Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives
Abstract
1. Introduction
2. Rabbit Genome Information
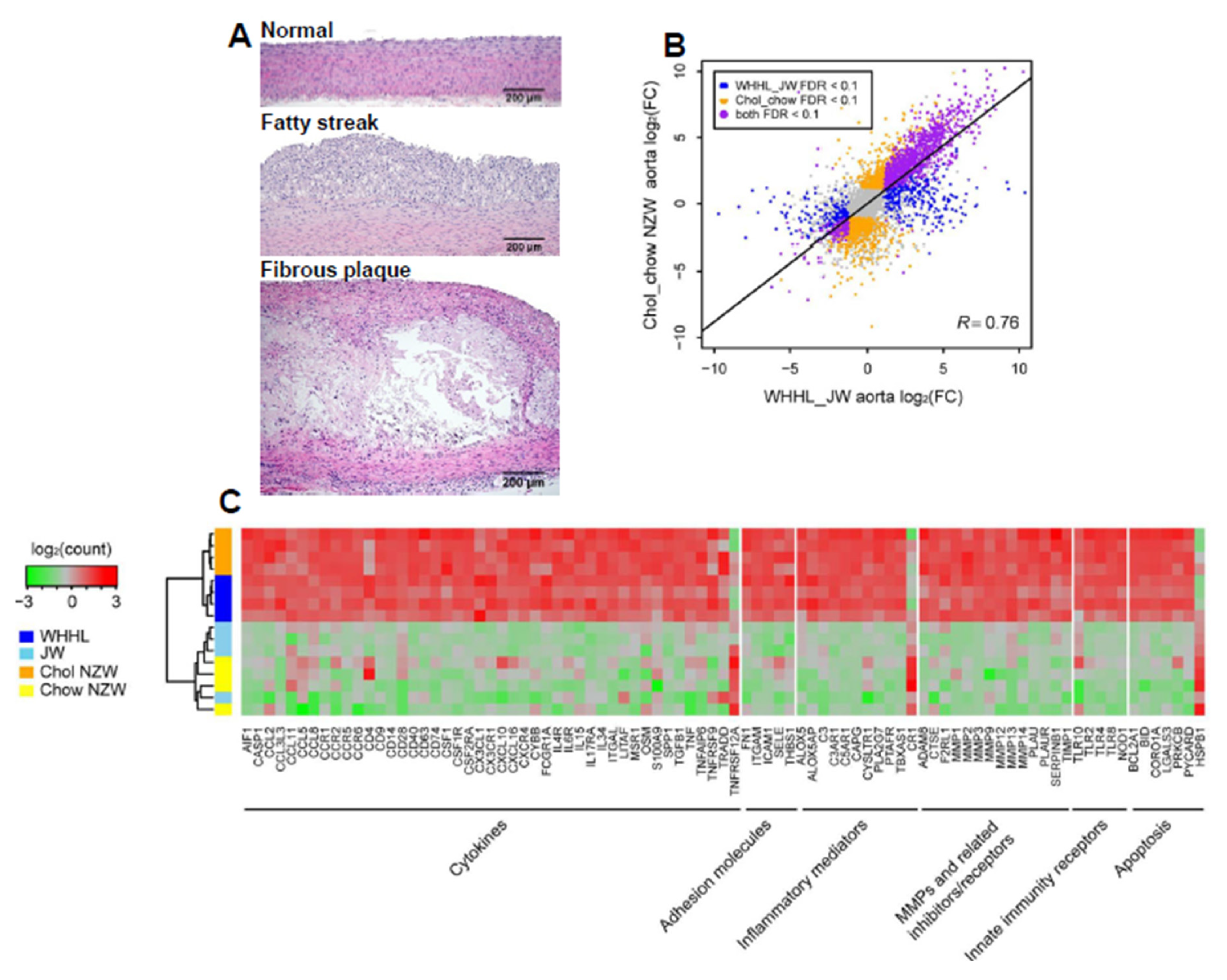
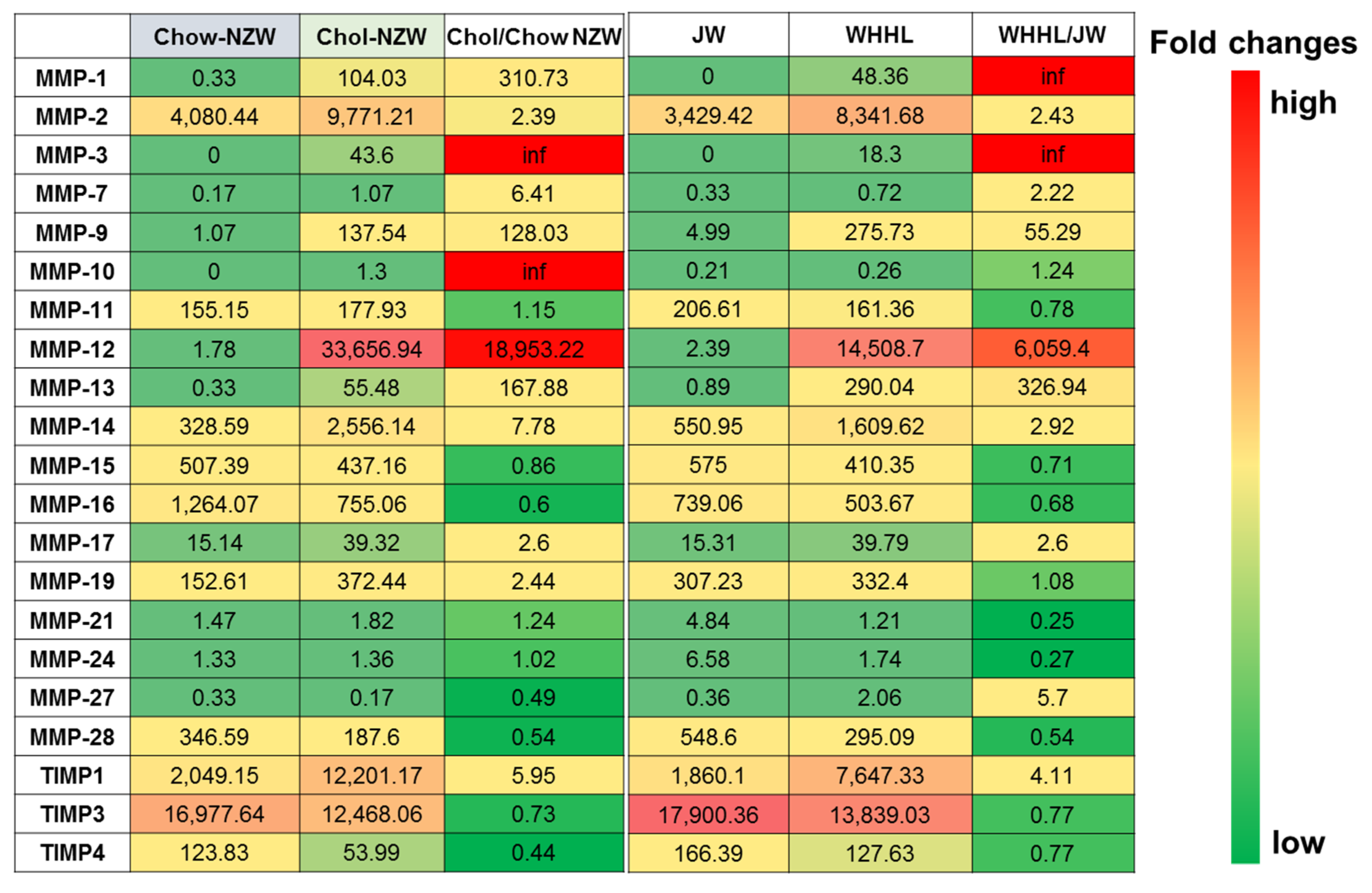
3. Transcriptome Profiling of Rabbit Atherosclerotic Lesions
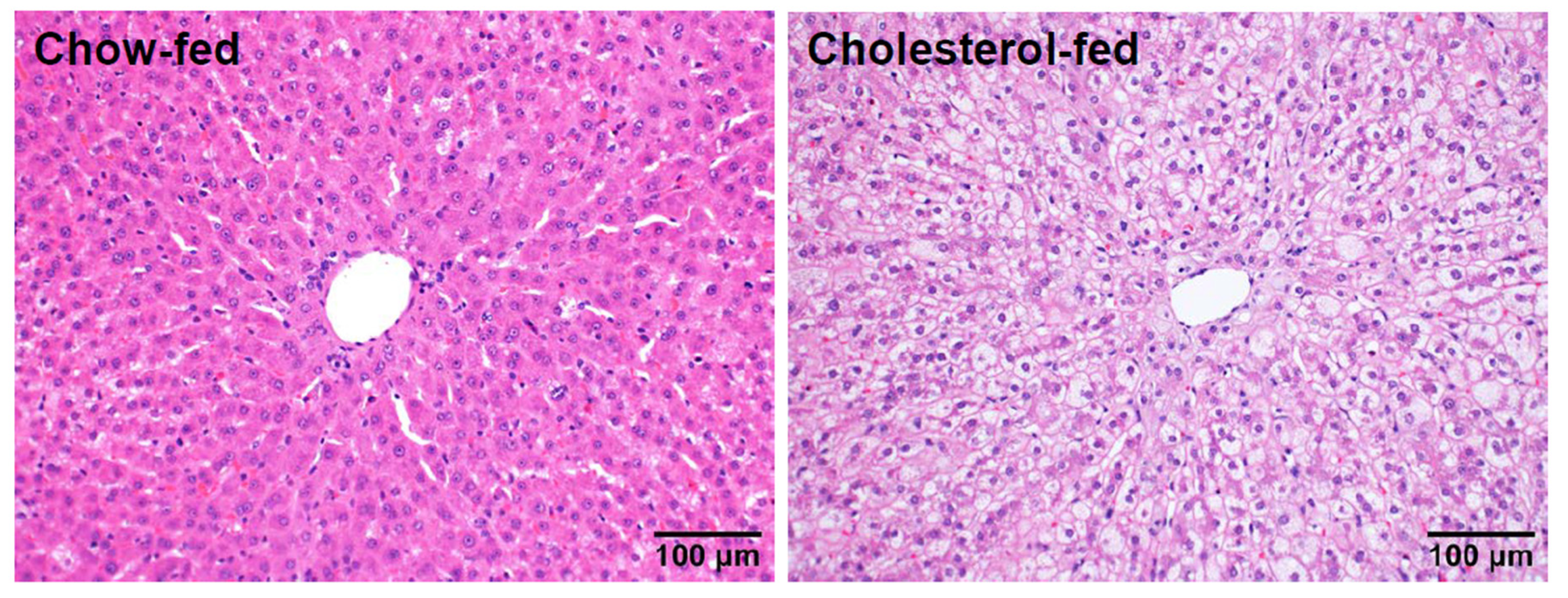
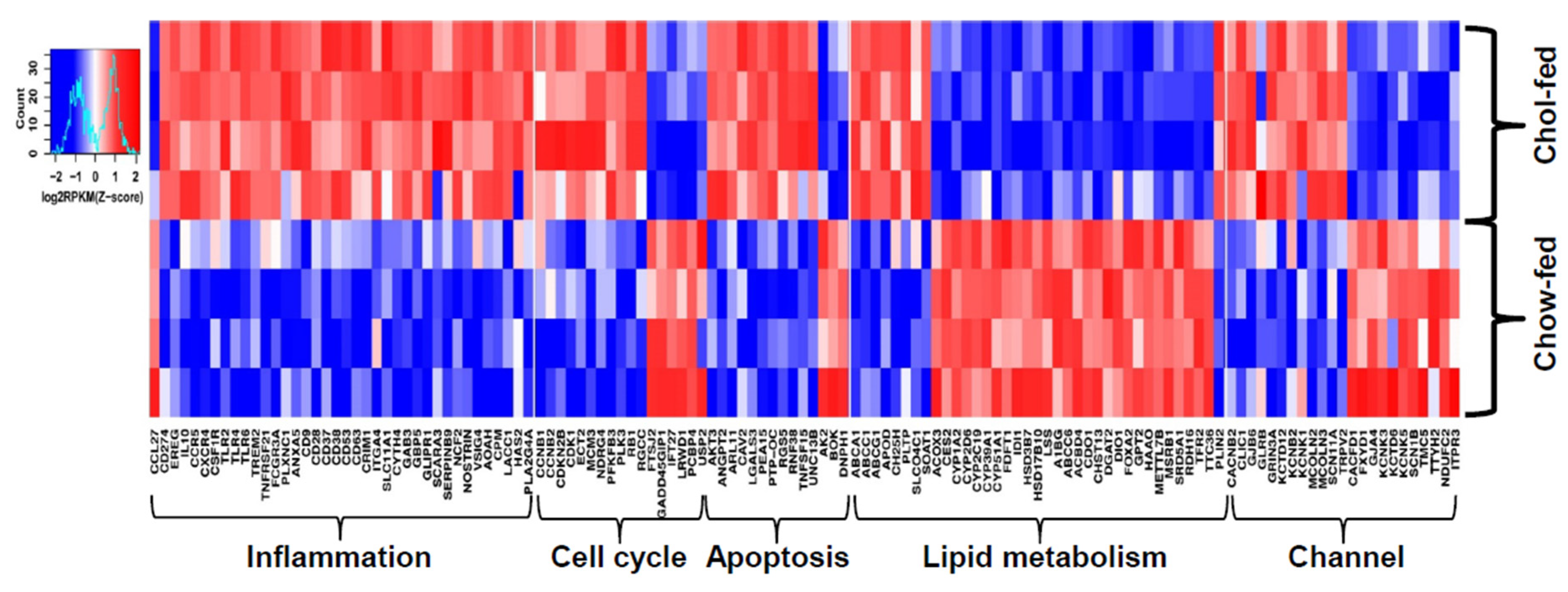
4. Transcriptomic Profiling of Livers
5. Concluding Remarks
Author Contributions
Funding
Abbreviations
| DEG | differentially expressed gene |
| KO | knock-out |
| LDL | low density lipoproteins |
| MMP | matrix metalloproteinase |
| TIMP | tissue inhibitor of metalloproteinase |
| VLDL | very low density lipoproteins |
| WHHL | Watanabe heritable hyperlipidemic |
References
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of nafld and nash: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Wouters, K.; van Gorp, P.J.; Bieghs, V.; Gijbels, M.J.; Duimel, H.; Lutjohann, D.; Kerksiek, A.; van Kruchten, R.; Maeda, N.; Staels, B.; et al. Dietary cholesterol, rather than liver steatosis, leads to hepatic inflammation in hyperlipidemic mouse models of nonalcoholic steatohepatitis. Hepatology 2008, 48, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Volzke, H.; Robinson, D.M.; Kleine, V.; Deutscher, R.; Hoffmann, W.; Ludemann, J.; Schminke, U.; Kessler, C.; John, U. Hepatic steatosis is associated with an increased risk of carotid atherosclerosis. World J. Gastroenterol. 2005, 11, 1848–1853. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Kitajima, S.; Watanabe, T.; Xu, J.; Zhang, J.; Liu, E.; Chen, Y.E. Rabbit models for the study of human atherosclerosis: From pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015, 146, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Kitajima, S.; Higaki, Y.; Morimoto, M.; Sun, H.; Watanabe, T.; Yamada, N.; Fan, J. High lipoprotein lipase activity increases insulin sensitivity in transgenic rabbits. Metab. Clin. Exp. 2005, 54, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Waqar, A.B.; Koike, T.; Yu, Y.; Inoue, T.; Aoki, T.; Liu, E.; Fan, J. High-fat