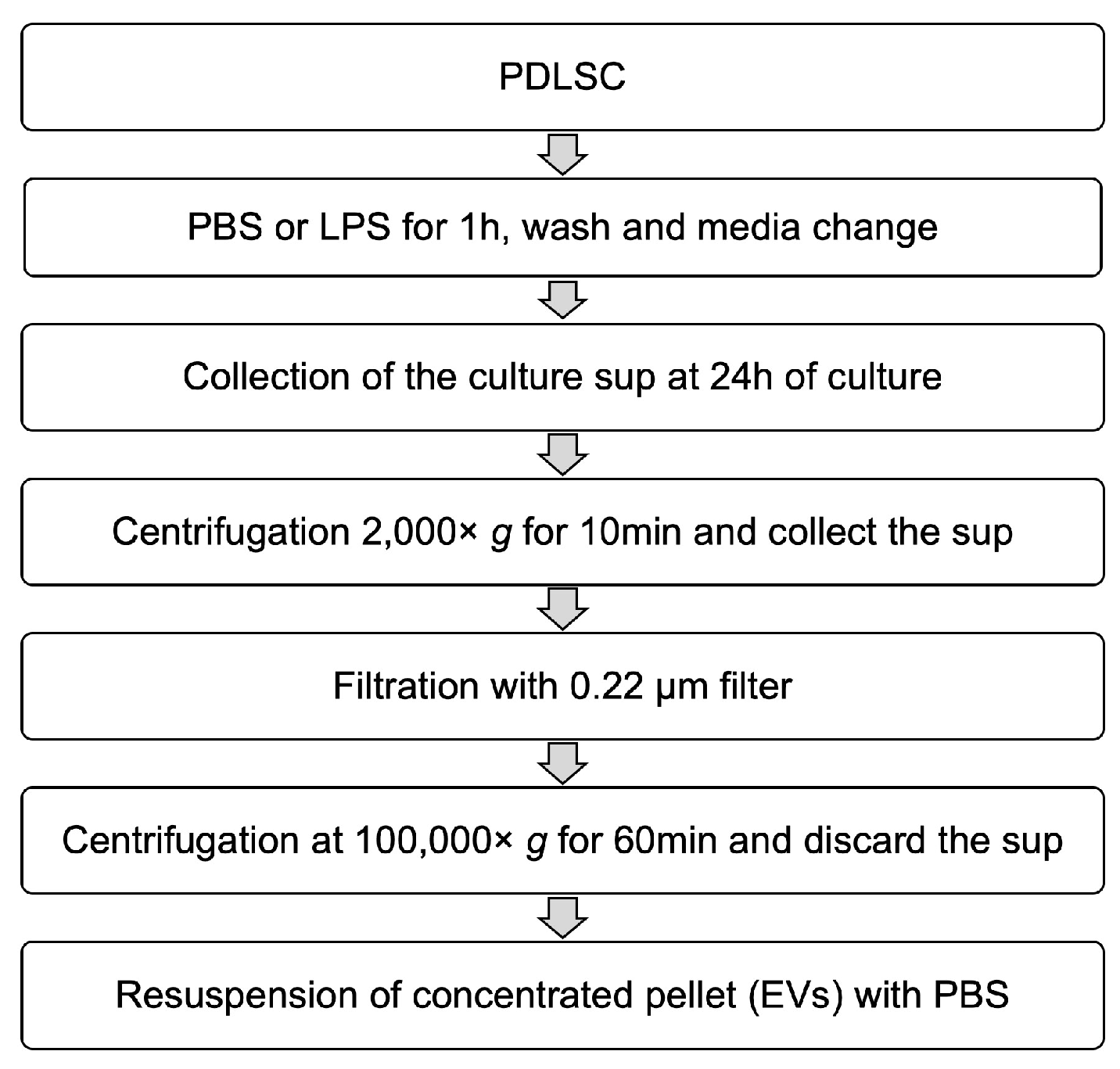
2.5. Isolation of EVs from PDLSCs and Their Characterization
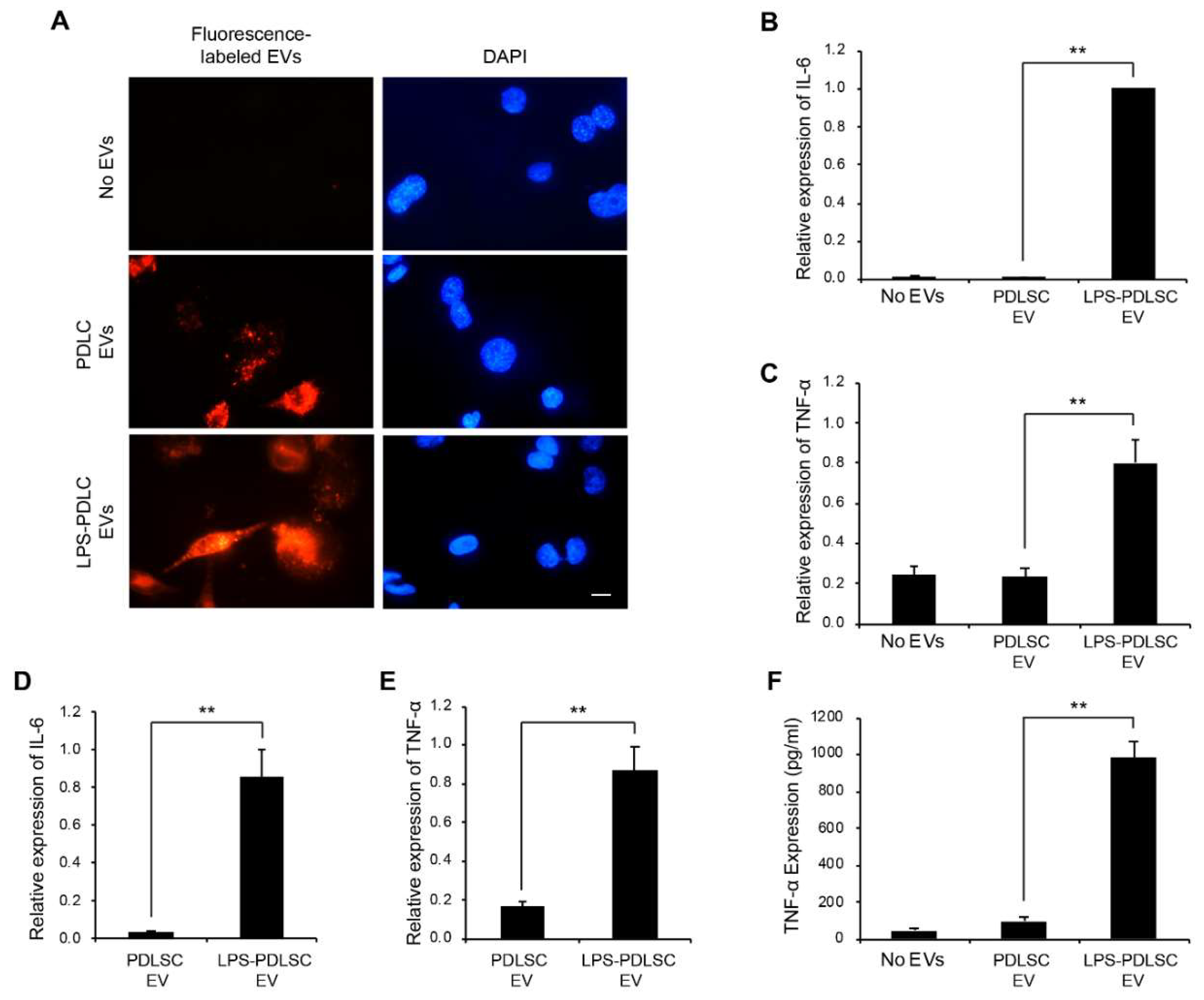
2.6. EVs from LPS-Preconditioned PDLSCs Synergize with the Action of IFN-γ on Macrophages
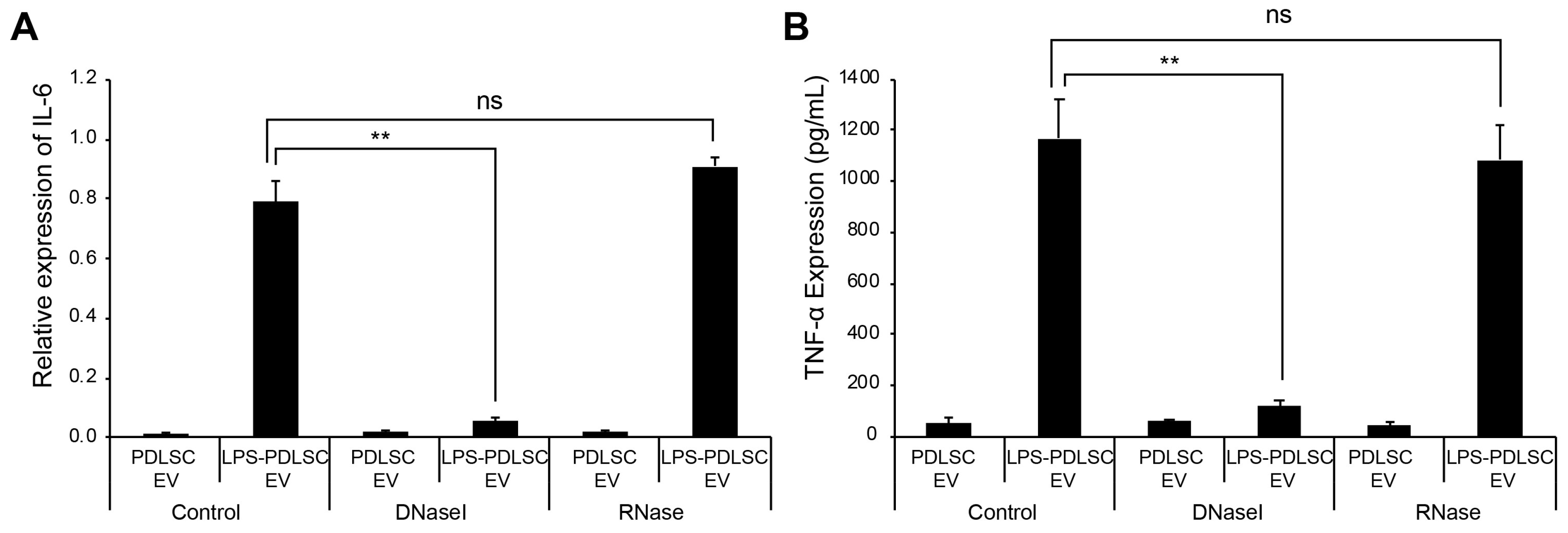
2.7. DNase I Treatment Abolishes M1 Polarization Effect of EVs
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Flow Cytometry
4.3. Osteogenic Differentiation of PDLSCs
4.4. Quantitative Real-Time Reverse Transcription PCR (RT-qPCR)
4.5. Induction of M1 and M2 Phenotype of THP-1 Cells
4.6. Isolation of EVs from PDLSCs
4.7. Nano-Particle Tracking Analysis
4.8. Labeling and Tracking of the Entry of EVs
4.9. Remove of Endotoxin from EVs
4.10. TNF-α ELISA
4.11. Statistical Analyzes
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eke, P.I.; Dye, B.A.; Wei, L.; Slade, G.D.; Thornton-Evans, G.O.; Borgnakke, W.S.; Taylor, G.W.; Page, R.C.; Beck, J.D.; Genco, R.J. Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 to 2012. J. Periodontol. 2015, 86, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Offenbacher, S.; Barros, S.P.; Beck, J.D. Rethinking periodontal inflammation. J. Periodontol. 2008, 79, 1577–1584. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, M.; Garcia, J.; Dutzan, N.; Henriquez, L.; Puente, J.; Carvajal, P.; Hernandez, M.; Gamonal, J. Interferon-gamma, interleukins-6 and -4, and factor XIII-A as indirect markers of the classical and alternative macrophage activation pathways in chronic periodontitis. J. Periodontol. 2014, 85, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Allam, J.P.; Duan, Y.; Heinemann, F.; Winter, J.; Gotz, W.; Deschner, J.; Wenghoefer, M.; Bieber, T.; Jepsen, S.; Novak, N. IL-23-producing CD68(+) macrophage-like cells predominate within an IL-17-polarized infiltrate in chronic periodontitis lesions. J. Clin. Periodontol. 2011, 38, 879–886. [Google Scholar] [CrossRef]
