A Mutation in the Mesorhizobium loti oatB Gene Alters the Physicochemical Properties of the Bacterial Cell Wall and Reduces Survival inside Acanthamoeba castellanii
Abstract
1. Introduction
2. Results
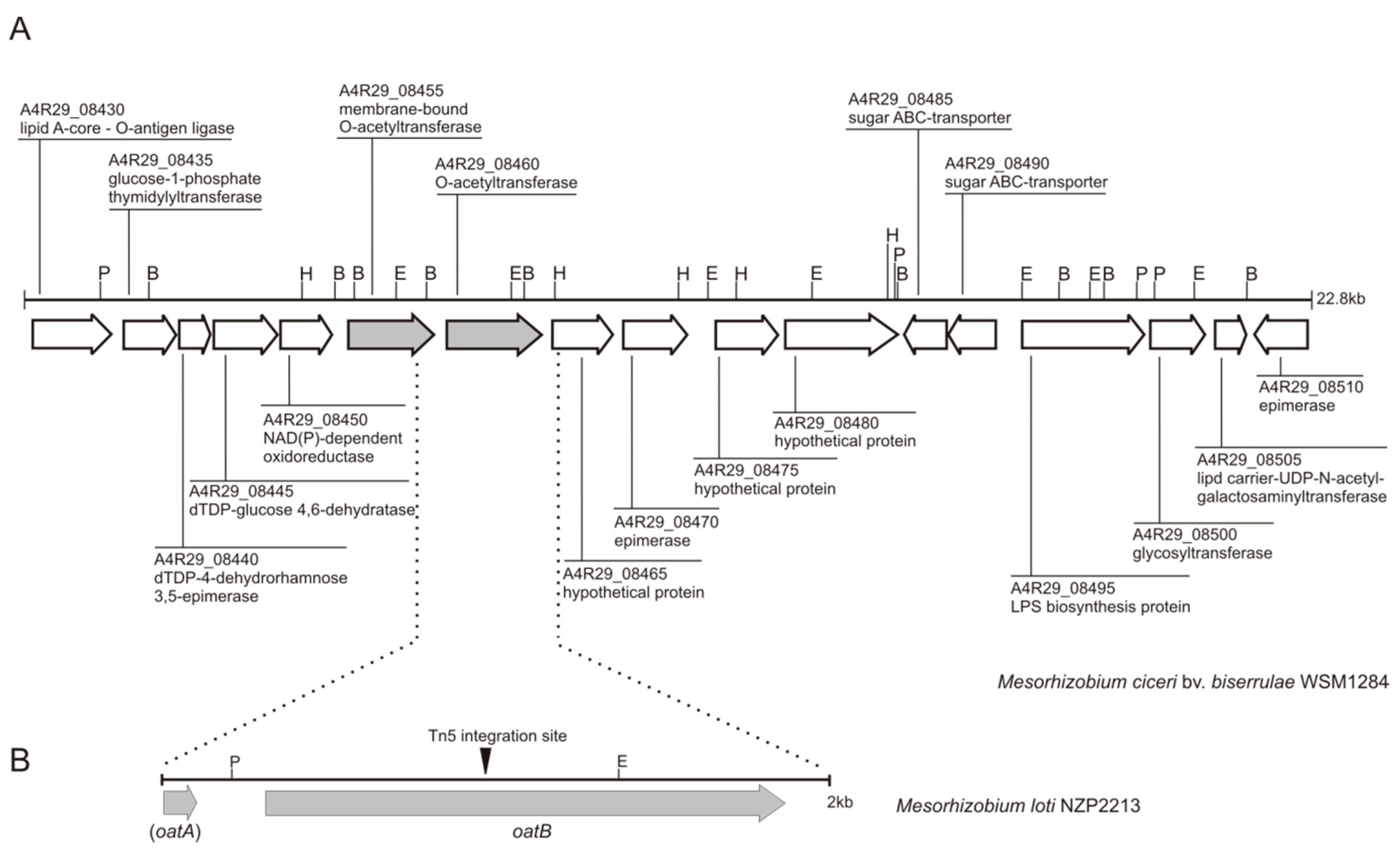
2.1. Clarification of the Localization of the Tn5 Insertion in the NZP2213.1 (oatB) Mutant in the Light of New Data
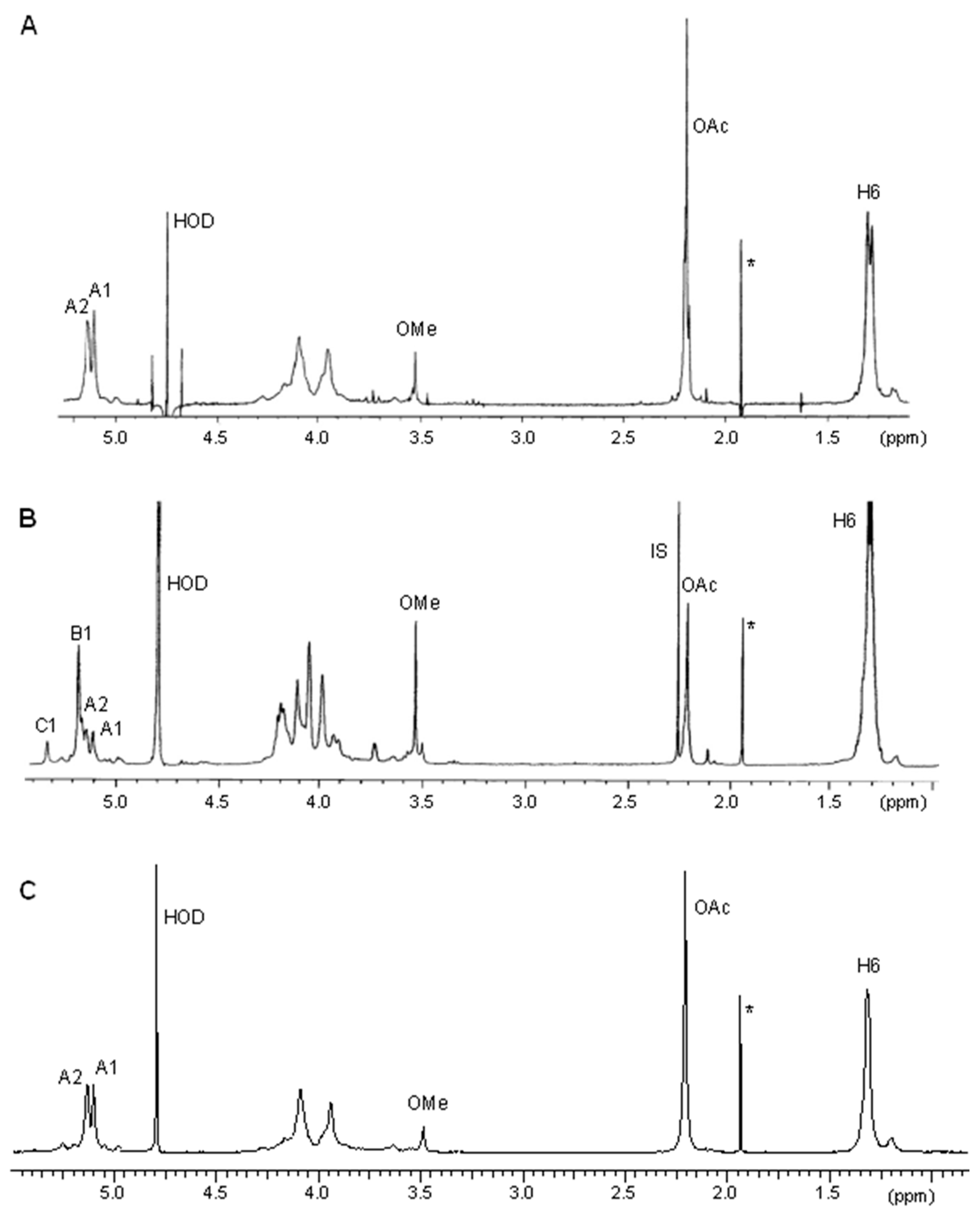
2.2. Restoration of O-acetyl Transferase Function in the Complemented Strain. LPS and O-PS Phenotype
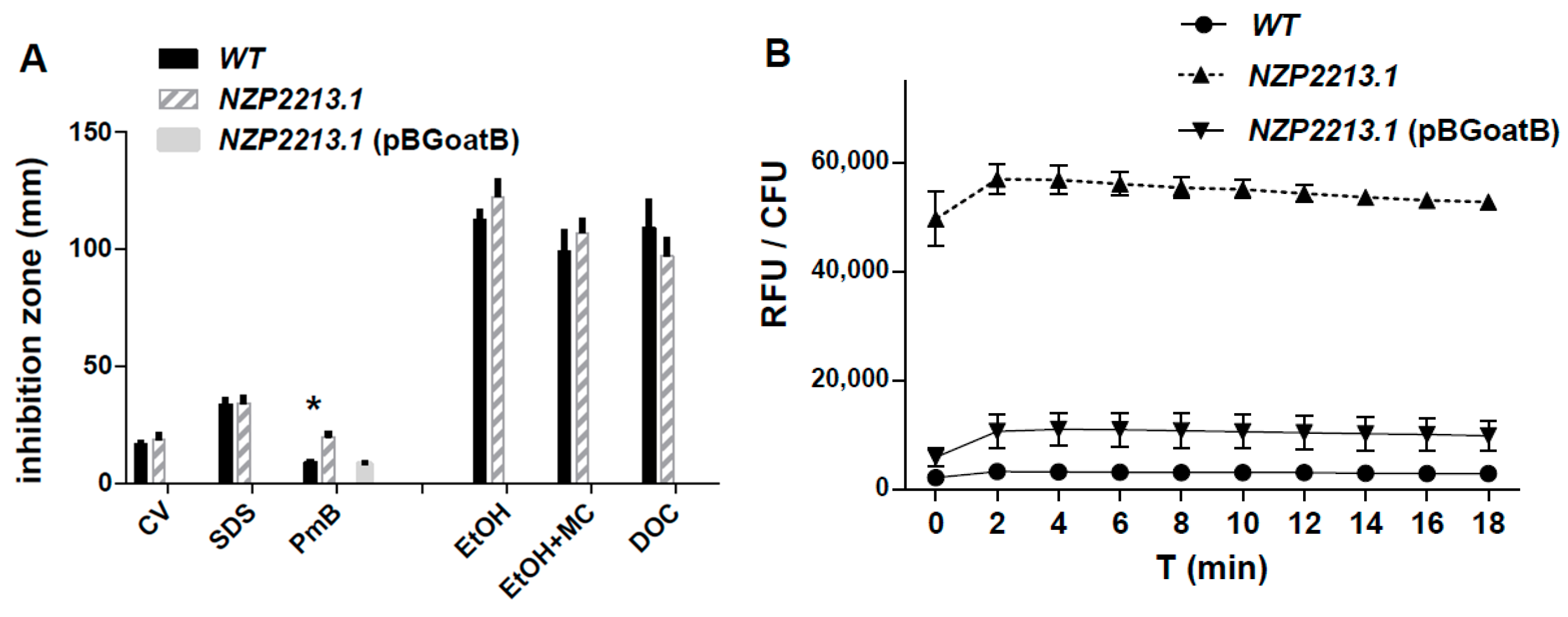
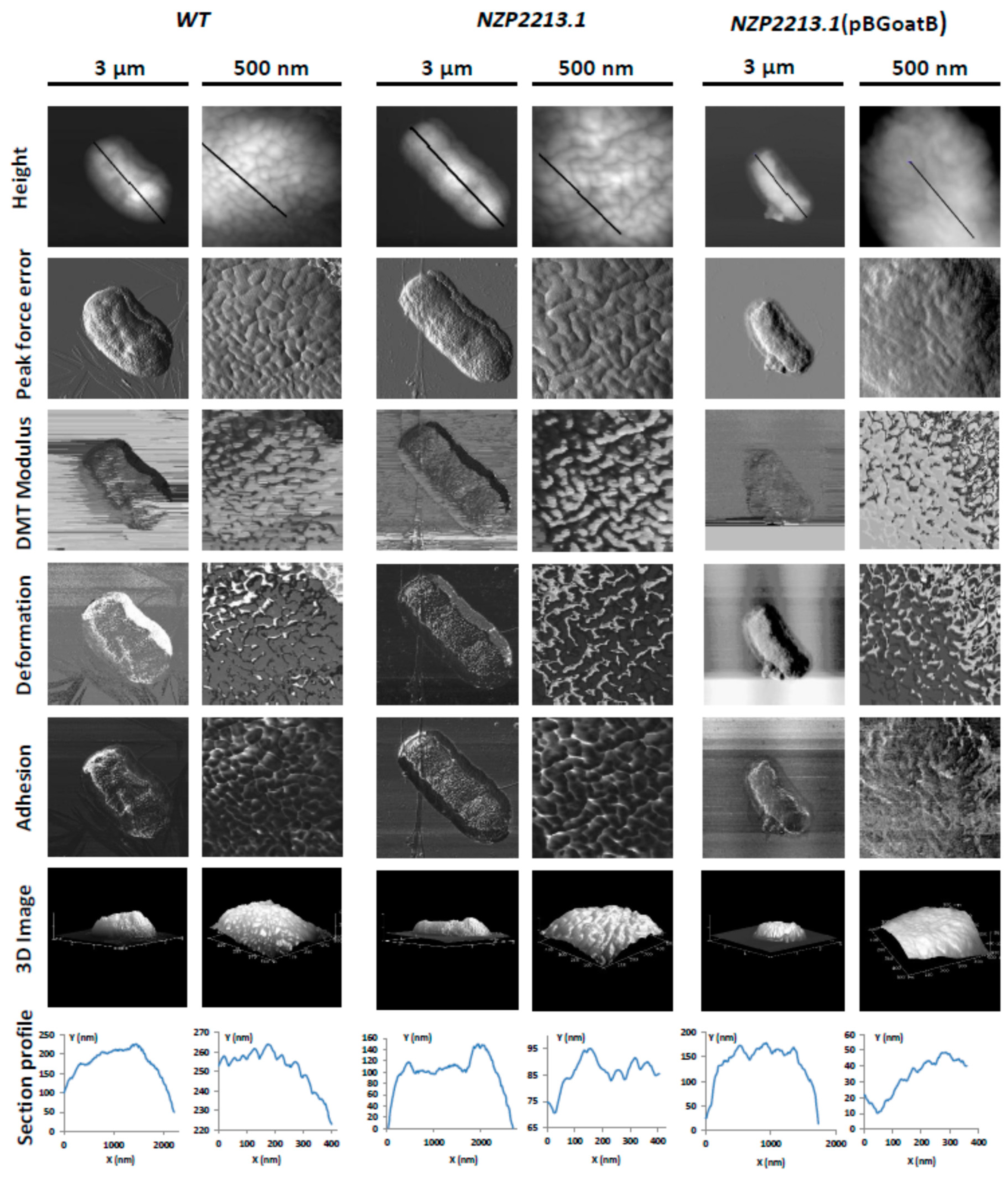
2.3. Physicochemical and Surface Properties of the NZP2213.1 (oatB) Mutant
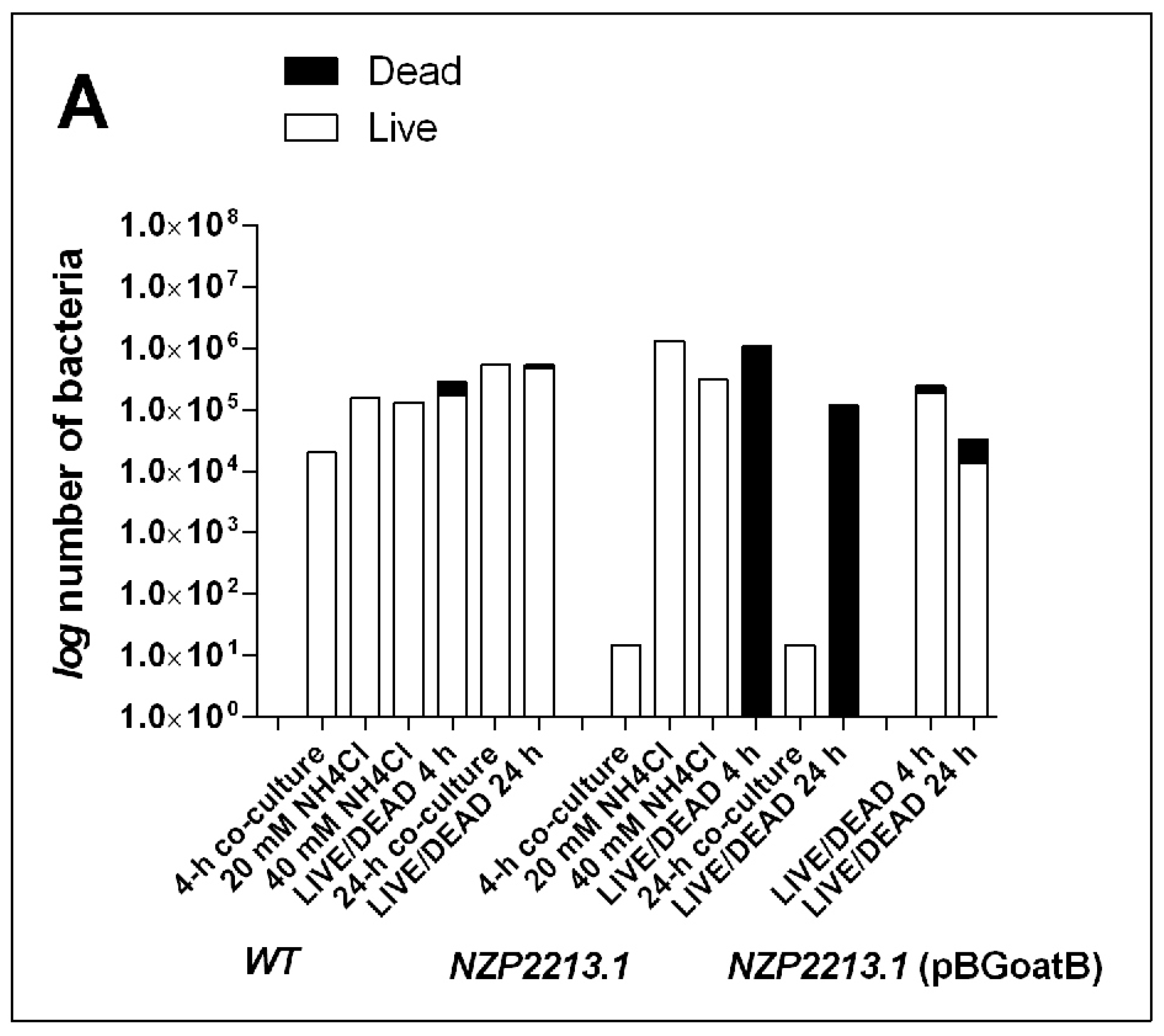
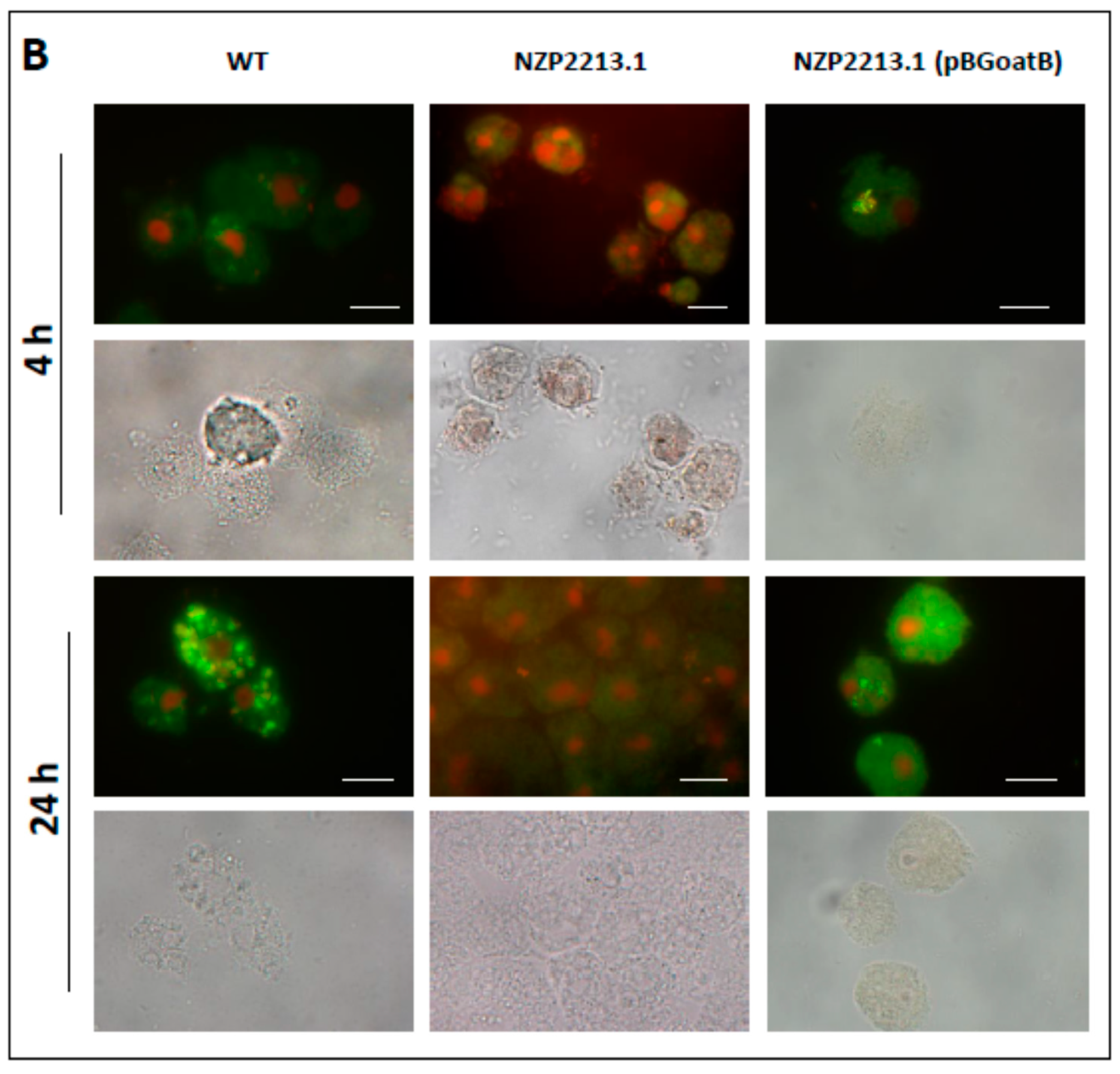
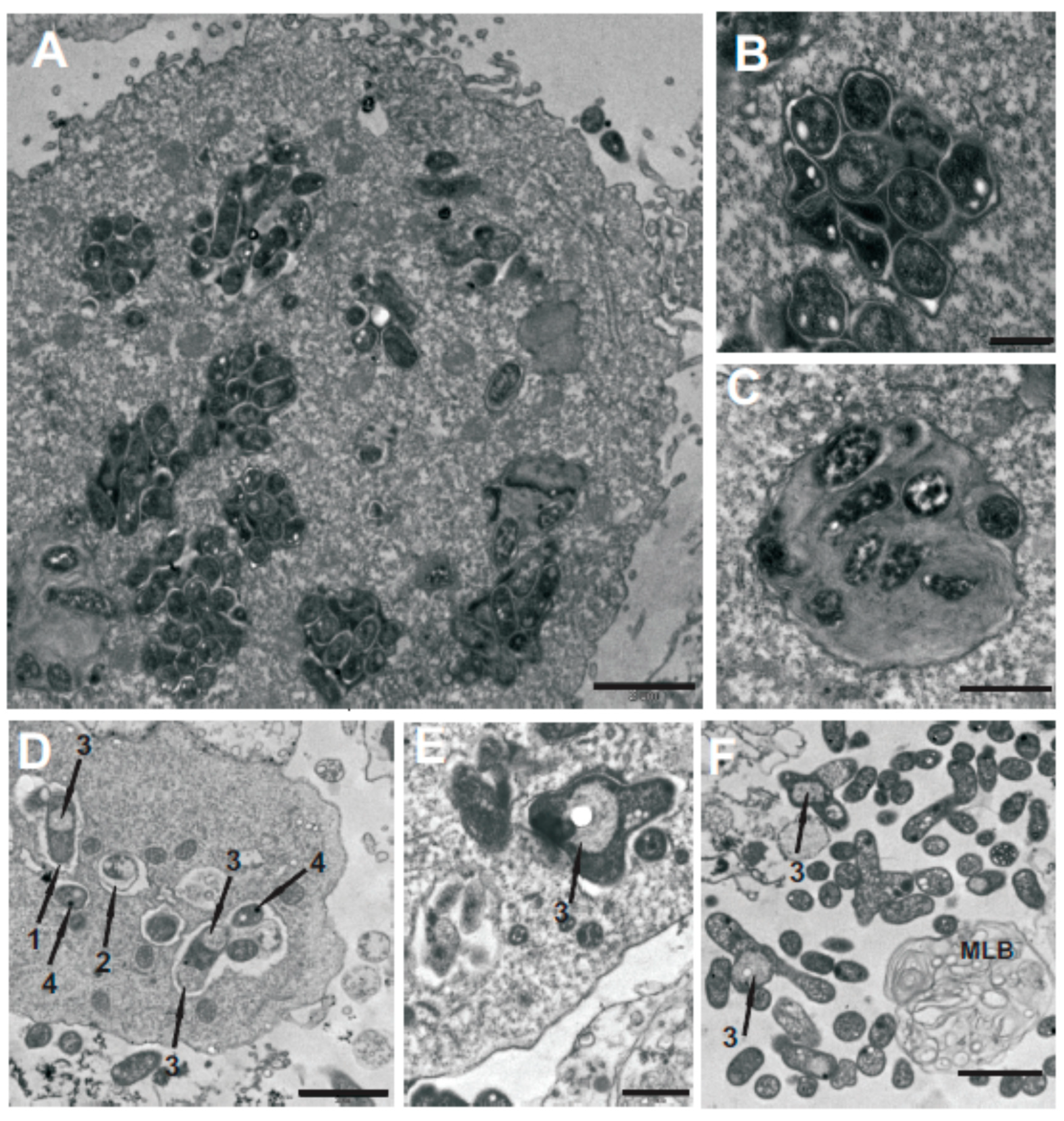
2.4. The Influence of Mutation in the oatB Gene on the Interactions between M. loti and A. castellanii
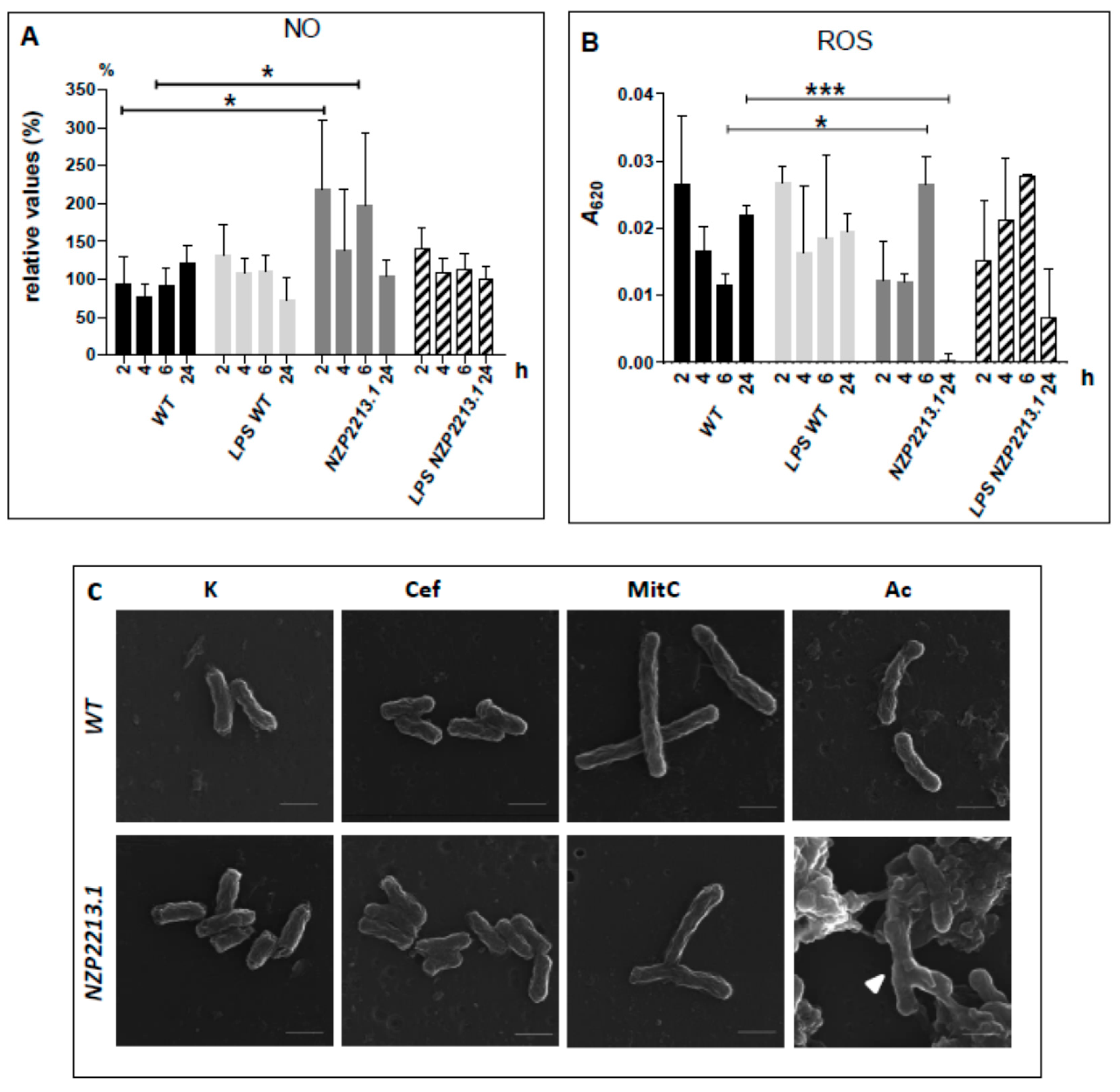
2.5. Bactericidal Mechanism of Clearance of the NZP2213.1 (oatB) Mutant by A. castellanii
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Construction of Plasmid for Complementation
4.3. Lipopolysaccharide Extraction and Purification and the Sugar Composition of LPS and O-PS
4.4. Hydrophobicity Test—Microbial Adhesion to Hydrocarbons (MATH)
4.5. 1-N-Phenyl-1-Naphthylamine Uptake Assay
4.6. Sensitivity of Bacteria to Membrane Disruption Agents, Acidic pH, and Hydrogen Peroxide
4.7. Co-Culture Experiments
4.8. LIVE/DEAD Assay
4.9. Nitrosative and Oxidative Stress Assays
4.10. The Effect of Amoeba Lysate and Antibiotics Treatment on Bacterial Cell Morphology
4.11. 1H NMR Spectroscopy
4.12. Atomic Force Microscopy (AFM)
4.13. Electron Microscopy
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | atomic force microscopy |
| Cef | cephalexin |
| DOC | sodium deoxycholate |
| EtOH | ethanol |
| iNOS | inducible nitric oxide synthase |
| LPS | lipopolysaccharide |
| MCVs | Mesorhizobium-containing vacuoles |
| MES | (N-morpholino)ethanesulfonic acid |
| MitC | mitomycin C |
| MLBs | multilamellar bodies |
| MOI | multiplicity of infection |
| NBT | nitro blue tetrazolium |
| NMR | nuclear magnetic resonance |
| NO | nitric oxide |
| iNOS | inducible nitric oxide synthase |
| NOX | NADPH oxidase |
| NPN | 1-N-phenylnaphthylamine |
| OM | outer membrane |
| O-PS | O-antigen, O-polysaccharide chain |
| PAS | Page’s amoeba saline |
| PBS | phosphate buffer saline |
| PGPR | plant growth promoting bacteria |
| PI | propidium iodide |
| PmB | polymyxin B |
| RFU | relative fluorescent units |
| RMS | root-mean-square roughness |
| RNS | reactive nitrogen species |
| ROS | reactive oxigen species |
| SDS | sodium dodecyl sulfate |
| SEM | scanning electron microscopy |