diet without excess calories induces metabolic disorders and enhances atherosclerosis in rabbits. Atherosclerosis 2010, 213, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ning, B.; Wang, X.; Yu, Y.; Waqar, A.B.; Yu, Q.; Koike, T.; Shiomi, M.; Liu, E.; Wang, Y.; Fan, J. High-fructose and high-fat diet-induced insulin resistance enhances atherosclerosis in watanabe heritable hyperlipidemic rabbits. Nutr. Metab. 2015, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Su, X.; Yu, Q.; Chen, Y.; Guan, H.; Liu, E.; Fan, J. Establishment of a novel nonalcoholic fatty liver disease model using cholesterolfed rabbits with reference to the potential role of endoplasmic reticulum stress. Mol. Med. Rep. 2018, 18, 2898–2904. [Google Scholar] [PubMed]
- Zhang, J.; Niimi, M.; Yang, D.; Liang, J.; Xu, J.; Kimura, T.; Mathew, A.V.; Guo, Y.; Fan, Y.; Zhu, T.; et al. Deficiency of cholesteryl ester transfer protein protects against atherosclerosis in rabbits. Arterioscler. Thromb. Vas. Biol. 2017, 37, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [PubMed]
- Gibbs, R.A.; Weinstock, G.M.; Metzker, M.L.; Muzny, D.M.; Sodergren, E.J.; Scherer, S.; Scott, G.; Steffen, D.; Worley, K.C.; Burch, P.E.; et al. Genome sequence of the brown norway rat yields insights into mammalian evolution. Nature 2004, 428, 493–521. [Google Scholar] [PubMed]
- Carneiro, M.; Albert, F.W.; Afonso, S.; Pereira, R.J.; Burbano, H.; Campos, R.; Melo-Ferreira, J.; Blanco-Aguiar, J.A.; Villafuerte, R.; Nachman, M.W.; et al. The genomic architecture of population divergence between subspecies of the european rabbit. PLoS Genet. 2014, 10, e1003519. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.; Rubin, C.J.; Di Palma, F.; Albert, F.W.; Alfoldi, J.; Barrio, A.M.; Pielberg, G.; Rafati, N.; Sayyab, S.; Turner-Maier, J.; et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 2014, 345, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Li, H.; Li, J.; Niimi, M.; Ding, G.; Chen, H.; Xu, J.; Zhang, H.; Xu, Z.; et al. Hyperlipidemia-associated gene variations and expression patterns revealed by whole-genome and transcriptome sequencing of rabbit models. Sci. Rep. 2016, 6, 26942. