1. Introduction
In the mid-1990s, nuclear factor-erythroid 2 (EF-E2) p45 subunit-related factor 1 (Nrf1, also called Nfe2L1, with accession NM_001130450 in GenBank, but it herein does not stand for the abbreviation of the nuclear respiratory factor) was identified as a key member of the cap’n’collar (CNC) basic-region leucine zipper (bZIP) family, which comprises Nrf2, Nrf3, Bach1, Bach2, Skn-1 and Cnc, in addition to p45 and Nrf1 [
1,
2,
3,
4]. Within this CNC-bZIP family, Nrf1 and Nrf2 are two principal master regulators of cognate target genes in mammals and are also widely expressed in various distinct tissues. Ever-increasing experimental evidence has demonstrated that Nrf1 exerts its unique functions, which are distinctive from Nrf2 [
5]. This notion has been supported by the fact that Nrf1, but not Nrf2, is indispensable for maintaining cellular homoeostasis and organ integrity during normal development and growth, although both factors are required for diverse adaptive responses to a vast variety of intracellular and environmental stresses, which are also involved in most pathophysiological processes [
6,
7]. It is important to note that the essential function of Nrf1 is finely tuned by a steady-state balance between its production and concomitant processing through both post-transcriptional and post-translational pathways before being turned over. Notably, the processing of the
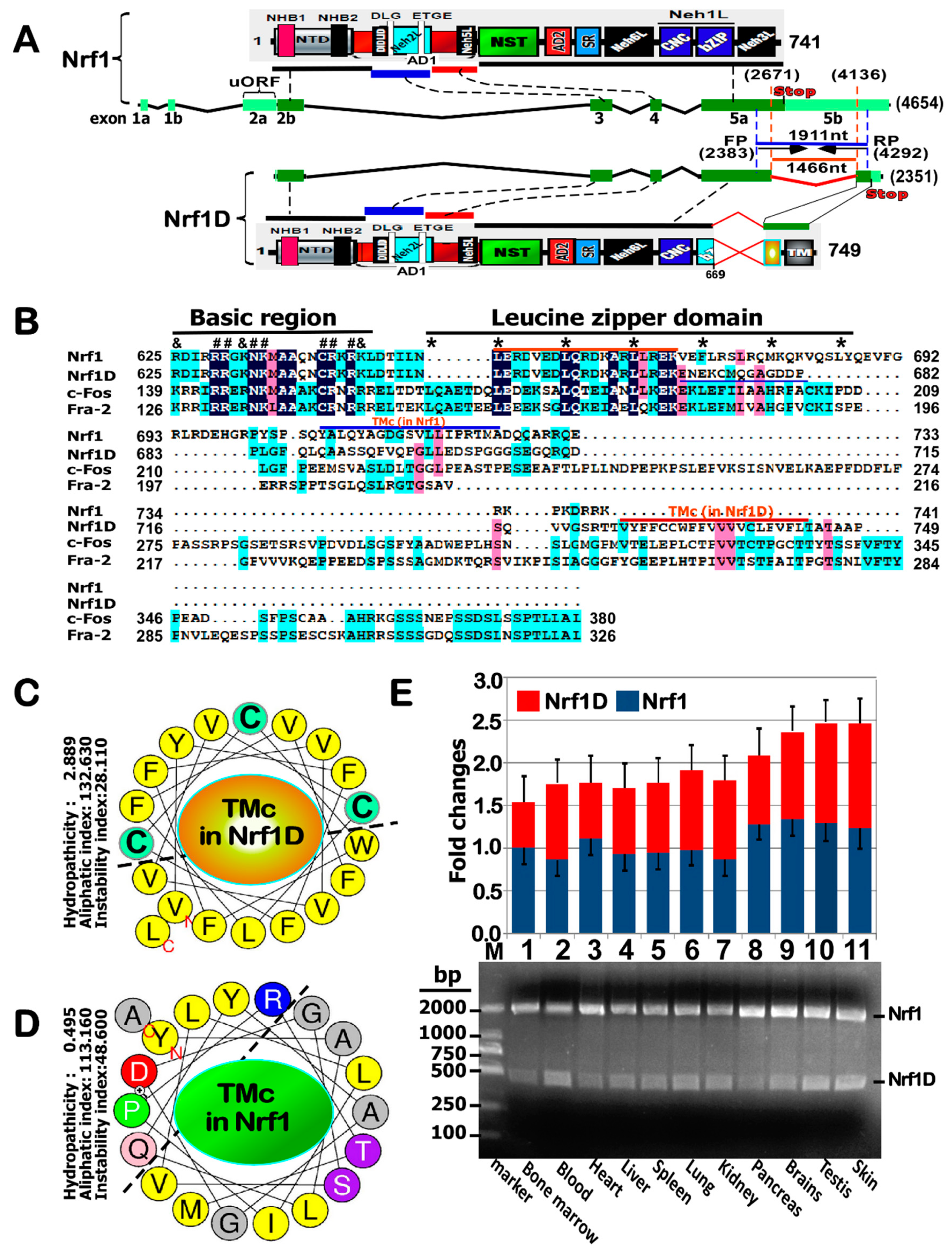
Nfe2l1 gene products ultimately leads to generating various lengths of mRNA transcripts and protein isoforms (with different and even opposing abilities) [
8,
9]. Overall, distinct Nrf1 isoforms are postulated together to confer cytoprotection on the host robust against cellular stress through coordinated regulation of distinct subsets of target genes. Transcriptional expression of these genes, particularly their basal expression, is predominantly driven by Nrf1 through binding to antioxidant response elements (AREs) or other homologous consensus sequences (i.e., AP-1 binding site) in those gene promoter regions.
In early studies using the consensus NF-E2/AP1-binding sites as a probe to clone the cDNA sequence of Nrf1, it was identified to consist of 742 aa in humans [
1] or 741 aa in mice [
10]. Similar cloning strategies were also employed to identify LCR-F1 [
4] and TCF11 [
2] that comprise 447 and 772 aa (with GenBank accession NO. U08853.1 and NM_003204.2), respectively. With the exception of length variations, both the nucleotide and amino acid sequences of LCR-F1 and TCF11 are fully identical with equivalents of Nrf1, and they are thus viewed as different length isoforms [
5]. In fact, the prototypic Nrf1 (i.e., its full-length protein Nrf1α) is generated by translation from alternative splicing of mRNA to remove exon 4 that encodes
242VPSGEDQTALSLEECLRLLEATCPFGENAE
271, called the Neh4L region, from human TCF11 [
2]. Since Neh4L is lost in Nrf1, it was shown to exhibit similar transactivation activity to that of TCF11 [
11], but this long TCF11 is not found in mice [
10]. In addition, the post-synthetic processing of Nrf1/TCF11 may also yield multiple distinct polypeptides of between 140-kDa and 25-kDa, which together determine its overall activity to differentially regulate different target genes [
8,
9,
12]. Further comparison of amino acid sequences demonstrates that LCR-F1 is a shorter form of Nrf1 (i.e., Nrf1β) [
13], which is translated by its in-frame perfect Kozak initiation signal (5′-puCCATGG-3′) that exists around the methionine codons at between positions 289-297 in mice [
1,
2,
10]. Thus, relative to Nrf1α, Nrf1β/LCR-F1 lacks the N-terminal acidic domain 1 (AD1) [
11,
14] and hence exhibits only a weak transactivation activity [
4,
8,
15,
16]. As such, Nrf1β/LCR-F1 activity may also be differentially induced in responses to distinct stressors [
15,
16,
17]. In addition, Nrf1β/LCR-F1 is unstable because it may be rapidly processed to give rise to two small isoforms of 36-kDa Nrf1γ and 25-kDa Nrf1δ [
8,
9]; both may also be generated by additional in-frame translation. Among them, it is important to note that these two small dominant-negative Nrf1γ and Nrf1δ, when over-expressed, have a capability to competitively interfere with a functional assembly of the putative active CNC-bZIP transcription factors, so as to down-regulate expression of NF-E2/ AP1-like ARE-driven genes [
8,
15].