| TEM | transmission electron microscopy |
| TLRs | Toll-like receptors |
References
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.L.; Krishnamurthy, L. Plant growth promoting rhizobia: Challenges and opportunities. 3 Biotech 2015, 5, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Parnell, J.J.; Berka, R.; Young, H.A.; Sturino, J.M.; Kang, Y.; Barnhart, D.M.; DiLeo, M.V. From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 2016, 7, 1110. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, K.; Bertaux, J.; Krome, K.; Hartman, A.; Scheu, S.; Bonkowski, M. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 2009, 3, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.R.; MacAdam, J.W.; Reeve, J.R. Establishment of birdsfoot trefoil (Lotus corniculatus) pastures on organic dairy farms in the Mountain West USA. Org. Agric. 2014, 5, 63–77. [Google Scholar] [CrossRef]
- Kaplan, M.; Atalay, A.I.; Medjekal, S. Potential nutritive value of wild birdsfoot trefoil (Lotus corniculatus) plants grown in different sites. LRRD 2009, 21, 99. [Google Scholar]
- Chandra, S.; Choure, K.; Dubey, R.C.; Maheshwari, D.K. Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris). Braz. J. Microbiol. 2007, 38, 124–130. [Google Scholar] [CrossRef]
- Turska-Szewczuk, A.; Łotocka, B.; Kutkowska, J.; Król, J.; Urbanik-Sypniewska, T.; Russa, R. The incomplete substitution of lipopolysaccharide with O-chain prevents the establishment of effective symbiosis between Mesorhizobium loti 2213.1 and Lotus corniculatus. Microbiol. Res. 2009, 164, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Karaś, M.A.; Turska-Szewczuk, A.; Trapska, D.; Urbanik-Sypniewska, T. Growth and survival of Mesorhizobium loti inside Acanthamoeba enhanced its ability to develop more nodules on Lotus corniculatus. Microb. Ecol. 2015, 70, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Trent, M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, I.; Vanderleyden, J. O-antigen structural variation: Mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 2002, 26, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Goebel, E.M.; Wolfe, D.N.; Elder, K.; Stibitz, S.; Harvill, E.T. O antigen protects Bordetella parapertussis from complement. Infect. Immun. 2008, 76, 1774–1780. [Google Scholar] [CrossRef] [PubMed]
- Rittig, M.G.; Kaufmann, A.; Robins, A.; Shaw, B.; Sprenger, H.; Gemsa, D.; Foulongne, V.; Rouot, B.; Dornand, J. Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes. J. Leukoc. Biol. 2003, 74, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.C.; McGhee, G.C.; Zhao, Y.; Sundin, G.W. Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora. FEMS Microbiol. Lett. 2009, 291, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, Y.; Shai, Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: Role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 2006, 1758, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Jousse, A. Rhizotrophs: Plant. Growth Promotion to Bioremediation, Microorganisms for Sustainability 2; Springer Nature Singapore Private Limited: Singapore, 2017; pp. 263–273. ISBN 9789811048623. [Google Scholar]