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y. Serial inbreeding of rabbits with hereditary hyperlipidemia (whhl-rabbit). Atherosclerosis 1980, 36, 261–268. [Google Scholar] [CrossRef]
- Zhang, B.; Saku, K.; Hirata, K.; Liu, R.; Tateishi, K.; Yamamoto, K.; Arakawa, K. Insulin resistance observed in whhl rabbits. Atherosclerosis 1991, 91, 277–278. [Google Scholar] [CrossRef]
- Shiomi, M.; Kobayashi, T.; Kuniyoshi, N.; Yamada, S.; Ito, T. Myocardial infarction-prone watanabe heritable hyperlipidemic rabbits with mesenteric fat accumulation are a novel animal model for metabolic syndrome. Pathobiology 2012, 79, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, M.; Ito, T. The watanabe heritable hyperlipidemic (whhl) rabbit, its characteristics and history of development: A tribute to the late dr. Yoshio watanabe. Atherosclerosis 2009, 207, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Yokota, M.; Yamamoto, K.; Nakashima, E.; Katsuya, T.; Asano, H.; Isono, M.; Nabika, T.; Sugiyama, T.; Fujioka, A.; et al. Genome-wide association study of coronary artery disease in the japanese. Eur. J. Hum. Genet. 2012, 20, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Weiner, H.; Crabb, D.W. The mutation in the mitochondrial aldehyde dehydrogenase (aldh2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J. Clin. Investig. 1996, 98, 2027–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, L.; Zhang, Y.L.; Zhang, L.; Lu, J.; Guo, S.; Liang, N.; Ge, J.; Zhu, M.; Tao, Y.; et al. Acetaldehyde dehydrogenase 2 interactions with ldlr and ampk regulate foam cell formation. J. Clin. Investig. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Chen, Y.; Yan, H.; Niimi, M.; Wang, Y.; Liang, J. Principles and applications of rabbit models for atherosclerosis research. J. Atheroscler. Thromb. 