Distinct other isoforms of Nrf1 have been determined to arise from multiple variants of mRNA transcripts, most of which are deposited in GenBank and Ensembl (i.e., ENSMUSG00000038615 and ENSG00000082641, representing mouse and human
Nef2l1 products, respectively). For example, those variants within the 3′- and 5′-untranslated regions were found to yield four different types of mRNA transcripts, that are hence consequently translated into distinct lengths of Nrf1 isoforms with different
trans-acting potentials [
2]. Additional isoforms were also identified to be generated from the alternative splicing of Nrf1 mRNA transcripts [
15,
16]. Among these variants, two that were originally designated as Nrf1 clones Δ767 and D are the most peculiar, with a deletion of its original translational start or stop codons, respectively. Removal of the first translation start codon results in the production of an N-terminally-truncated isoform designated as Nrf1
ΔN [
12,
13], within which the first N-terminal 181-aa region of Nrf1/TCF11 is replaced by an extra dodecylpeptide MGWESRLT- AASA. By striking contrast, the variant clone D of Nrf1 (with accession No. AF071084.1 in Genbank) does not only lack the intact original translation stop codon, but also acquires a substitutive change in the second half of its leucine zipper domain alongside with an extended C-terminal region, which is hence renamed Nrf1D (
Figure 1A). However, both the biochemical characteristics and biological behaviors of Nrf1D have remained elusive to date.
To address the above issues on Nrf1D, we herein examine whether: (i) it has an unique C-terminal transmembrane (TMc) region, which is distinctive from the counterpart of intact Nrf1, involved in the topovectorial process; (ii) the TMc enables this variant to become more sensitive to oxidants and reducing agents, when compared with wild-type protein; and (iii) it is allowed for its putative processing to yield a putative specific isoform that is retained in the cytoplasm or secreted in the blood plasma, although most of this variant exists widely in somatic tissues.
3. Discussion
In the present study, Nrf1D is identified as a candidate secretory transcription factor, with its precursor existing as a unique redox-sensitive transmembrane-bound CNC-bZIP protein detected in several somatic tissues (e.g., heart, lung, liver and testis, as well as bone borrow), before being unleashed into the blood plasma. Collectively, a salient feature of Nrf1D is principally dictated by its C-terminal extra 80-aa region, allowing this variant protein to entail a similar, but different, membrane-topology from that of the prototypic Nrf1 (
Figure 8C,D). In addition to the C-terminal distinction, it should also be noted that Nrf1D shares the longer N-termial 670-aa portion with Nrf1 (
Figure 1). From this, it is inferred that both possess many common biochemical characteristics with some similar biological functions.
To give a better explicit interpretation of Nrf1 topobiology, an integral model (
Figure 8C) is proposed on the basis of ever-accumulating evidence obtained from previous studies by both us [
9,
18,
25,
26,
27] and other groups [
28,
29,
30]. Upon translation of Nrf1, its newly-synthesized polypeptide is targeted and anchored within ER membranes through its NHB1 signal peptide, spinning in an N
cyto/C
lum orientation (i.e., its N-terminal and C-terminal ends face the cytoplasmic and luminal sides, respectively). The NHB1-adjoining portions of Nrf1 are successively translocated into the ER lumen, in which it is
N-glycosylated by oligosaccharyltransferases to yield an inactive glycoprotein, but its DNA-binding (CNC and bZIP) domains are retained on the cytoplasmic side of membranes (
Figure 8C). When required for biological cues, portions of ER luminal-resident transactivation domain (TAD) elements will be dynamically repositioned through p97-fueled retrotranslocation [
31] into extra-ER cyto/nucleoplasmic sides of membrane, whereupon Nrf1 is allowed for deglycosylation by
N-glycosidases (e.g., NGLY1) and ubiquitination by Hrd1. Such modified Nrf1 protein is further subjected to the selective proteolytic processing to remove distinct lengths of its N-terminal portions by cytosolic DDI-1/2 protease-mediated cleavage and/or limited proteolysis by 26S proteasomes or calpains. In turn, transcriptional expression of p97, DDI-1, and proteasomal genes is predominantly regulated by processed mature Nrf1 factor through coupled positive and negative feedback circuits [
26,
27]. In addition, Nrf1 is also likely sorted out of the ER through putative membrane-ferrying to the outer nuclear membranes and then transported in the inner nuclear membrane to gain a direct access to target genes [
25].