- Kreuzer, K.; Adamczyk, J.; Iijima, M.; Wagner, M.; Scheu, S.; Bonkowski, M. Grazing of a common species of soil protozoa (Acanthamoeba castellanii) affects rhizosphere bacterial community composition and root architecture of rice (Oryza sativa L.). Soil Biol. Biochem. 2006, 38, 1665–1672. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Gomes, K.X.; Cortines, J.R.; Peralta, J.M.; Peralta, R.H. Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol. Res. 2016, 193, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Wildschutte, H.; Wolfe, D.M.; Tamewitz, A.; Lawrence, J.G. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. USA 2004, 11, 10644–10649. [Google Scholar] [CrossRef] [PubMed]
- Turska-Szewczuk, A.; Pietras, H.; Borucki, W.; Russa, R. Alteration of O-specific polysaccharide structure of symbiotically defective Mesorhizobium loti mutant 2213.1 derived from strain NZP2213. Acta Biochim. Pol. 2008, 55, 191–199. [Google Scholar] [PubMed]
- Russa, R.; Urbanik-Sypniewska, T.; Lindström, K.; Mayer, H. Chemical characterization of two lipopolysaccharide species isolated from Rhizobium loti NZP2213. Arch. Microbiol. 1995, 163, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Turska-Szewczuk, A.; Russa, R.; Karaś, M.A.; Danikiewicz, W.; Spólnik, G. Structural elucidation of the outer core tetrasaccharide isolated from the LPS of Rhizobium leguminosarum bv. trifolii strain 24. Carbohydr. Res. 2015, 409, 1–8. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, R.R.L.; Albuquerque, D.A.C.; Cruz, T.G.S.; Yamaji, F.M.; Leite, F.L. Atomic Force Microscopy. Imaging, Measuring and Manipulating Surfaces at the Atomic Scale; InTechOpen: London, UK, 2012; pp. 147–174. ISBN 9535104144. [Google Scholar]
- Abu-Lail, N.I.; Camesano, T.A. The effect of solvent polarity on the molecular surface properties and adhesion of Escherichia coli. Colloids. Surf. B Biointerfaces 2006, 51, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, N.D.; Thiagarajah, J.R.; Verkman, A.S. Chloride concentration in endosomes measured using a ratioable fluorescent Cl- Indicator. Evidence for chloride accumulation during acidification. J. Biol. Chem. 2002, 277, 5506–5513. [Google Scholar] [CrossRef] [PubMed]