2018, 25, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xiao, Q.; Bi, J.; Wang, Z.; Li, Y. Rabgtd: A comprehensive database of rabbit genome and transcriptome. Database 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Wuttge, D.M.; Sirsjo, A.; Eriksson, P.; Stemme, S. Gene expression in atherosclerotic lesion of apoe deficient mice. Mol. Med. 2001, 7, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Yechoor, V.; Raja, R.; Li, L.; Chan, L. Microarray gene profiling of laser-captured cells: A new tool to study atherosclerosis in mice. Atherosclerosis 2008, 200, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Schadt, E.E.; Wang, H.; Wang, X.P.; Ingram-Drake, L.; Shi, W.; Drake, T.A.; Lusis, A.J. Identification of pathways for atherosclerosis in mice—Integration of quantitative trait locus analysis and global gene expression data. Circ. Res. 2007, 101, E11–E30. [Google Scholar] [CrossRef] [PubMed]
- Perisic, L.; Hedin, E.; Razuvaev, A.; Lengquist, M.; Osterholm, C.; Folkersen, L.; Gillgren, P.; Paulsson-Berne, G.; Ponten, F.; Odeberg, J.; et al. Profiling of atherosclerotic lesions by gene and tissue microarrays reveals pcsk6 as a novel protease in unstable carotid atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. Collagenases and cracks in the plaque. J. Clin. Investig. 2013, 123, 3201–3203. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Koike, T.; Kitajima, S.; Liu, E.; Morimoto, M.; Shiomi, M.; Hatakeyama, K.; Asada, Y.; Wang, K.Y.; Sasaguri, Y.; et al. Temporal and quantitative analysis of expression of metalloproteinases (mmps) and their endogenous inhibitors in atherosclerotic lesions. Histol. Histopathol. 2008, 23, 1503–1516. [Google Scholar] [PubMed]
- Liang, J.; Liu, E.; Yu, Y.; Kitajima, S.; Koike, T.; Jin, Y.; Morimoto, M.; Hatakeyama, K.; Asada, Y.; Watanabe, T.; et al. Macrophage metalloelastase accelerates the progression of atherosclerosis in transgenic rabbits. Circulation 2006, 113, 1993–2001. [Google Scholar] [CrossRef] [PubMed]