- Pussinen, P.J.; Vilkuna-Rautiainen, T.; Alfthan, G.; Palosuo, T.; Jauhiainen, M.; Sundvall, J.; Vesanen, M.; Mattila, K.; Asikainen, S. Severe periodontitis enhances macrophage activation via increased serum lipopolysaccharide. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 2174–2180. [Google Scholar] [CrossRef]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Francois, M.; Romieu-Mourez, R.; Li, M.; Galipeau, J. Human MSC suppression correlates with cytokine induction of indoleamine 2,3-dioxygenase and bystander M2 macrophage differentiation. Mol. Ther. 2012, 20, 187–195. [Google Scholar] [CrossRef]
- Cho, D.I.; Kim, M.R.; Jeong, H.Y.; Jeong, H.C.; Jeong, M.H.; Yoon, S.H.; Kim, Y.S.; Ahn, Y. Mesenchymal stem cells reciprocally regulate the M1/M2 balance in mouse bone marrow-derived macrophages. Exp. Mol. Med. 2014, 46, e70. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Kong, E.F.; Meiller, T.F.; Jabra-Rizk, M.A. Periodontal Diseases: Bug Induced, Host Promoted. PLoS Pathog. 2015, 11, e1004952. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Shirane, T.; Anamura, S.; Nakanishi, K.; Okamoto, H. A study of the effects of bacterial endotoxin on periodontal tissue in vivo (author’s transl). Nihon Shishubyo Gakkai Kaishi 1980, 22, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Daly, C.G.; Seymour, G.J.; Kieser, J.B. Bacterial endotoxin: A role in chronic inflammatory periodontal disease? J. Oral Pathol. 1980, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nagata, M.; Iwasaki, K.; Akazawa, K.; Komaki, M.; Yokoyama, N.; Izumi, Y.; Morita, I. Conditioned Medium from Periodontal Ligament Stem Cells Enhances Periodontal Regeneration. Tissue Eng. Part A 2017, 23, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Iwata, T.; Yamato, M.; Zhang, Z.; Mukobata, S.; Washio, K.; Ando, T.; Feijen, J.; Okano, T.; Ishikawa, I. Validation of human periodontal ligament-derived cells as a reliable source for cytotherapeutic use. J. Clin. Periodontol. 2010, 37, 1088–1099. [Google Scholar] [CrossRef]
