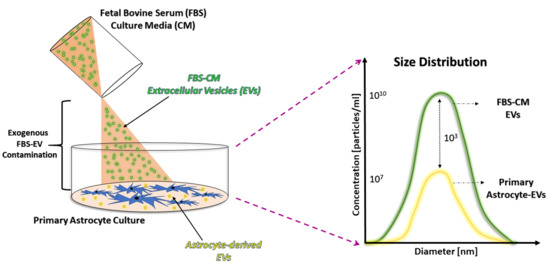
Fetal Bovine Serum-Derived Extracellular Vesicles Persist within Vesicle-Depleted Culture Media
Abstract
1. Introduction
2. Results
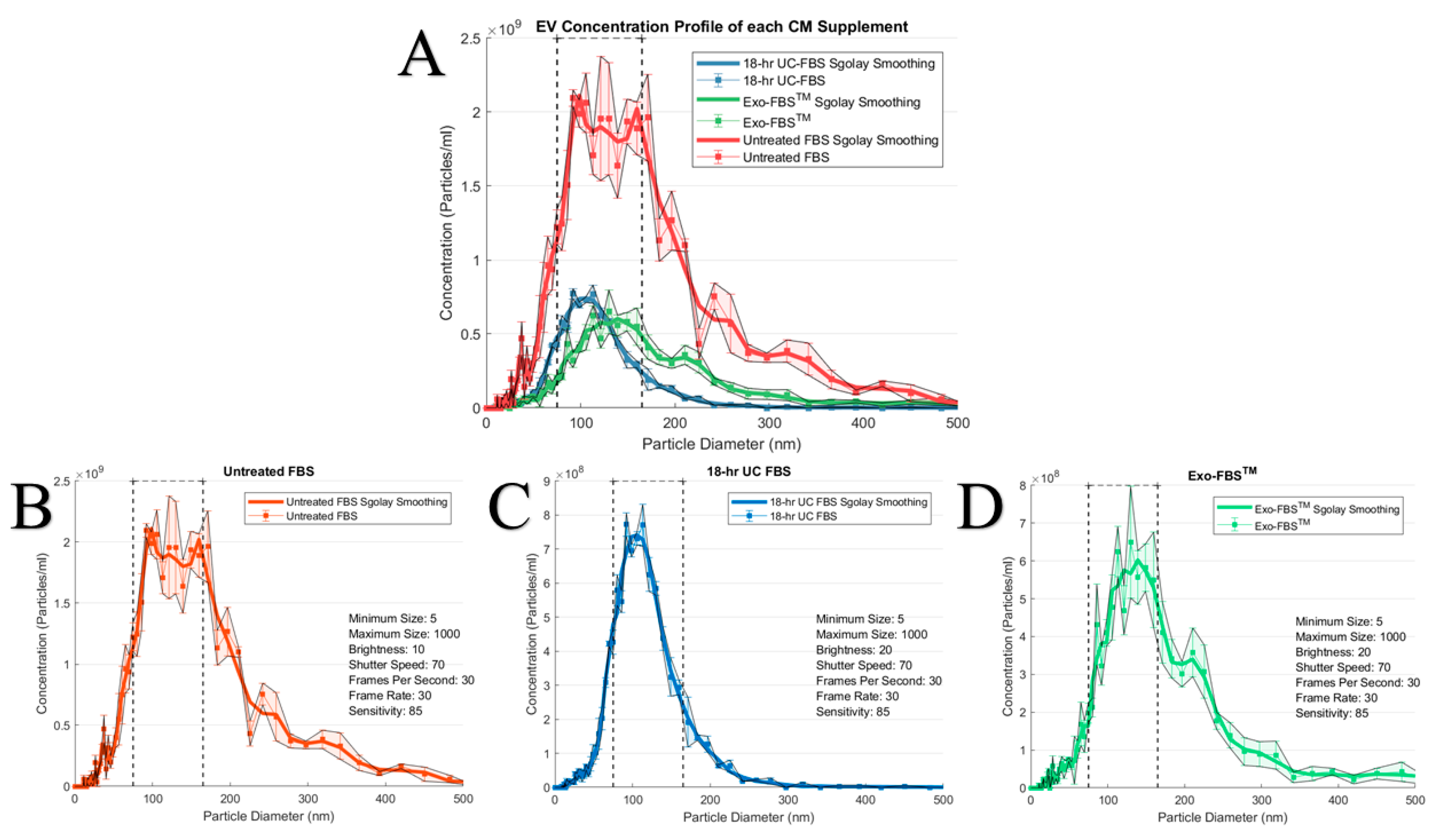
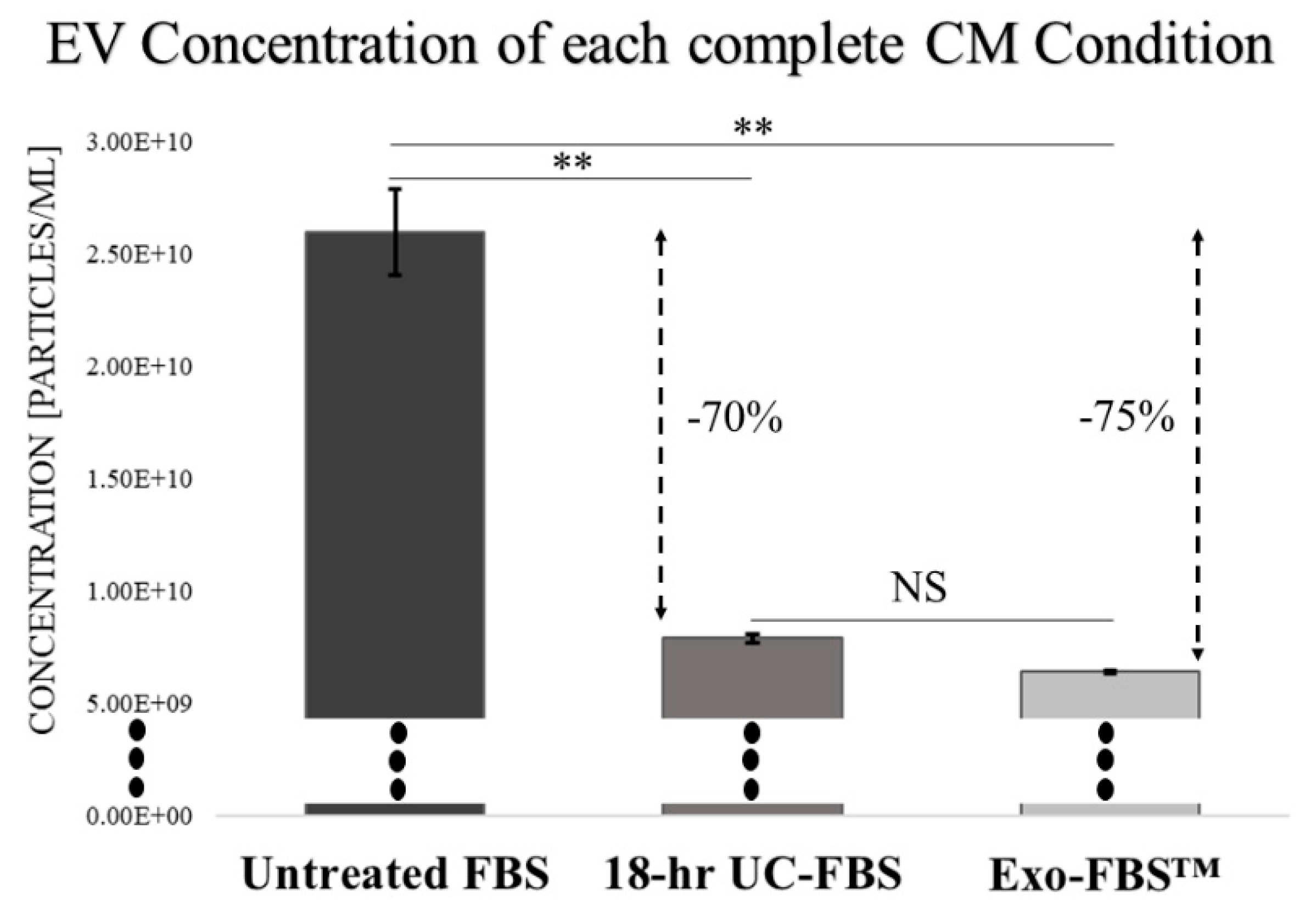
2.1. An Abundant Population of EVs Remains within EV-Depleted Complete CM
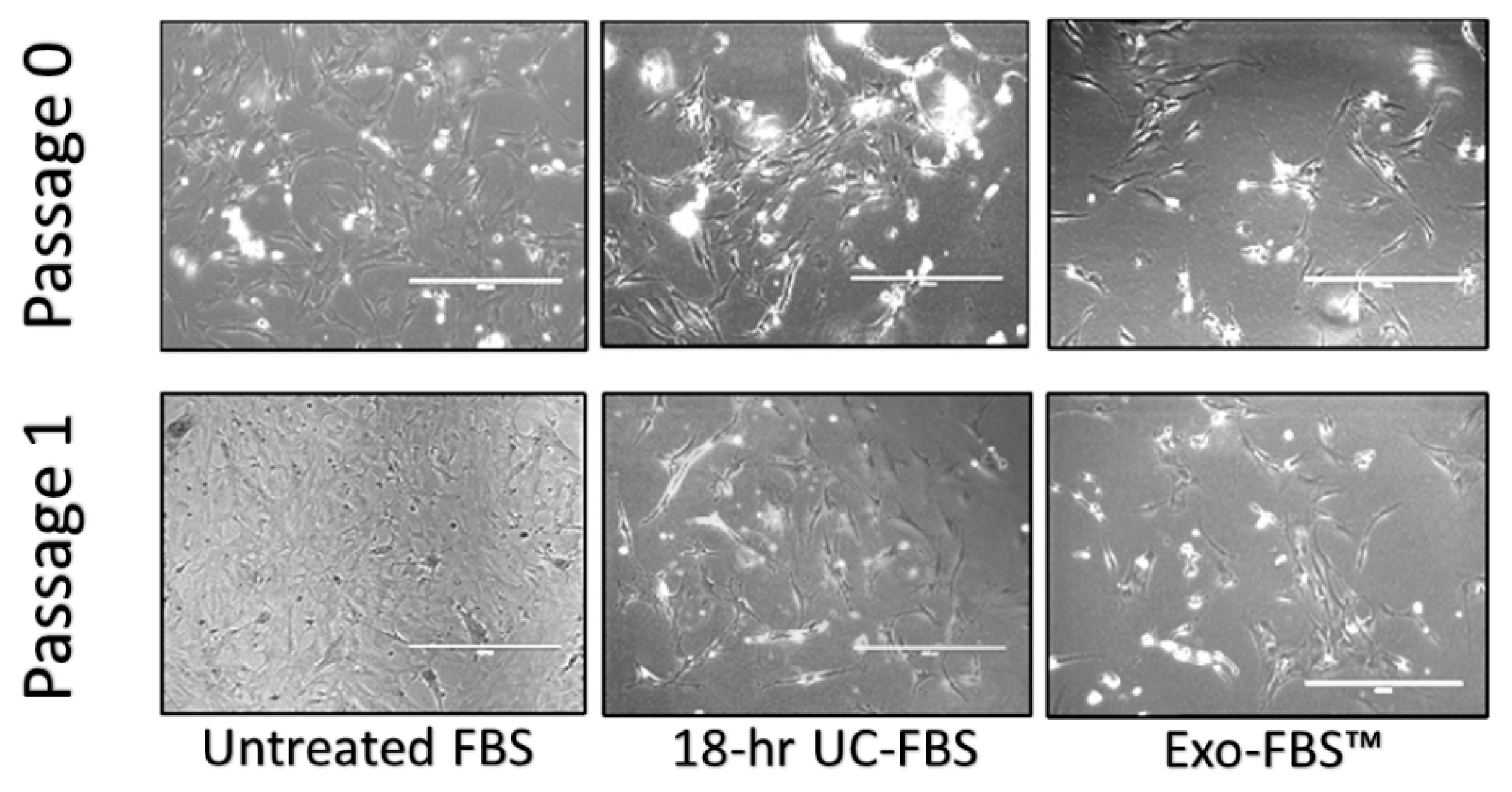
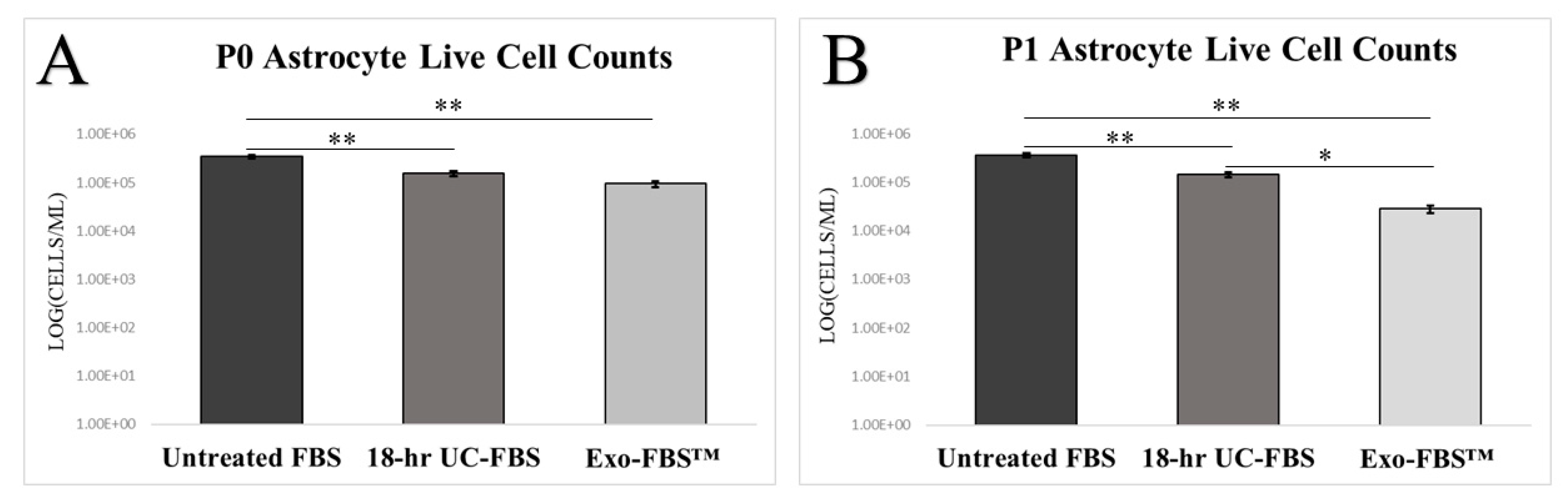
2.2. Astrocyte Growth and Viability Is Reduced in EV-Depleted Serum Conditions
3. Discussion
4. Materials and Methods
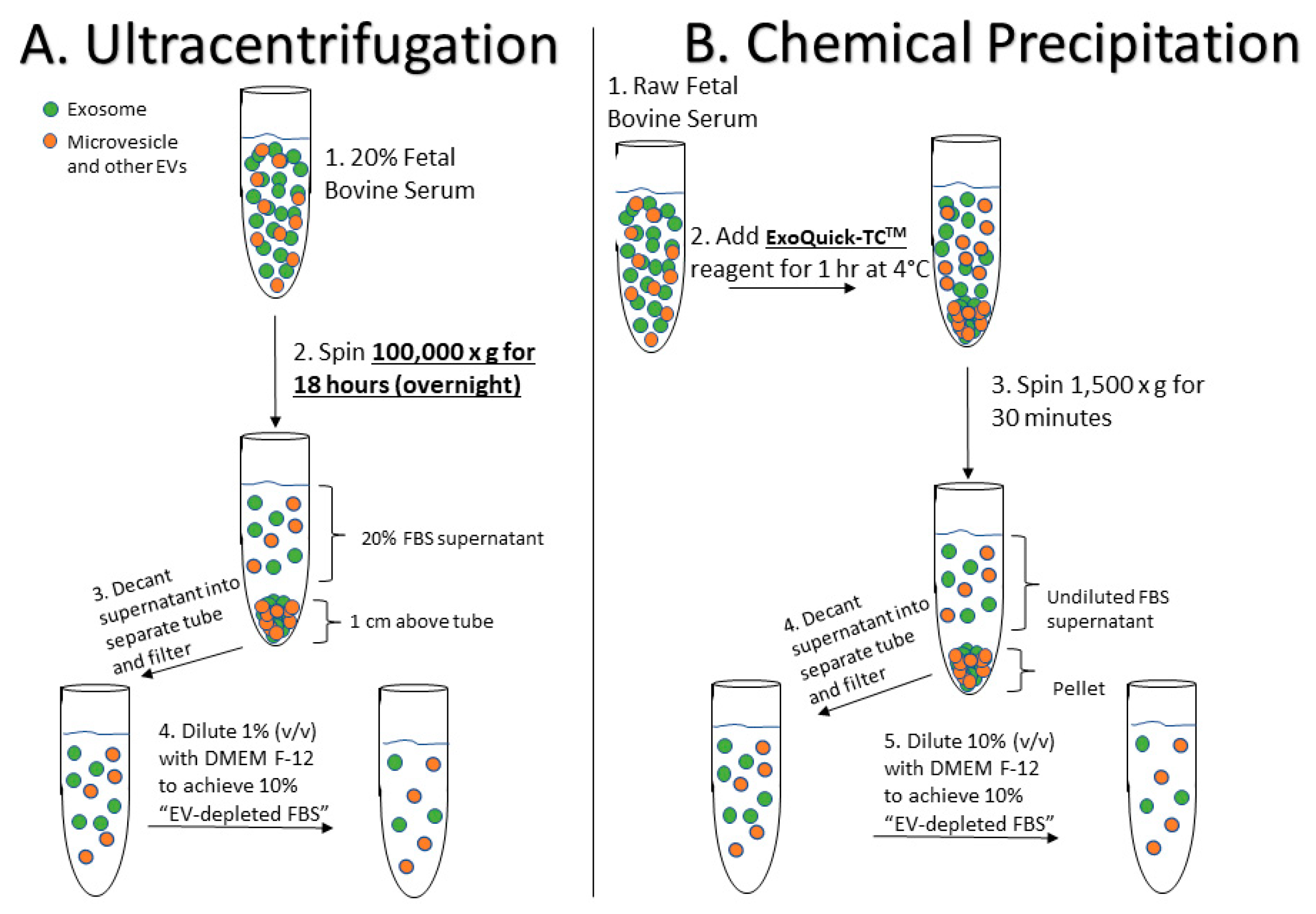
4.1. Preparation of EV-Depleted Serum for Testing
4.2. Primary Rat Astrocyte Cell Cultures
4.3. EV Size and Concentration Determinations via Nanoparticle Tracking Analytics
4.4. Cell Counts and Time-Lapse Photo-Microscopy
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CM | Culture Media |
| FBS | Fetal Bovine Serum |
| EV | Extracellular Vesicle |
| ECF | Extracellular Fluid |