- Sukhova, G.K.; Schonbeck, U.; Rabkin, E.; Schoen, F.J.; Poole, A.R.; Billinghurst, R.C.; Libby, P. Evidence for increased collagenolysis by interstitial collagenases-1 and -3 in vulnerable human atheromatous plaques. Circulation 1999, 99, 2503–2509. [Google Scholar] [CrossRef] [PubMed]
- Quillard, T.; Araujo, H.A.; Franck, G.; Tesmenitsky, Y.; Libby, P. Matrix metalloproteinase-13 predominates over matrix metalloproteinase-8 as the functional interstitial collagenase in mouse atheromata. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Quillard, T.; Tesmenitsky, Y.; Croce, K.; Travers, R.; Shvartz, E.; Koskinas, K.C.; Sukhova, G.K.; Aikawa, E.; Aikawa, M.; Libby, P. Selective inhibition of matrix metalloproteinase-13 increases collagen content of established mouse atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2464–2472. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, V.; O′Byrne, T.K.; Borczuk, A.C.; Okada, Y.; Tall, A.R.; D’Armiento, J. Apoe knockout mice expressing human matrix metalloproteinase-1 in macrophages have less advanced atherosclerosis. J. Clin. Investig. 2001, 107, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Dwivedi, A.; Somerville, M.; George, S.J.; Newby, A.C. Matrix metalloproteinase (mmp)-3 activates mmp-9 mediated vascular smooth muscle cell migration and neointima formation in mice. Arterioscler. Thromb. Vasc. Biol. 2011, 31, e35–e44. [Google Scholar] [CrossRef] [PubMed]
- Galis, Z.S.; Sukhova, G.K.; Lark, M.W.; Libby, P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J. Clin. Investig. 1994, 94, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Loftus, I.M.; Naylor, A.R.; Goodall, S.; Crowther, M.; Jones, L.; Bell, P.R.; Thompson, M.M. Increased matrix metalloproteinase-9 activity in unstable carotid plaques. A potential role in acute plaque disruption. Stroke 2000, 31, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Luttun, A.; Lutgens, E.; Manderveld, A.; Maris, K.; Collen, D.; Carmeliet, P.; Moons, L. Loss of matrix metalloproteinase-9 or matrix metalloproteinase-12 protects apolipoprotein e-deficient mice against atherosclerotic media