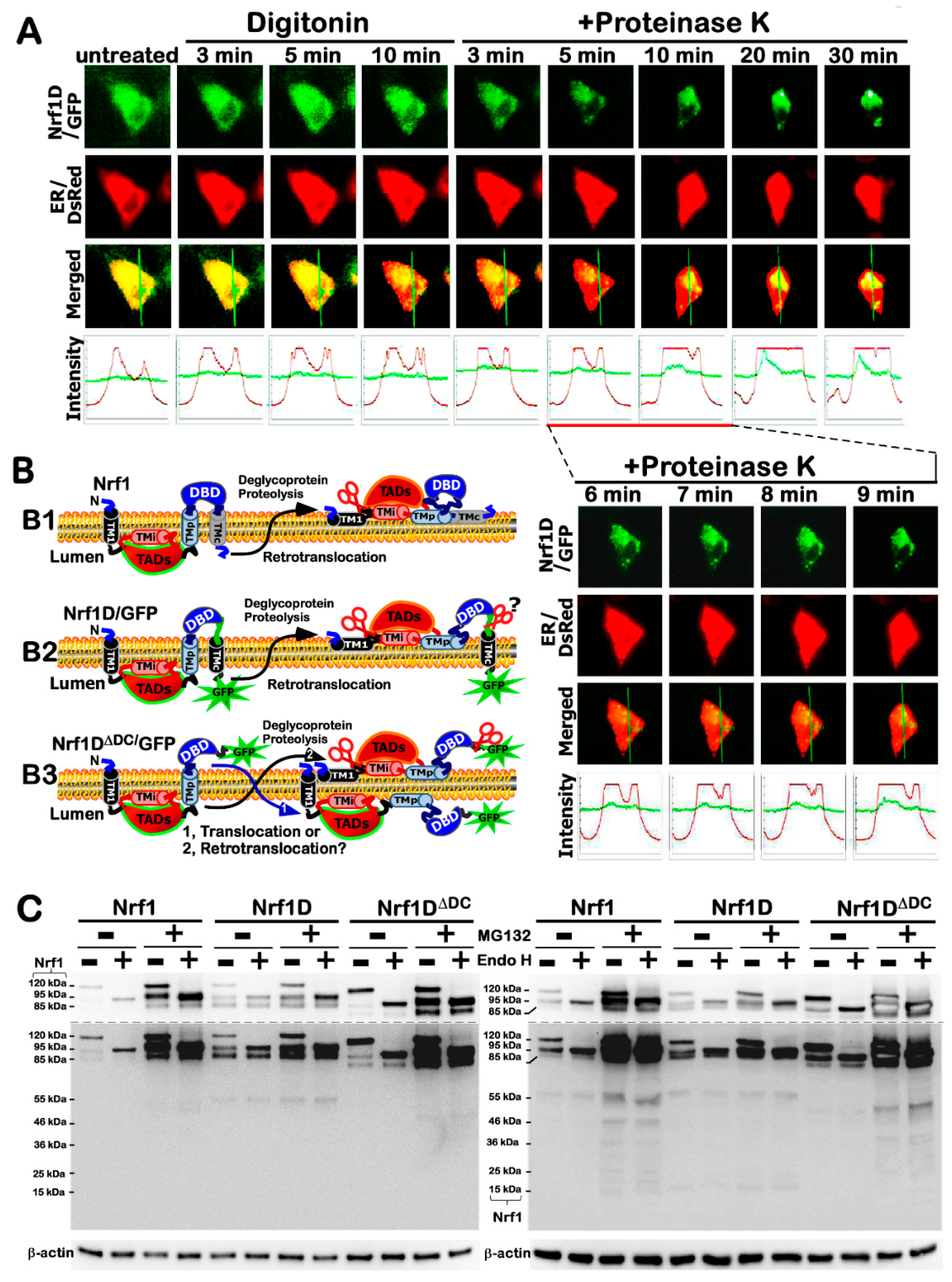
By contrast, Nrf1D is also a moving membrane-protein that entails dynamic topologies, which are much similar to, but slightly distinctive from, the folding of prototypic Nrf1 integrated within and around the ER membranes (
Figure 8D). This distinction is determined by the unique C-terminal 80-aa region of Nrf1D, which only substitutes the C-terminal 72-aa residues of wild-type Nrf1 (
Figure 1A). The C-terminal 80-aa region of Nrf1D is composed of a variant ZIP domain (that is certainly conserved with c-Fos and Fra-2,
Figure 1B) and another integral membrane-spinning TMc stretch (
Figure 1C). The variant bZIP of Nrf1D enables it to make distinctive dimerization from that made by the original bZIP of Nrf1, leading it to exert discrepant activity to mediate transcriptional expression of cognate target genes. However, the transcriptional activity of Nrf1D is tightly monitored by its distinct topovectorial processes, which are also determined by its unique redox- sensitive TMc module (
Figure 8D), in addition to the NHB1 signal sequence, thus enabling it to be topologically folded, post-translationally modified and proteolytically processed in very similar, but different, fashions to account for the prototypic Nrf1 (
Figure 8C).
In a combination with our previous studies of Nrf1 [
9,
18,
25,
26,
27], the evidence that has been presented here reveals that Nrf1D is integrated in dynamic membrane-topologies within and around the ER (
Figure 8D), which are also predominantly determined by both of its N-terminal signal anchor (i.e., NHB1) and its C-terminal redox-sensitive TMc module. Within Nrf1D, its amphipathic NHB1 peptide adapts an N
cyto/C
lum orientation spinning cross ER membranes, whereas the relative hydrophobic TMc peptide is inferable to orientate in an N
cyto/C
lum fashion across membranes. The acidic NST-containing TADs of Nrf1D are
cis-translocated in the ER lumen, allowing it to be N-glycosylated, while its basic aa-enriched CNC and bZIP domains are retained on extra-luminal cyto/nucleoplasmic sides of membranes. Once some parts of the luminal TAD-adjoining portions of Nrf1D are dynamically repartitioned and repositioned by p97-driven retrotranslocation pathway from the ER lumen across membranes into extra-ER side [
26,
27,
31], in which it should be allowed for deglycosylation digestion and further proteolytic processing by similar mechanisms accounting for Nrf1 (for the detailed descriptions, please see the references [
26,