- Doyscher, D.; Fieseler, L.; Dons, L.; Loessner, M.J.; Schuppler, M. Acanthamoeba feature a unique backpacking strategy to trap and feed on Listeria monocytogenes and other motile bacteria. Environ. Microbiol. 2013, 15, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Hammer, M.U.; Brauser, A.; Olak, C.; Brezesinski, G.; Goldmann, T.; Gutsmann, T.; Andrä, J. Lipopolysaccharide interaction is decisive for the activity of the antimicrobial peptide NK-2 against Escherichia coli and Proteus mirabilis. Biochem. J. 2010, 427, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Flores-Martin, S.; Fonseca, B.; Otth, C.; Fernandez, H. Mechanisms associated with phagocytosis of Arcobacter butzleri by Acanthamoeba castellanii. Parasitol. Res. 2014, 113, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.M.; Lohan, A.J.; Liu, B.; Lagkouvardos, I.; Roy, S.; Zafar, N.; Bertelli, C.; Schilde, C.; Kianianmomeni, A.; Bürglin, T.R.; et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013, 14, R11. [Google Scholar] [CrossRef] [PubMed]
- Declerck, P.; Behets, J.; de Keersmaecker, B.; Ollevier, F. Receptor-mediated uptake of Legionella pneumophila by Acanthamoeba castellanii and Naegleria lovaniensis. J. Appl. Microbiol. 2007, 103, 2697–2703. [Google Scholar] [CrossRef] [PubMed]
- Akya, A.; Pointon, A.; Thomas, C. Mechanism involved in phagocytosis and killing of Listeria monocytogenes by Acanthamoeba polyphaga. Parasitol. Res. 2009, 105, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.; Chowdhury, I.H.; Szczesny, B.; Wan, X.; Garg, N.J. Macrophages promote oxidative metabolism to drive nitric oxide generation in response to Trypanosoma cruzi. Infect. Immun. 2016, 84, 3527–3541. [Google Scholar] [CrossRef] [PubMed]
- Halablab, M.A.; Bazin, M.; Richards, L.; Pacy, J. Ultra-structure and localisation of formazan formed by human neutrophils and amoebae phagocytosing virulent and avirulent Legionella pneumophila. FEMS Microbiol. Immunol. 1990, 2, 295–301. [Google Scholar] [CrossRef]
- Rosenthal, S.; Reed, E.J.; Weisman, R.A. Effect of lytic enzymes of Acanthamoeba castellanii on bacterial cell walls. J. Bacteriol. 1969, 98, 182–189. [Google Scholar] [PubMed]
- Latch, J.N.; Margolin, W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J. Bacteriol. 1997, 179, 2373–2381. [Google Scholar] [CrossRef] [PubMed]
- McNeil, P.L.; Tanasugarn, L.; Meigs, J.B.; Taylor, D.L. Acidification of phagosomes is initiated before lysosomal enzyme activity is detected. J. Cell Biol. 1983, 97, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Attridge, S.R.; Morona, R. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 2006, 188, 2735–2739. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, L.E.; Chadfield, M.S.; Bispham, J.; Wallis, T.S.; Olsen, J.E.; Ingmer, H. Reduced amounts of LPS affects both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 2003, 288, 225–231. [Google Scholar] [CrossRef]