destruction but differentially affects plaque growth. Circulation 2004, 109, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; George, S.J.; Newby, A.C.; Jackson, C.L. Divergent effects of matrix metalloproteinases 3, 7, 9, and 12 on atherosclerotic plaque stability in mouse brachiocephalic arteries. Proc. Natl. Acad. Sci. USA 2005, 102, 15575–15580. [Google Scholar] [CrossRef] [PubMed]
- Zaltsman, A.B.; George, S.J.; Newby, A.C. Increased secretion of tissue inhibitors of metalloproteinases 1 and 2 from the aortas of cholesterol fed rabbits partially counterbalances increased metalloproteinase activity. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 1700–1707. [Google Scholar] [CrossRef] [PubMed]
- Rouis, M.; Adamy, C.; Duverger, N.; Lesnik, P.; Horellou, P.; Moreau, M.; Emmanuel, F.; Caillaud, J.M.; Laplaud, P.M.; Dachet, C.; et al. Adenovirus-mediated overexpression of tissue inhibitor of metalloproteinase-1 reduces atherosclerotic lesions in apolipoprotein e-deficient mice. Circulation 1999, 100, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, V.; Soloway, P.D.; D’Armiento, J. Increased medial degradation with pseudo-aneurysm formation in apolipoprotein e-knockout mice deficient in tissue inhibitor of metalloproteinases-1. Circulation 2003, 107, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K. Inflammation, atherosclerosis, and coronary artery disease—Reply. N. Engl. J. Med. 2005, 353, 429–430. [Google Scholar]
- Hansson, G.K. Mechanisms of disease—Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Kainuma, M.; Fujimoto, M.; Sekiya, N.; Tsuneyama, K.; Cheng, C.; Takano, Y.; Terasawa, K.; Shimada, Y. Cholesterol-fed rabbit as a unique model of nonalcoholic, nonobese, non-insulin-resistant fatty liver disease with characteristic fibrosis. J. Gastroenterol. 