- Steinert, A.F.; Kunz, M.; Prager, P.; Gobel, S.; Klein-Hitpass, L.; Ebert, R.; Noth, U.; Jakob, F.; Gohlke, F. Characterization of bursa subacromialis-derived mesenchymal stem cells. Stem Cell Res. Ther. 2015, 6, 114. [Google Scholar] [CrossRef]
- Mildmay-White, A.; Khan, W. Cell Surface Markers on Adipose-Derived Stem Cells: A Systematic Review. Curr. Stem Cell Res. Ther. 2017, 12, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, J.Q.; Kruger, A.; Gallogly, S.; Hanley, S.A.; Hogan, M.C.; Ward, C.J.; Joshi, L.; Griffin, M.D. Surface glycosylation profiles of urine extracellular vesicles. PLoS ONE 2013, 8, e74801. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bardi, G.T.; Smith, M.A.; Hood, J.L. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine 2018, 105, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.J.; Kim, H.S.; Hwang, E.H.; Woo, J.; Zhang, M.; Moon, W.K. Breast cancer cell-derived exosomes and macrophage polarization are associated with lymph node metastasis. Oncotarget 2018, 9, 7398–7410. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef]
- Lian, Q.; Xu, J.; Yan, S.; Huang, M.; Ding, H.; Sun, X.; Bi, A.; Ding, J.; Sun, B.; Geng, M. Chemotherapy-induced intestinal inflammatory responses are mediated by exosome secretion of double-strand DNA via AIM2 inflammasome activation. Cell Res. 2017, 27, 784–800. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Bernabe, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.; Marcenes, W. Global burden of severe periodontitis in 1990-2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef]