27]). However, in a striking contrast with Nrf1, Nrf1D possesses its specific C-terminal redox-sensitive TMc module that enables this CNC-bZIP protein to exhibit its unique membrane-topological behavior. Thereby, this variant Nrf1D is also specifically processed in an additional way, that are distinctive from the Nrf1 processing. This is also supported by the data of live-cell imaging of Nrf1D (attached C-terminally to the GFP ectope) demonstrates that the local membrane-topology of TMc, though it has a higher hydrophobicity than the amphipathic NHB1 peptide, is still dynamically repositioned out of the ER, such that it is then dynamically dislocated into extra-ER cyto/nucleoplasmic compartments. Once Nrf1D enters these subcellular locations, it loses the membrane protection against digestion by cytosolic proteases (e.g., DDI-1/2, calpains and 26S proteasomes), in order to yield minor 95-kDa and major 85-kDa, along with several smaller isoforms of between 55-kDa and 25-kDa. This is also evidenced by subcellular fractionation revealing that among all Nrf1D isoforms, its full-length 120-kDa and 95-kDa isoforms were recovered in the non-nuclear, rather than nuclear, fractions, while other isoforms of between 85-kDa and 25-kDa were objectively present in both the nuclear and extra-nuclear fractions (
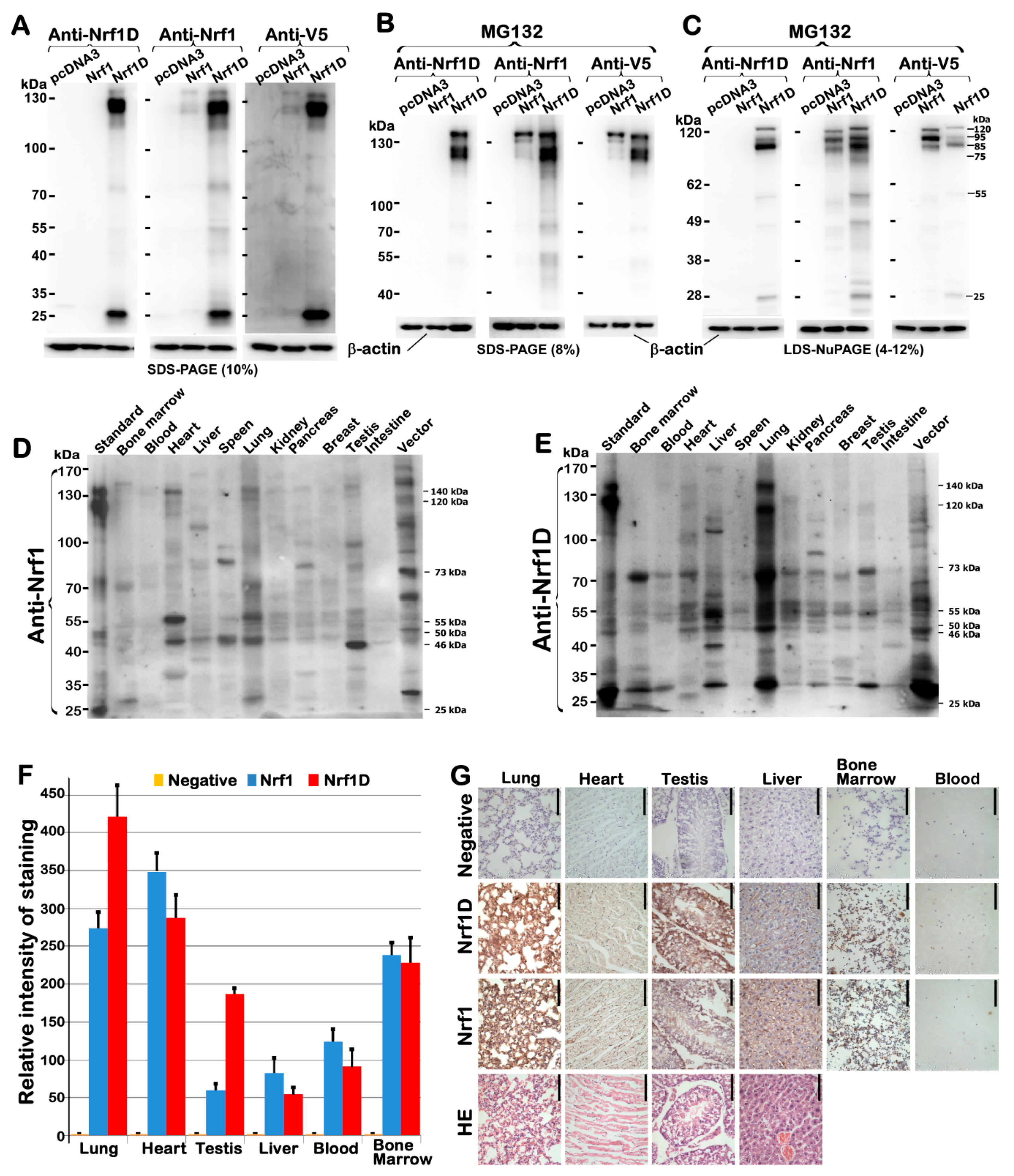
Figure 8A,B).
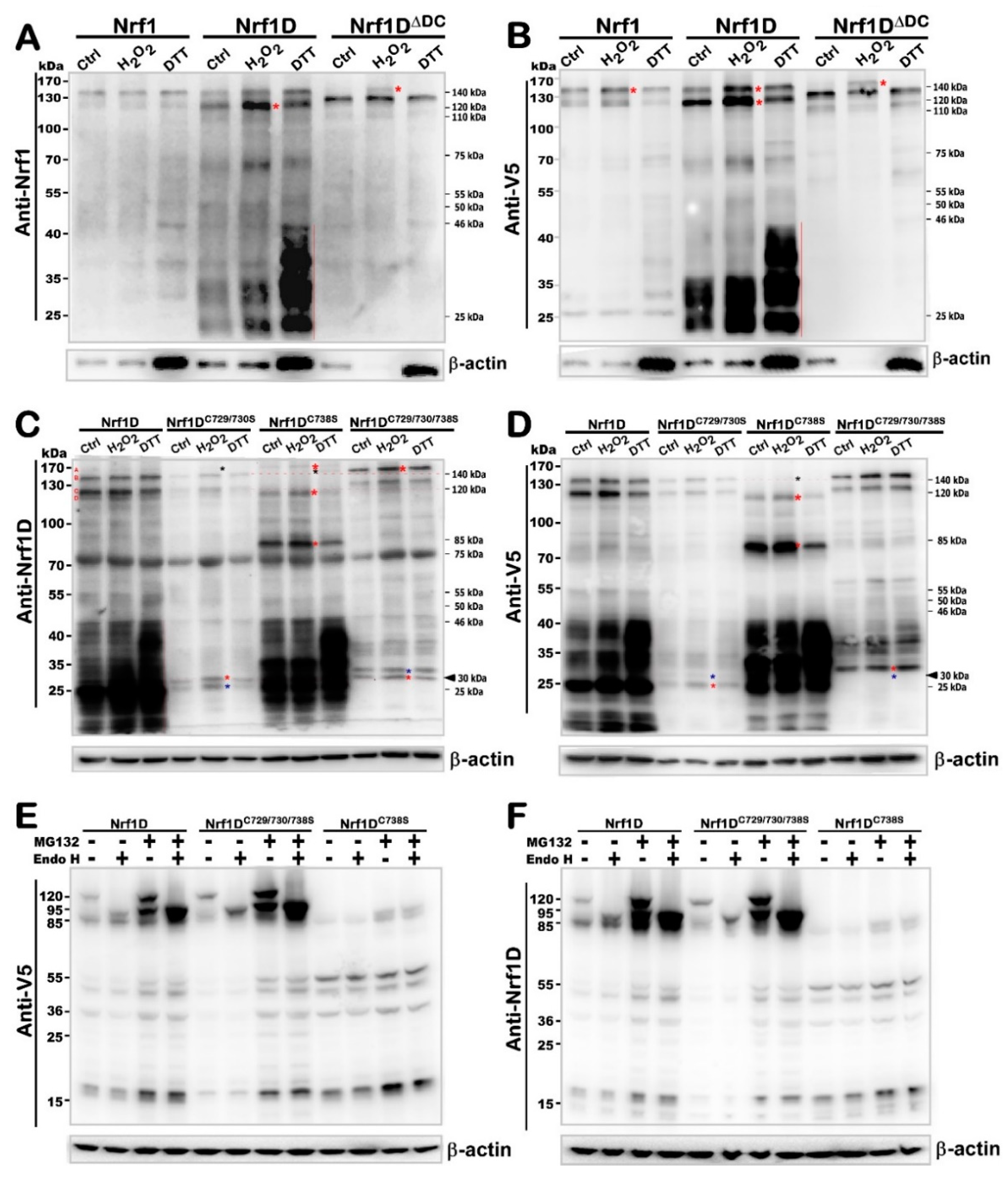
Specifically, further experimental evidence unravels that the repositioning of Nrf1D and its selective proteolytic processing are finely tuned through a strict quality control by the precision thiol-based redox-sensitive module of TMc peptide. For example, the Cys
738-to-Ser mutation of TMc enables it to give rise to an unstable protein of Nrf1D
C738S, which could be allowed for fast dynamic repositioning into the extra-ER sides of membranes and rapidly proteolytic processing insomuch as that the putative glycoprotein and deglycoprotein of this mutant disappear abruptly and is replaced by several processed isoforms distinctive from equivalents arising from Nrf1D (
Figure 6 and
Figure S4). Furthermore, the di-cysteine-to-Ser mutant Nrf1D
C729/730S displays a relative slower electrophoretic mobility of glycoprotein and deglycoprotein, when compared with those of Nrf1D. This being the case, the redox-sensitive TMc of Nrf1D still enables this variant CNC-bZIP factor to be endowed with a less activity than wild-type Nrf1 to mediate ARE-driven reporter gene upon stimulation by redox stressors.