2006, 41, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Li, H.; Wang, K.; Lin, J.; Wang, Q.; Zhao, G.; Jia, W.; Zhang, Q. Analysis of transcriptome and metabolome profiles alterations in fatty liver induced by high-fat diet in rat. Metabolism 2010, 59, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Kleemann, R.; Verschuren, L.; van Erk, M.J.; Nikolsky, Y.; Cnubben, N.H.P.; Verheij, E.R.; Smilde, A.K.; Hendriks, H.F.J.; Zadelaar, S.; Smith, G.J.; et al. Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: A combined transcriptomics and metabolomics analysis. Genome Biol. 2007, 8, R200. [Google Scholar] [CrossRef] [PubMed]
- Kirpich, I.A.; Gobejishvili, L.N.; Homme, M.B.; Waigel, S.; Cave, M.; Arteel, G.; Barve, S.S.; McClain, C.J.; Deaciuc, I.V. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J. Nutr. Biochem. 2011, 22, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Y.; Bai, L.; Zhao, S.; Wang, R.; Liu, B.; Zhang, Y.; Fan, J.; Liu, E. Transcriptomic analysis of the liver of cholesterol-fed rabbits reveals altered hepatic lipid metabolism and inflammatory response. Sci. Rep. 2018, 8, 6437. [Google Scholar] [CrossRef] [PubMed]
- Teufel, A.; Itzel, T.; Erhart, W.; Brosch, M.; Wang, X.Y.; Kim, Y.O.; von Schonfels, W.; Herrmann, A.; Bruckner, S.; Stickel, F.; et al. Comparison of gene expression patterns between mouse models of nonalcoholic fatty liver disease and liver tissues from patients. Gastroenterology 2016, 151, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Loke, S.Y.; Wong, P.T.; Ong, W.Y. Global gene expression changes in the prefrontal cortex of rabbits with hypercholesterolemia and/or hypertension. Neurochem. Int. 2017, 102, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Fillios, L.C.; Mann, G.V. The importance of sex in the variability of the cholesteremic response of rabbits fed cholesterol. Circ. Res. 1956, 4, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Overturf, M.L.; Smith, S.A.; Hewett-Emmett, D.; Loose-Mitchell, D.S.; Soma, M.R.; Gotto, A.M., Jr.; Morrisett, J.D. Development and partial metabolic characterization of a dietary cholesterol-resistant colony of rabbits. J. Lipid Res. 1989, 30, 263–273. [Google Scholar] [PubMed]