- Noel, K.D.; Forsberg, L.S.; Carlson, R.W. Varying the abundance of O antigen in Rhizobium etli and its effect on symbiosis with Phaseolus vulgaris. J. Bacteriol. 2000, 182, 5317–5324. [Google Scholar] [CrossRef] [PubMed]
- Alsam, S.; Jeong, S.R.; Sissons, J.; Dudley, R.; Kim, K.S.; Khan, N.A. Escherichia coli interactions with Acanthamoeba: A symbiosis with environmental and clinical implications. J. Med. Microbiol. 2006, 55, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, S.; Varas, M.; Valenzuela, C.; Velozo, P.; Chahin, N.; Aguilera, P.; Santiviago, C.A. Relevant genes linked to virulence are required for Salmonella Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum. Front. Microbiol. 2016, 7, 1305. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmied, P.; Chai, T.; Flury, P.; Blom, J.; Smits, T.H.; Maurhofer, M.; Keel, C. Specific surface glycan decorations enable antimicrobial peptide resistance in plant-beneficial pseudomonads with insect-pathogenic properties. Environ. Microbiol. 2016, 18, 4265–4281. [Google Scholar] [CrossRef] [PubMed]
- Sheu, B.C.; Lin, C.C.; Fu, Y.H.; Lee, S.Y.; Lai, H.C.; Wu, R.S.; Liu, C.H.; Tsai, J.C.; Lin, S. Unraveling the role of the rssC gene of Serratia marcescens by atomic force microscopy. Microsc. Microanal. 2010, 16, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Signo, K.S.; Vanderlinde, E.M.; Yost, C.K.; Dahms, T.E. Atomic force microscopy of a ctpA mutant in Rhizobium leguminosarum reveals surface defects linking CtpA function to biofilm formation. Microbiology 2011, 157, 3049–3058. [Google Scholar] [CrossRef] [PubMed]
- Amro, N.A.; Kotra, L.P.; Wadu-Mesthrige, K.; Bulychev, A.; Mobashery, S.; Liu, G.-Y. High-resolution Atomic Force Microscopy studies of the Escherichia coli outer membrane: structural basis for permeability. Langmuir 2000, 16, 2789–2796. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Prokhorov, N.S.; Shashkov, A.S.; Ovchinnikova, O.G.; Zdorovenko, E.L.; Liu, B.; Kostryukova, E.S.; Larin, A.K.; Golomidova, A.K.; Letarov, A.V. Variations in O-Antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J. Bacteriol. 2015, 197, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.G.; Jeon, H.E.; So, J.S.; Chang, W.S. Effects of the Bradyrhizobium japonicum waaL (rfaL) Gene on hydrophobicity, motility, stress tolerance, and symbiotic relationship with soybeans. Int. J. Mol. Sci. 2015, 16, 16778–16791. [Google Scholar] [CrossRef] [PubMed]
- Hölzer, S.U.; Schlumberger, M.C.; Jäckel, D.; Hensel, M. Effect of the O-antigen length of lipopolysaccharide on the functions of type III secretion systems in Salmonella enterica. Infect. Immun. 2009, 77, 5458–5470. [Google Scholar] [CrossRef] [PubMed]