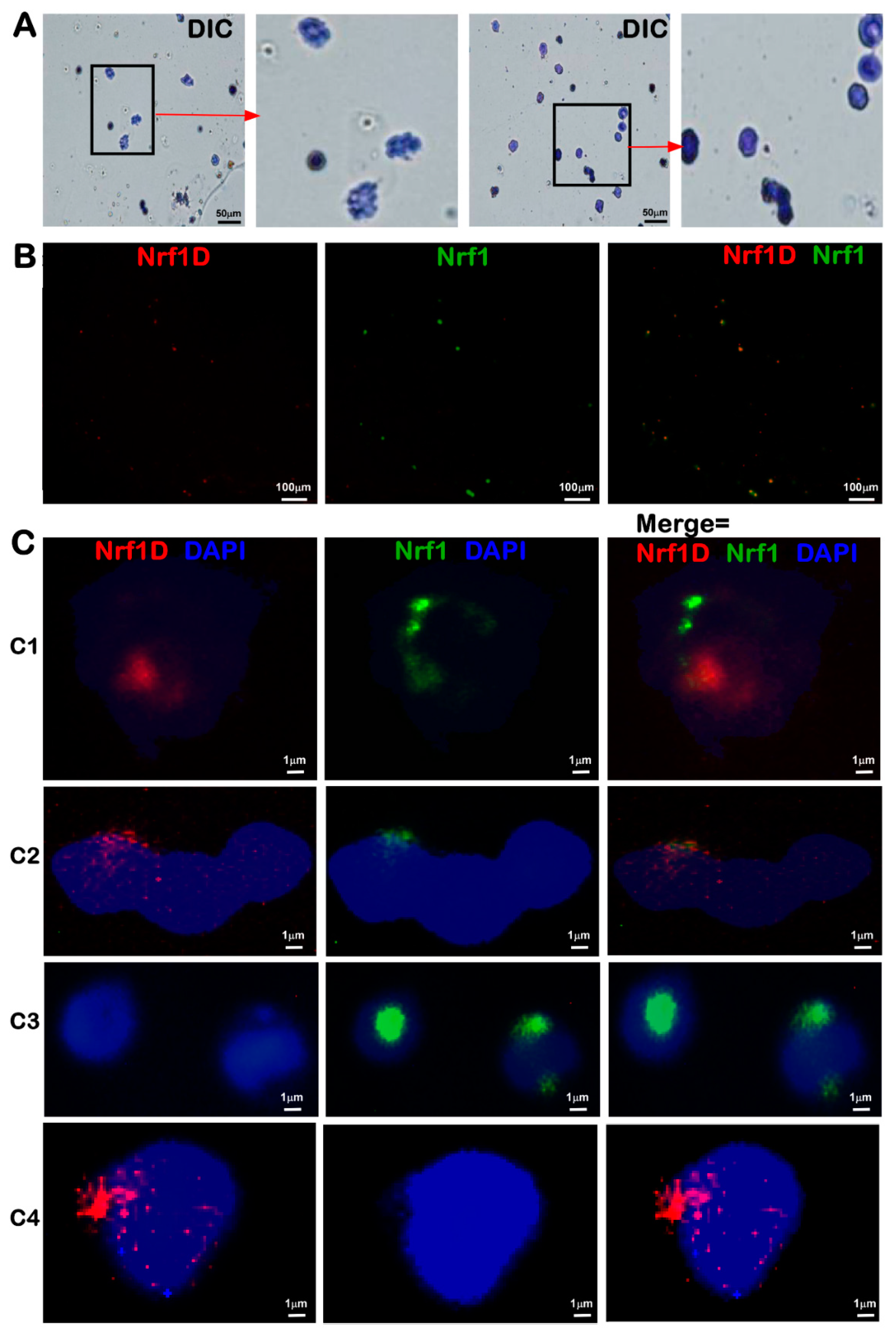
The low activity of Nrf1D is also attributable to a possible restriction of its juxtanuclear location detected in the bone marrow-derived cells (
Figure 3). Importantly, further subcellular fractionation reveals that the low tranactivation activity of Nrf1D is much likely due to the fact that only a small fraction of the mature processed 85-kDa Nrf1D protein was translocated in the nucleus, but the majority of the 85-kDa fraction was still dominantly retained in other extra-nuclear compartments (
Figure 8A,B). This finding therefore raises another possibility; much to our surprise, we discovered that some isoforms of Nrf1D are present in the blood plasma (
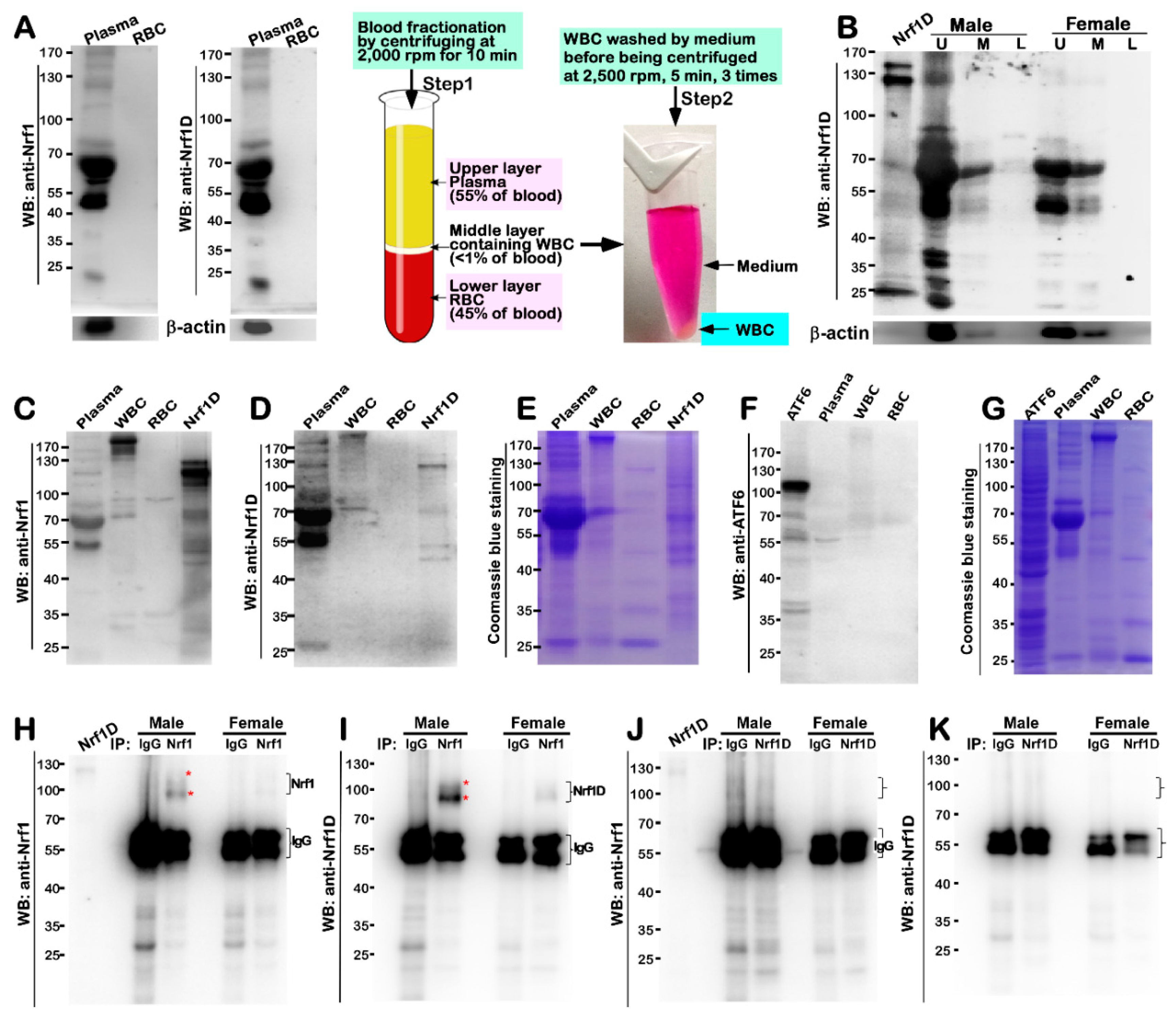
Figure 4), although how it reaches the blood vessel is unclear. This exciting discovery demonstrates that Nrf1D has more than one possibly secretory isoform arising from the proteolytic processing of this variant CNC-bZIP transcription factor, which is released to enter the blood vessels (
Figure 8D), albeit its precursor protein is existing as a unique transmembrane-bound way in hemopoietic and somatic tissues (
Figure 2). However, the detailed topovectorial mechanisms whereby Nrf1D is proteolytically processed and then unleashed from membranes in order to be secreted into the blood plasma remains elusive. This is a major and difficult question to address where the intracellular Nrf1D is released and how it is delivered to the blood plasma. The plasma isoforms of Nrf1D may be derived from its founding mast cells and/or dendritic cells in the blood and other hematopoietic tissues [
15], but the non-hematopoietic tissues cannot also be ruled out.
Apart from the classic secretory pathways, whether Nrf1D may also be postulated to act as an unconventional protein to be secreted into the blood plasma [
32,
33,
34] should be taken into account. If Nrf1D is subjected to an unconventional protein secretory pathway, it will be unleashed from the ER membrane-tethered compartments and then enclosed within exosomes or other extracellular vesicles [
35,
36] before being released into the blood. This speculation also appears to be, at least in part, supported by the evidence that Nrf1D-derived isoforms of between 85-kDa and 25-kDa were stable in the nuclear and extra-nuclear fractions, whereas similar lengths of Nrf1-derived isoforms seemed to be rapidly degraded in the non-nuclear compartments, but this did not occur in the nucleus (
Figure 8A,B). Such nuance indicates that Nrf1D isoforms of from 85-kDa to 25-kDa could be encompassed in a membrane-protected vesicles (e.g., exosomes) that will be secreted into the blood possibly by a not-as-yet identified pathway. As such, the existence of these vehicles involved in potential unconventional protein secretory pathways may also, indeed, partially support the idea of a transcription factor as Nrf1D released in the plasma to reach other peripheral tissues [
32]. Once reaching target cells, the putative Nrf1D-containing vesicles will be internalized by fusion with the recipient plasma membranes so as to generate endosomes and then elicit its functionality in distinct target cells.