- Dhadse, P.; Gattani, D.; Mishra, R. The link between periodontal disease and cardiovascular disease: How far we have come in last two decades? J. Indian Soc. Periodontol. 2010, 14, 148–154. [Google Scholar] [CrossRef]
- Teeuw, W.J.; Kosho, M.X.; Poland, D.C.; Gerdes, V.E.; Loos, B.G. Periodontitis as a possible early sign of diabetes mellitus. BMJ Open Diabetes Res. Care 2017, 5, e000326. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Menicanin, D.; Shi, S.; Bartold, P.M.; Gronthos, S. Immunomodulatory properties of human periodontal ligament stem cells. J. Cell. Physiol. 2009, 219, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Pelaez, D.; Dominguez-Bendala, J.; Garcia-Godoy, F.; Cheung, H.S. Plasticity of stem cells derived from adult periodontal ligament. Regen. Med. 2009, 4, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Z.; Su, W.R.; Shi, S.H.; Wilder-Smith, P.; Xiang, A.P.; Wong, A.; Nguyen, A.L.; Kwon, C.W.; Le, A.D. Human gingiva-derived mesenchymal stem cells elicit polarization of m2 macrophages and enhance cutaneous wound healing. Stem Cells 2010, 28, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, N.; Okubo, N.; Kamo, M.; Chosa, N.; Mikami, T.; Suzuki, K.; Yokota, S.; Ibi, M.; Ohtsuka, M.; Taira, M.; et al. Bone marrow-derived mesenchymal stem cells propagate immunosuppressive/anti-inflammatory macrophages in cell-to-cell contact-independent and -dependent manners under hypoxic culture. Exp. Cell Res. 2017, 358, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Ti, D.; Hao, H.; Tong, C.; Liu, J.; Dong, L.; Zheng, J.; Zhao, Y.; Liu, H.; Fu, X.; Han, W. LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. J. Transl. Med. 2015, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Takasugi, M. Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell 2018, 17, e12734. [Google Scholar] [CrossRef]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef]