- Mares, J.; Kumaran, S.; Gobbo, M.; Zerbe, O. Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. J. Biol. Chem. 2009, 284, 11498–11506. [Google Scholar] [CrossRef] [PubMed]
- De Maagd, R.A.; Rao, A.S.; Mulders, I.H.; Goosen-de Roo, L.; Van Loosdrecht, M.C.; Wijffelman, C.A.; Lugtenberg, B.J. Isolation and characterization of mutants of Rhizobium leguminosarum bv. viciae 248 with altered lipopolysaccharides: Possible role of surface charge or hydrophobicity in bacterial release from the infection thread. J. Bacteriol. 1989, 171, 1143–1150. [Google Scholar] [PubMed]
- Kannenberg, E.L.; Carlson, R.W. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharide to become hydrophobic during bacteroid development. Mol. Microbiol. 2001, 39, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.W.; Spacht, D.; Koudelka, G.B. Determinants that govern the recognition and uptake of Escherichia coli O157 : H7 by Acanthamoeba castellanii. Cell Microbiol. 2016, 18, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- March, C.; Cano, V.; Moranta, D.; Llobet, E.; Pérez-Gutiérrez, C.; Tomás, J.M.; Bengoechea, J.A. Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS ONE 2013, 8, e56847. [Google Scholar] [CrossRef] [PubMed]
- Matz, C.; Jürgens, K. Effects of hydrophobic and electrostatic cell surface properties of bacteria on feeding rates of heterotrophic nanoflagellates. Appl. Environ. Microbiol. 2001, 67, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Denoncourt, A.M.; Paquet, V.E.; Charette, S.J. Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 2014, 5, 240. [Google Scholar] [CrossRef] [PubMed]
- De Bagüés, M.P.J.; Terraza, A.; Gross, A.; Dornand, J. Different responses of macrophages to smooth and rough Brucella spp.: Relationship to virulence. Infect. Immun. 2004, 72, 2429–2433. [Google Scholar] [CrossRef]
- Slauch, J.M. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol. 2011, 80, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Jaszek, M.; Janczarek, M.; Kuczyński, K.; Piersiak, T.; Grzywnowicz, K. The response of the Rhizobium leguminosarum bv. trifolii wild type and mutants defective in exopolysaccharide production to oxidative stress. Plant Soil J. 2014, 376, 75–94. [Google Scholar] [CrossRef]
- Paiva, C.N.; Bozza, M.T. Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 2014, 20, 1000–1037. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. A Manual for the Practical Study of Root Nodule Bacteria, 1st ed.; UK Blackwell Scientific Publications Ltd.: Oxford, UK, 1970; pp. 153–159. [Google Scholar]
- Sambrook, J.; Russell, D.W. Preparation and transformation of competent E. coli using calcium chloride. CSH Protoc. 2006, 2006. [Google Scholar] [CrossRef]