Notably, all the Nrf family proteins, such as NF-E2 p45, Nrf1 (with distinct lengths of isoforms Nrf1D, TCF11, LCR-F1/Nrf1β, and Nrf1
ΔN) Nrf2 and Nrf3 were originally identified in the erythroid hematopoietic cells [
1,
2,
3,
4]. Importantly, the reticulocyte unravels a novel pathway of secretion during maturation into erythrocytes (reviewed in ref. [
35]). Relevant multivesicular endosomes are found to associate with some small internal bodies, which are released after fusion of the endosomes with the plasma membrane of target cells. The novel form of putative secretion is the way by which the components of plasma membrane are discarded from maturing reticulocytes. Collectively, the exosome is defined for small membrane vesicles formed by vesiculation of intracellular endosomes and, thereafter, released by exocytosis [
36]. However, it is unknown whether Nrf1D is secreted by exosomes during maturation and enucleation of erythrocytes and also internalized by endosomes in other recipient cells.
Moreover, it is crucial important to note that Nrf1D is highly expressed in the male testis rather than female ovary (
Figure 2D–G and
Figure S5, along with the preprinted version of this paper that had been posted at bioRxiv 342428,
https://doi.org/10.1101/342428), and that the plasma existence of this variant CNC-bZIP transcription factor also implicates a possible gender difference, i.e., principally in male rather than female mice (
Figure 4H,I). This finding thus raises another big open question of whether Nrf1D is secreted along with the release of the cytoplasmic contents during spermatogenesis.
4. Materials and Methods
4.1. Chemicals, Antibodies and Plasmids
All chemicals were of the highest quality commercially available. Among them, proteinase K (PK), Digitonin and Endo H were purchased from New England Biolabs, whereas MG132, CHX, DMSO, VitC, tBHQ, TPA and DTT were obtained from Sigma-Aldrich (St. Louis, MO, USA). Mouse monoclonal antibodies against the V5 epitope was from Invitrogen Ltd (Carlsbad, CA, USA). Of note, two specific antibodies against Nrf1 (aa 192–741) or Nrf1D-specific peptide (nearly at its C-terminus) were produced in rabbits by our laboratory in collaboration with commercial companies. Two expression constructs for Nrf1 and ER/DsRed had been described previously [
14,
25]. Here, Nrf1D was engineered by inserting the XbaI/SacII fragment (encoding aa 1–669 of Nrf1) with a Nrf1D-specific cDNA sequence (encoding its unique aa 670–749, which was synthesized by a commercial company) into the pcDNA3.1/V5-His B vector. Additional 3 cysteine-to-serine mutants Nrf1D
C729/730/738S, Nrf1D
C729/730S and Nrf1D
C738S were created separately. The C-terminally-truncated Nrf1D
ΔDC was also made by only inserting the cDNA sequence encoding the N-terminal 669 aa of Nrf1. Furthermore, other expression constructs for two GFP-fusion proteins (i.e., Nrf1D-GFP and Nrf1D
ΔDC-GFP) were generated by ligating their encoding sequences to the 3′-end of GFP-encoding cDNA into the KpnI/BstBI site of pcDNA3.1/V5-His B vector, respectively. The fidelity of all cDNA products was confirmed by sequencing.