| iPSCs | Induced-Pluripotent Stem Cells |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| UC | Ultracentrifugation |
| NTA | Nanoparticle Tracking Analytics |
| ICC | Immunocytochemistry |
| FACS | Fluorescence-Activated Cell Sorting |
| DLS | Dynamic Light Scattering |
| RPS | Resistive Pulse Sensing |
| ANOVA | One-way Analysis of Variance |
References
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 766. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.R. Q&A: What are exosomes, exactly? BMC Biol. 2016, 14, 46. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Thery, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Brunner, D.; Frank, J.; Appl, H.; Gstraunthaler, G. Serum-free cell culture: The serum-free media interactive online database. Altex 2010, 27, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- Aswad, H.; Jalabert, A.; Rome, S. Depleting extracellular vesicles from fetal bovine serum alters proliferation and differentiation of skeletal muscle cells in vitro. BMC Biotechnol. 2016, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Zhang, S.; Witwer, K.W.; Mattson, M.P. Extracellular vesicle–depleted fetal bovine and human sera have reduced capacity to support cell growth. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Muth, D.C.; Eitan, E.; Travers, M.; Learman, L.N.; Lehrmann, E.; Witwer, K.W. Serum extracellular vesicle depletion processes affect release and infectivity of HIV-1 in culture. Sci. Rep. 2017, 7, 2558. [Google Scholar] [CrossRef] [PubMed]
- Beninson, L.A.; Fleshner, M. Exosomes in fetal bovine serum dampen primary macrophage IL-1beta response to lipopolysaccharide (LPS) challenge. Immunol. Lett. 2015, 163, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Shelke, G.V.; Lässer, C.; Gho, Y.S.; Lötvall, J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J.Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Batagov, A.O.; Carter, D.R.; Krichevsky, A.M. Fetal Bovine Serum RNA Interferes with the Cell Culture derived Extracellular RNA. Sci. Rep. 2016, 6, 31175. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, E.; Coumans, F.A.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Ovstebo, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef] [PubMed]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Kim, H.; Kim, T. Exosomes Secreted from Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Accelerate Skin Cell Proliferation. Int. J. Mol. Sci. 2018, 19, 3119. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef] [PubMed]
- Kornilov, R.; Puhka, M.; Mannerstrom, B.; Hiidenmaa, H.; Peltoniemi, H.; Siljander, P.; Seppanen-Kaijansinkko, R.; Kaur, S. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2018, 7, 1422674. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lee, Y.; Johansson, H.J.; Mager, I.; Vader, P.; Nordin, J.Z.; Wiklander, O.P.; Lehtio, J.; Wood, M.J.; Andaloussi, S.E. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015, 11, 600–607. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.D.; de Vellis, J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980, 85, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Hamrick, M.W.; Liu, Y. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef] [PubMed]
- BPM ExoPlot: MATLAB App for Nanoparticle Tracking Analytics, version 1.22.0.0; MATLAB 9.7 (R2017b); Mathworks: Natick, MA, USA, 2018.
- Cadena-Herrera, D.; Esparza-De Lara, J.E.; Ramirez-Ibanez, N.D.; Lopez-Morales, C.A.; Perez, N.O.; Flores-Ortiz, L.F.; Medina-Rivero, E. Validation of three viable-cell counting methods: Manual, semi-automated, and automated. Biotechnol. Rep. 2015, 7, 9–16. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lehrich, B.M.; Liang, Y.; Khosravi, P.; Federoff, H.J.; Fiandaca, M.S. Fetal Bovine Serum-Derived Extracellular Vesicles Persist within Vesicle-Depleted Culture Media. Int. J. Mol. Sci. 2018, 19, 3538. https://doi.org/10.3390/ijms19113538
Lehrich BM, Liang Y, Khosravi P, Federoff HJ, Fiandaca MS. Fetal Bovine Serum-Derived Extracellular Vesicles Persist within Vesicle-Depleted Culture Media. International Journal of Molecular Sciences. 2018; 19(11):3538. https://doi.org/10.3390/ijms19113538
Chicago/Turabian StyleLehrich, Brandon M., Yaxuan Liang, Pooya Khosravi, Howard J. Federoff, and Massimo S. Fiandaca. 2018. "Fetal Bovine Serum-Derived Extracellular Vesicles Persist within Vesicle-Depleted Culture Media" International Journal of Molecular Sciences 19, no. 11: 3538. https://doi.org/10.3390/ijms19113538
APA StyleLehrich, B. M., Liang, Y., Khosravi, P., Federoff, H. J., & Fiandaca, M. S. (2018). Fetal Bovine Serum-Derived Extracellular Vesicles Persist within Vesicle-Depleted Culture Media. International Journal of Molecular Sciences, 19(11), 3538. https://doi.org/10.3390/ijms19113538