- Karaś, M.A.; Russa, R. New long chain bases in lipophosphonoglycan of Acanthamoeba castellanii. Lipids 2013, 48, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering, transposon mutagenesis in gram-negative bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Lindström, K.; Lehtomäki, S. Metabolic properties, maximum growth temperature and phage sensitivity of Rhizobium sp. (Galega) compared with other fast-growing rhizobia. FEMS Microbiol. Lett. 1988, 50, 277–287. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Meth. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Rachwał, K.; Boguszewska, A.; Kopcińska, J.; Karaś, M.; Tchórzewski, M.; Janczarek, M. A regulatory protein encoded by rosR affects Rhizobium leguminosarum bv. trifolii protein profiles, cell surface properties and symbiosis with Trifolium pretense. Front. Microbiol. 2016, 7, 1302. [Google Scholar] [CrossRef]
- Ferguson, G.P.; Roop, R.M.; Walker, G.C. Deficiency of a Sinorhizobium meliloti BacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 2002, 184, 5625–5632. [Google Scholar] [CrossRef] [PubMed]
- Steinert, M.; Birkness, K.; White, E.; Fields, B.; Quinn, F. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphag and survive within cyst walls. App. Environ. Microbiol. 1998, 64, 2256–2261. [Google Scholar]
- Inoue, S.; Itagaki, S.; Amano, F. Intracellular killing of Listeria monocytogenes in the J774.1 macrophage-like cell line and the lipopolysaccharide (LPS)-resistant mutant LPS1916 cell line defective in the generation of reactive oxygen intermediates after LPS treatment. Infect. Immun. 1995, 63, 1876–1886. [Google Scholar] [PubMed]
- Choi, H.S.; Kim, J.W.; Cha, Y.N.; Kim, C. A quantitative nitroblue tetrazolium assay for determining intracellular superoxide anion production in phagocytic cells. J. Immunoass. Immunochem. 2006, 27, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Januszanis, B.; Mak, P.; Cytryńska, M. An atomic force microscopy study of Galleria mellonella apolipophorin III effect on bacteria. Biochim. Biophys. Acta 2011, 1808, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Lesse, A.J.; Campagnari, A.A.; Bittner, W.E.; Apicella, M.A.J. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 1990, 126, 109–117. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Biosca, E.G.; Oliver, J.D.; Amaro, C. Phenotypic characterization of Vibrio vulnificus biotype 2 a lipopolysaccharide- based homogenous serogroup within Vibrio vulnificus. Appl. Environ. Microbiol. 1996, 62, 918–927. [Google Scholar] [PubMed]
| Glycosyl Residue | Molar Ratio a | ||
|---|---|---|---|
| S-LPS from Phenol Phase | |||
| WT | NZP2213.1 | NZP2213.1(pBGoatB) | |