- Wilson, K.M.; Jagger, A.M.; Walker, M.; Seinkmane, E.; Fox, J.M.; Kroger, R.; Genever, P.; Ungar, D. Glycans modify mesenchymal stem cell differentiation to impact on the function of resulting osteoblasts. J. Cell Sci. 2018. [Google Scholar] [CrossRef]
- Thibault, R.A.; Scott Baggett, L.; Mikos, A.G.; Kasper, F.K. Osteogenic differentiation of mesenchymal stem cells on pregenerated extracellular matrix scaffolds in the absence of osteogenic cell culture supplements. Tissue Eng. Part A 2010, 16, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.E.; To, J.; O’Brien, B.A.; Donnelly, S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods 2016, 430, 64–70. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Lee, M.-J.; Park, S.J.; Lee, M.-S. Lipopolysaccharide-Preconditioned Periodontal Ligament Stem Cells Induce M1 Polarization of Macrophages through Extracellular Vesicles. Int. J. Mol. Sci. 2018, 19, 3843. https://doi.org/10.3390/ijms19123843
Kang H, Lee M-J, Park SJ, Lee M-S. Lipopolysaccharide-Preconditioned Periodontal Ligament Stem Cells Induce M1 Polarization of Macrophages through Extracellular Vesicles. International Journal of Molecular Sciences. 2018; 19(12):3843. https://doi.org/10.3390/ijms19123843
Chicago/Turabian StyleKang, Hyejong, Myung-Ju Lee, Sang June Park, and Myung-Shin Lee. 2018. "Lipopolysaccharide-Preconditioned Periodontal Ligament Stem Cells Induce M1 Polarization of Macrophages through Extracellular Vesicles" International Journal of Molecular Sciences 19, no. 12: 3843. https://doi.org/10.3390/ijms19123843
APA StyleKang, H., Lee, M.-J., Park, S. J., & Lee, M.-S. (2018). Lipopolysaccharide-Preconditioned Periodontal Ligament Stem Cells Induce M1 Polarization of Macrophages through Extracellular Vesicles. International Journal of Molecular Sciences, 19(12), 3843. https://doi.org/10.3390/ijms19123843