- Poorman, J.A.; Buck, R.A.; Smith, S.A.; Overturf, M.L.; Loose-Mitchell, D.S. Bile acid excretion and cholesterol 7 alpha-hydroxylase expression in hypercholesterolemia-resistant rabbits. J. Lipid Res. 1993, 34, 1675–1685. [Google Scholar] [PubMed]
- Li, J.; Yao, Q.; Feng, F.; He, S.; Lin, P.; Yang, L.; Yang, C.; Li, H.; Li, Y. Systematic identification of rabbit lncrnas reveals functional roles in atherosclerosis. Biochim. Biophys. Acta 2018, 1864, 2266–2273. [Google Scholar] [CrossRef] [PubMed]
- Johno, H.; Yoshimura, K.; Mori, Y.; Kimura, T.; Niimi, M.; Yamada, M.; Tanigawa, T.; Fan, J.; Takeda, S. Detection of potential new biomarkers of atherosclerosis by probe electrospray ionization mass spectrometry. Metabolomics 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Bai, L.; Li, Q.; Li, L.; Lin, Y.; Zhao, S.; Wang, W.; Wang, R.; Li, Y.; Yuan, J.; Wang, C.; et al. Plasma high-mannose and complex/hybrid n-glycans are associated with hypercholesterolemia in humans and rabbits. PLoS ONE 2016, 11, e0146982. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, J.; Chen, Y.; Yan, H.; Liu, B.; Wang, Y.; Zhang, J.; Chen, Y.E.; Liu, E.; Liang, J. Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives. Int. J. Mol. Sci. 2018, 19, 3512. https://doi.org/10.3390/ijms19113512
Fan J, Chen Y, Yan H, Liu B, Wang Y, Zhang J, Chen YE, Liu E, Liang J. Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives. International Journal of Molecular Sciences. 2018; 19(11):3512. https://doi.org/10.3390/ijms19113512
Chicago/Turabian StyleFan, Jianglin, Yajie Chen, Haizhao Yan, Baoning Liu, Yanli Wang, Jifeng Zhang, Y. Eugene Chen, Enqi Liu, and Jingyan Liang. 2018. "Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives" International Journal of Molecular Sciences 19, no. 11: 3512. https://doi.org/10.3390/ijms19113512
APA StyleFan, J., Chen, Y., Yan, H., Liu, B., Wang, Y., Zhang, J., Chen, Y. E., Liu, E., & Liang, J. (2018). Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives. International Journal of Molecular Sciences, 19(11), 3512. https://doi.org/10.3390/ijms19113512