| 2-O-methyl-6-deoxytalose | 0.40 | 1.10 | 0.93 |
| 6-deoxytalose | 15.00 | 7.5 | 13.4 |
| Rhamnose | 1.10 | 1.20 | 1.4 |
| N-acetylquinovosamine | 0.45 | 0.50 | 0.50 |
| Glucose | 1.30 | 1.20 | 2.0 |
| Glucuronic acid | 0.30 | 0.14 | 0.1 |
| Galactose | 1.00 | 0.70 | 0.16 c |
| Galacturonic acid | 0.10 | 0.10 | ND c |
| Glucosamine | 1.00 | 1.00 | 1.0 |
| Heptose | 2.80 | 3.00 | 2.0 |
| KDO b | 0.50 | 0.50 | 0.05 c |
| Bacterial Strains and Plasmids | Description | Reference or Source |
|---|---|---|
| E. colistrains | ||
| S17.1 | 294 derivative RP4-2-Tc::Mu-Km::Tn7 chromosomally integrated | [62] |
| DH5α | supE44ΔlacU169(ϕ80ΔlacZΔM15)hsdR17recA1endA1gyrA96thi-1relA1 | [60] |
| M. lotistrains | ||
| NZP2213(HAMBI1129) | Wild-type Rif R, Nod + Fix + | [63] |
| NZP2213.1 | NZP2213::Tn5Rif R, Km R, Nod+Fix − | [7] |
| NZP2213.1(pBGoatB) | Carrying pBGoatB complementation plasmid | This work |
| Plasmids | ||
| pBBR1MCS-5 | mob, Gm R | [64] |
| pBGoatB | pBBR1MCS-5 with 2005-bp fragment carrying 3′-end of oatA gene, PoatB, and the whole oatB gene, cloned into SmaI in the direction corresponding to Plac | This work |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaś, M.A.; Turska-Szewczuk, A.; Marczak, M.; Jaszek, M.; Janczarek, M.; Dworaczek, K.; Stefaniuk, D.; Wydrych, J. A Mutation in the Mesorhizobium loti oatB Gene Alters the Physicochemical Properties of the Bacterial Cell Wall and Reduces Survival inside Acanthamoeba castellanii. Int. J. Mol. Sci. 2018, 19, 3510. https://doi.org/10.3390/ijms19113510
Karaś MA, Turska-Szewczuk A, Marczak M, Jaszek M, Janczarek M, Dworaczek K, Stefaniuk D, Wydrych J. A Mutation in the Mesorhizobium loti oatB Gene Alters the Physicochemical Properties of the Bacterial Cell Wall and Reduces Survival inside Acanthamoeba castellanii. International Journal of Molecular Sciences. 2018; 19(11):3510. https://doi.org/10.3390/ijms19113510
Chicago/Turabian StyleKaraś, Magdalena Anna, Anna Turska-Szewczuk, Małgorzata Marczak, Magdalena Jaszek, Monika Janczarek, Katarzyna Dworaczek, Dawid Stefaniuk, and Jerzy Wydrych. 2018. "A Mutation in the Mesorhizobium loti oatB Gene Alters the Physicochemical Properties of the Bacterial Cell Wall and Reduces Survival inside Acanthamoeba castellanii" International Journal of Molecular Sciences 19, no. 11: 3510. https://doi.org/10.3390/ijms19113510
APA StyleKaraś, M. A., Turska-Szewczuk, A., Marczak, M., Jaszek, M., Janczarek, M., Dworaczek, K., Stefaniuk, D., & Wydrych, J. (2018). A Mutation in the Mesorhizobium loti oatB Gene Alters the Physicochemical Properties of the Bacterial Cell Wall and Reduces Survival inside Acanthamoeba castellanii. International Journal of Molecular Sciences, 19(11), 3510. https://doi.org/10.3390/ijms19113510