4.2. Organ Collection and Blood Fractionation from Mice Approved in this Study
All of experimental organs and blood samples were collected from 8-weeks-old male or female BALB/c mice. After these mice were narcotized by diethyl ether, the blood samples were obtained from their eyeballs, that were perfused with PBS (phosphate buffer saline) containing 10 mU/mL heparin. The bloods were fractionated by centrifuging at 2000 rpm for 10 min for a collection of upper layer (i.e., blood plasma), middle layer containing white blood cells (WBCs), and lower layer containing red blood cells (RBCs). After WBCs and RBCs were rinsed by a serum-free medium, they were collected by centrifuging at 2500 rpm for 5min (which were repeated for 3 times), prior to being stored at −80 °C for the further use. Thereafter, the mice were sacrificed by being decapitated to obtain relevant tissues and organs, which were harvested and immediately snap-frozen in liquid nitrogen for further analyses. In addition, the bone marrow was also obtained from the tibiae and femurs of these mice by washing with PBS.
All mice used here were maintained under standard animal housing conditions with a 12-h dark cycle and allowed access ad libitum to sterilized water and diet. All relevant studies were carried out on 8-week-old male mice (with the license No. PIL60/13167 approved by Home Office on the 29th July 2011) in accordance with the United Kingdom Animal (Scientific Procedures) Act (1986) and the guidelines of Animal Care and Use Committees of Chongqing University and the Third Military Medical University, both of which were subjected to the local ethical review in China. Of note, collection of mouse organs and blood, along with other relevant experimental procedures were all conducted completely according to the valid ethical regulations that have been approved by the University Laboratory Animal Welfare and Ethics Committee (with the two institutional licenses SCXK-PLA-20120011 and SYXK-PLA-2017002).
4.3. Real-Time qPCR Analysis
Total RNAs were extracted from examined tissues and then reverse-transcripted to yield the first single-stranded cDNAs, which served as temples of real-time quantitative PCR, according to the manufacturer’s protocol (TaKaRa, Dalian, China). To investigate the differential expression of both Nrf1D (Genbank No. AF071084.1) and Nrf1 (Genbank No. NM_008686.3) at their endogenous mRNA levels, a pair of optimal primers complementary to the indicated upstream (5′-GCTGACTTC CTGGACAAGCAGATGA-3′) and downstream (5′-GGAACCAACCACCTGGCTATC-3′) nucleotide sequences were designed and then added in the PCR. The reaction was conducted in the following conditions: Inactivation at 94 °C for 2 min followed by 34 cycles of 10 s at 98 °C denatured, 30 s at 58 °C annealed, and 2 min at 58 °C extended, with an additional extension of 7 min before being stopped. The resulting PCR products were isolated by 1% agarose gel electrophoresis and collected for cDNA sequencing. In addition, expression of β-actin or RPL13A mRNAs was used as an internal control for normalization.
4.4. Fluorescence In Situ Hybridization (FISH)
The bone marrow cells were fixed in 10 mL of 4% paraformaldehyde for 15 min at 4 °C, and pelleted by centrifuging at 1000 rpm for 5 min. After the supernatants were abandoned, the cells were permeabilized for 10 min by 0.1% Triton X-100 at normal temperature, and then collected by centrifuging at 1000 rpm for 5min. The cells were adjusted to an appropriate concentration of 3 × 106/mL of PBS and dropped on glass slides. Subsequently, the slides were immersed in a 2× SSC solution at 37 °C for 5 min, placed in a proper order of 70%, 85% and 100% ethanol at room temperature for 3 min each for dehydration, and then dried naturally. A hybridization solution was preheated at 58 °C for 2 h, and then added to the above slides at a total volume of 10 μL, which contained 1 μL of each probe of Nrf1 (5′-CGCACGATGGCTGACCAGCAGGCTC-3′, which retained within Nrf1 but lacked in Nrf1D, with its 5′-end labeled by 5-FAM, emitting a green fluorescence) and Nrf1D (5′-CGATTGCTTCGAGAAAAGGAAAATGAGAAGTGC-3′, which was labeled by Texas Red at its 5′-end so as to emit a red fluorescence). These probes were allowed to hybridize with indicated samples overnight at 37 °C in a hybridization chamber. The probe-hybridized sample slides were washed in 0.4× SSC plus 0.3% Tween-20 at 65 °C for 3 min, followed by 2× SSC plus 0.1% Tween-20 at room temperature for 30 min, and then stained with Wright-Giemsa and DAPI, respectively. All probes used in this study were synthesized by Invitrogen (Shanghai, China).
4.5. Cell Culture, Transfection, and Reporter Gene Assays
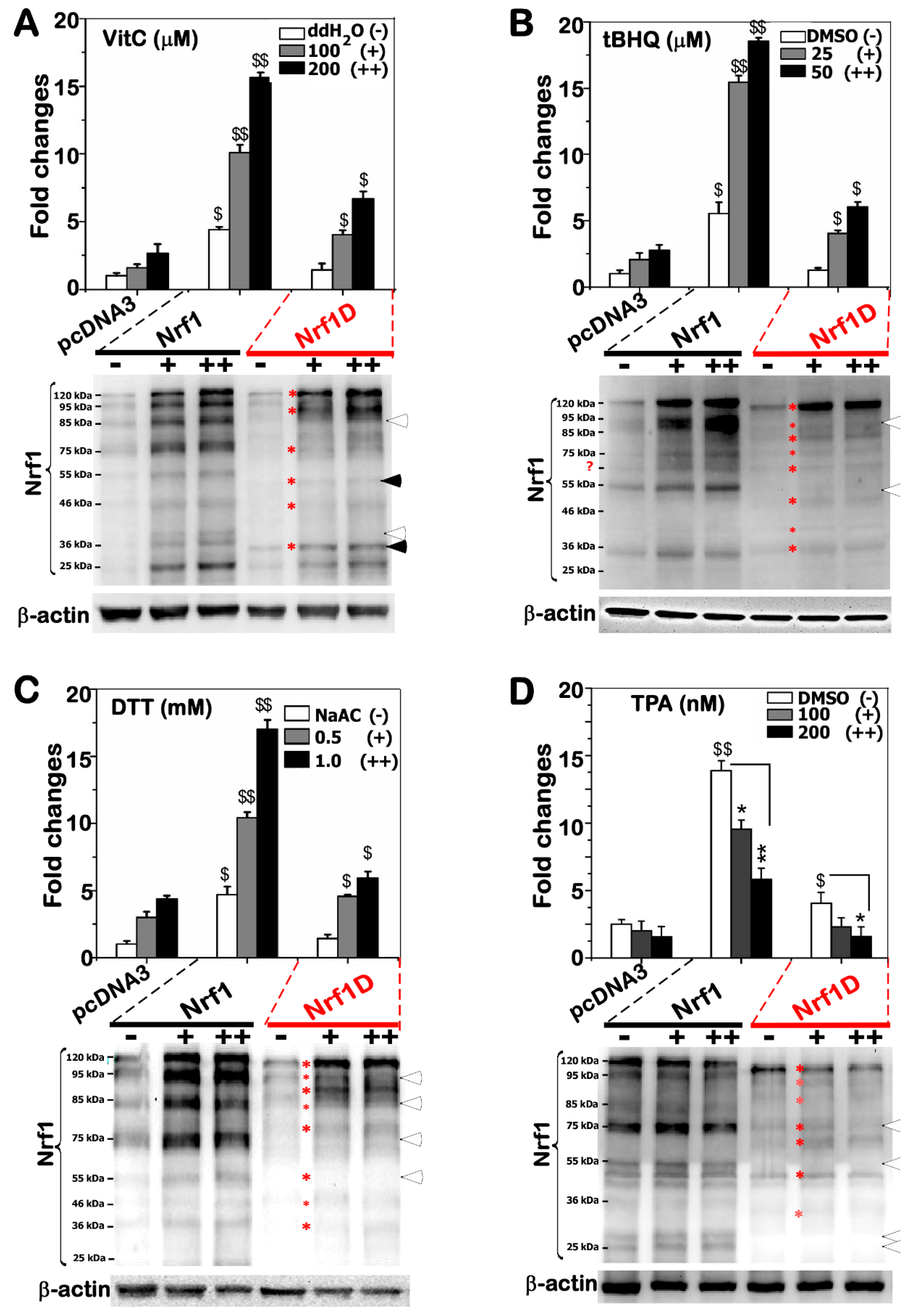
Unless otherwise indicated, monkey kidney COS-1 cells (3 × 105, which had been previously purchased from ATCC and maintained in our laboratory) were seeded in 6-well plates and grown for 24 h in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS). After reaching the 70% confluence, the cells were transfected with a Lipofectamine 3000 (Invitrogen) mixture that contained an expression construct for Nrf1, Nrf1D or mutants, together with two reporters PSV40Nqo1-ARE-Luc and pRL-TK (at a ratio of 10:5:1 of their cDNAs). The latter reporter was used as an internal control for transfection efficiency, whilst an additional mutant version of these reporter genes that lacked ARE sequences served as a negative control. At 24 h after transfection, the cells were further treated by different doses of DTT, tBHQ, VitC or TPA for an additional 24 h. Subsequently, ARE-driven luciferase reporter activity and western blotting were measured approximately 24 h after transfection alone or plus chemical treatments. Basal and stimulated reporter gene activity regulated by Nrf1, Nrf1D or mutants was calculated as a ratio of its values against the background activity (i.e., measured following co-transfection of empty pcDNA3.1/V5 His B vector and ARE-driven reporter in cellular response to indicated vehicle controls). Of note, the basal activity of empty vector was given the value of 1.0, and other data were calculated as fold changes (mean ± S.D.) relative to this value. The data presented each represent at least three independent experiments, each of which were performed in triplicate, followed by statistical analysis of significant differences.
4.6. Live-Cell Imaging Combined with In Vivo Membrane Protease Protection Assays
The live-cell imaging was performed as described by [
8,
9]. COS-1 cells (2 × 10
5) were seeded in 35-mm dishes and cultured overnight. The cells were then co-transfected for 6 h with 3 μg DNA of an expression construct for Nrf1D-GFP or Nrf1D
ΔDC-GFP and 0.5 μg DNA encoding ER/DsRed (as an ER luminal-resident protein marker [
14,
18]). Subsequently, the cells were allowed for a 16-h recovery from transfection, the plasma membranes of COS-1 cells were permeabilized by digitonin (20 μg/mL) for 10 min. Thereafter, the cells were subjected to in vivo membrane protection reactions against digestion by PK (50 μg/mL) for 35 min. During the experiment, live-cell images were acquired every minute on Leica DMI 6000 green and red fluorescence microscopes equipped with a high-sensitivity HAMAMATSH ORCAER camera. Relative fluorescence units were measured by using the Simulator SP5 Multi-Detection system for GFP with 488-nm excitation and 507-nm emission, and for DsRed2 with 570-nm excitation and 650-nm emission.
4.7. Deglycosylation Assay, Western Blotting and Coomassie Brilliant Blue Staining
Experimental cell lysates are subjected to in vitro deglycosylation digestion by Endo H, according to the manufacturer’s instruction (New England Biolabs, Ipswich, MA, USA). Equal amounts of proteins prepared from cell lysates were loaded into each of electrophoretic wells. They were subjected to protein separation by two distinct electrophoresis systems: SDS-PAGE and LDS-NuPAGE gels as described previously [
5,
18], followed by visualization by Western blotting with distinct antibodies against Nrf1, Nrf1D or its C-terminally-tagged V5 ectope. β-Actin served as an internal control to verify amounts of proteins loaded in each well. On some occasions, nitrocellulose membranes that had already been blotted with antibodies were rinsed for 30 min with a stripping buffer before being re-probed with additional primary antibody [
37]. Furthermore, some protein-blotted gels or nitrocellulose membranes were stained by 2.8 μg/mL of Coomassie brilliant Blue R-250 in 10% acetic acid. After the protein bands appeared in 30 min, the gels were de-stained in 10% acetic acid for over 2 h.
4.8. Immunoprecipitation and Immunohistochemistry
Blood samples were extracted in RIPA lysis buffer (Beyotime, China) and then clarified by pre-incubation with the pre-immunized serum, before being subjected to immunoprecipitation by incubation with specific antibodies against Nrf1 or Nrf1D at 4 °C overnight, along with Protein-G Sepharose 4 Fast Flow beads. The immunocomplexes-associated Protein-G Sepharose beads were washed 3 times with PBS. The resulting immunocomplexes with Nrf1 or Nrf1D antibodies were visualized by immunoblotting with the indicated antibodies. For immunohistochemistry analysis, distinct tissues were incubated with the primary antibodies against Nrf1 or Nrf1D and the secondary anti-IgG (Sangon Biotech, Shanghai, China), according to the instructions of manufacturer. The ‘negative’ images were obtained from pre-incubation of pre-immunize serum instead of the primary antibody in the immunohistochemistry.
4.9. Subcellular Fractionation
About COS-1 or HepG2 cells (3 × 105) were seeded in 6-well plates and allowed for growth for 24 h in DMEM containing 10% FBS. After reaching the 70% confluence, the cells were transfected with an expression construct for Nrf1 and Nrf1D, or empty pcDNA3.1 vector. Following 24-h recovery from transfection, the cells were harvested and then subjected to incubation with ice cold Nuclei EZ lysis buffer (0.5 mL added to each dish). The nuclei-containing pellets were collected by centrifuging at 500× g for 5 min at 4 °C. The supernatant were considered as the non-nuclear fractions, while the sediment nuclear fractions were saved. Subsequently, the sediment was re-suspended by adding 0.5 mL of the nuclei EZ lysis buffer (From Sigma-Aldrich, Saint Louis, MO, USA) and vortexing at moderate to high speeds. After being washed with the lysis buffer for 3 times, the clarified nuclei pellets were collected by centrifuging as described above. The resulting nuclear and non-nuclear fractions were diluted with RIPA lysis buffer (Beyotime, Shanghai, China), and determined by western blotting with distinct antibodies against Nrf1, Nrf1D-specific peptides or their C-terminal V5 tag.
4.10. Statistical Analysis
The significant differences in changes of Nrf1- or Nrf1D-mediated reporter gene activity, and the abundances of their transcript and protein expressed in different tissues were determined using the Student’s t-test or Multiple Analysis of Variations (MANOVA). The data are herein shown as a fold change (mean ± S.D.), each of which represents at least 3 independent experiments undertaken on separate occasions that were each performed in triplicate (n = 9).