A High-Resolution Proteomic Landscaping of Primary Human Dental Stem Cells: Identification of SHED- and PDLSC-Specific Biomarkers
Abstract
:1. Introduction
2. Results
2.1. In-Depth and High-Scale Proteomic Mapping of SHEDs and PDLSCs
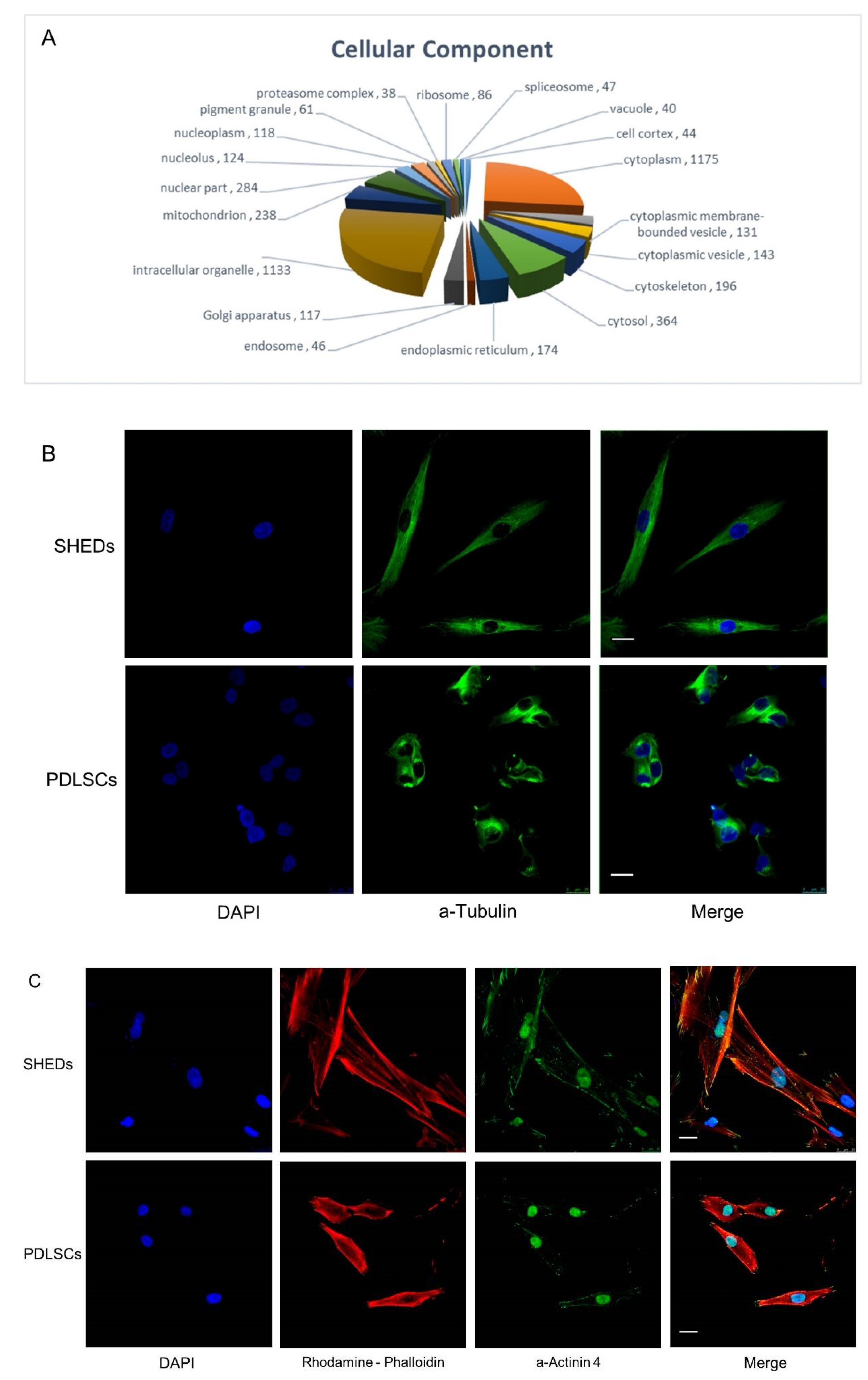
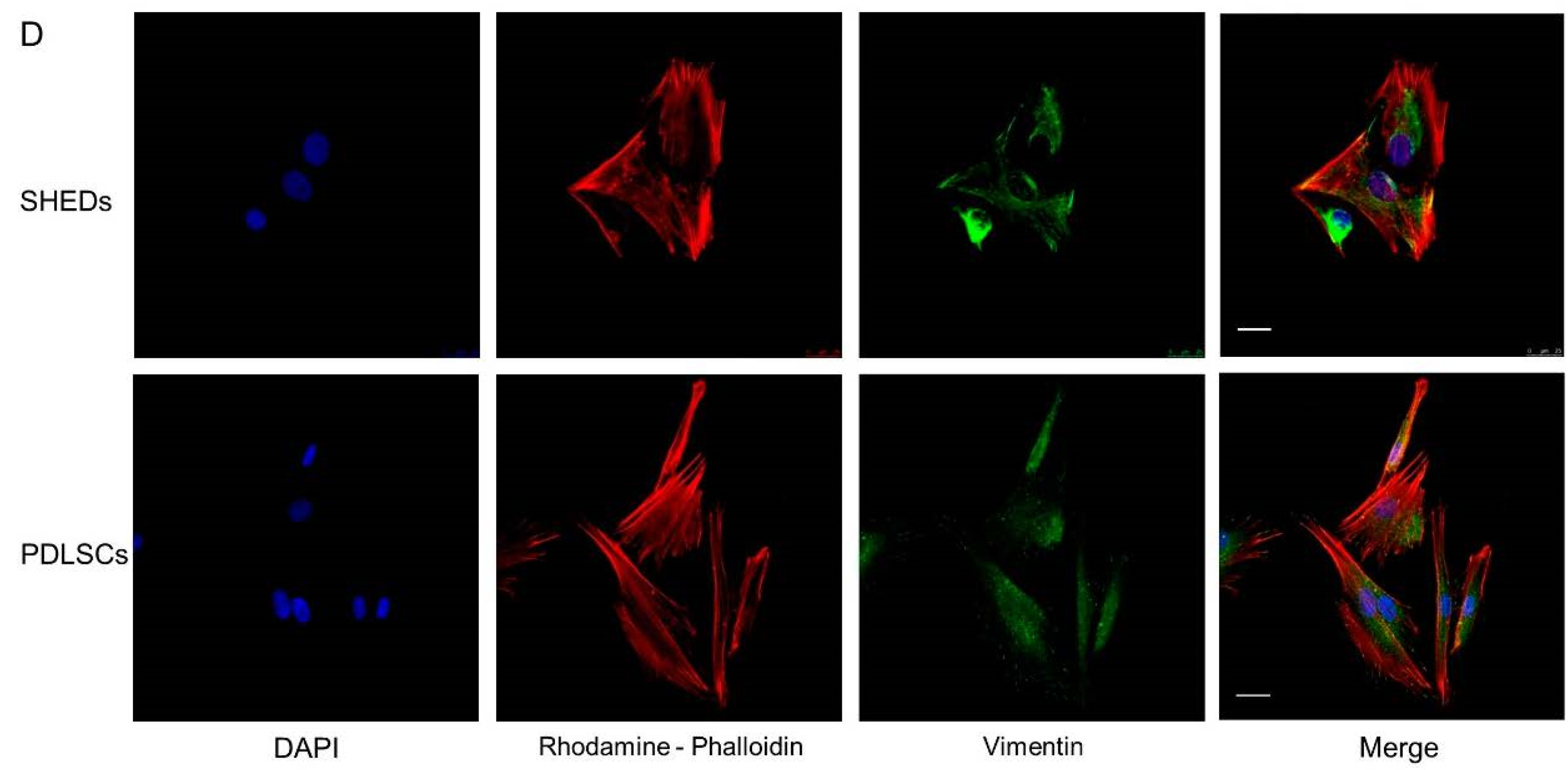
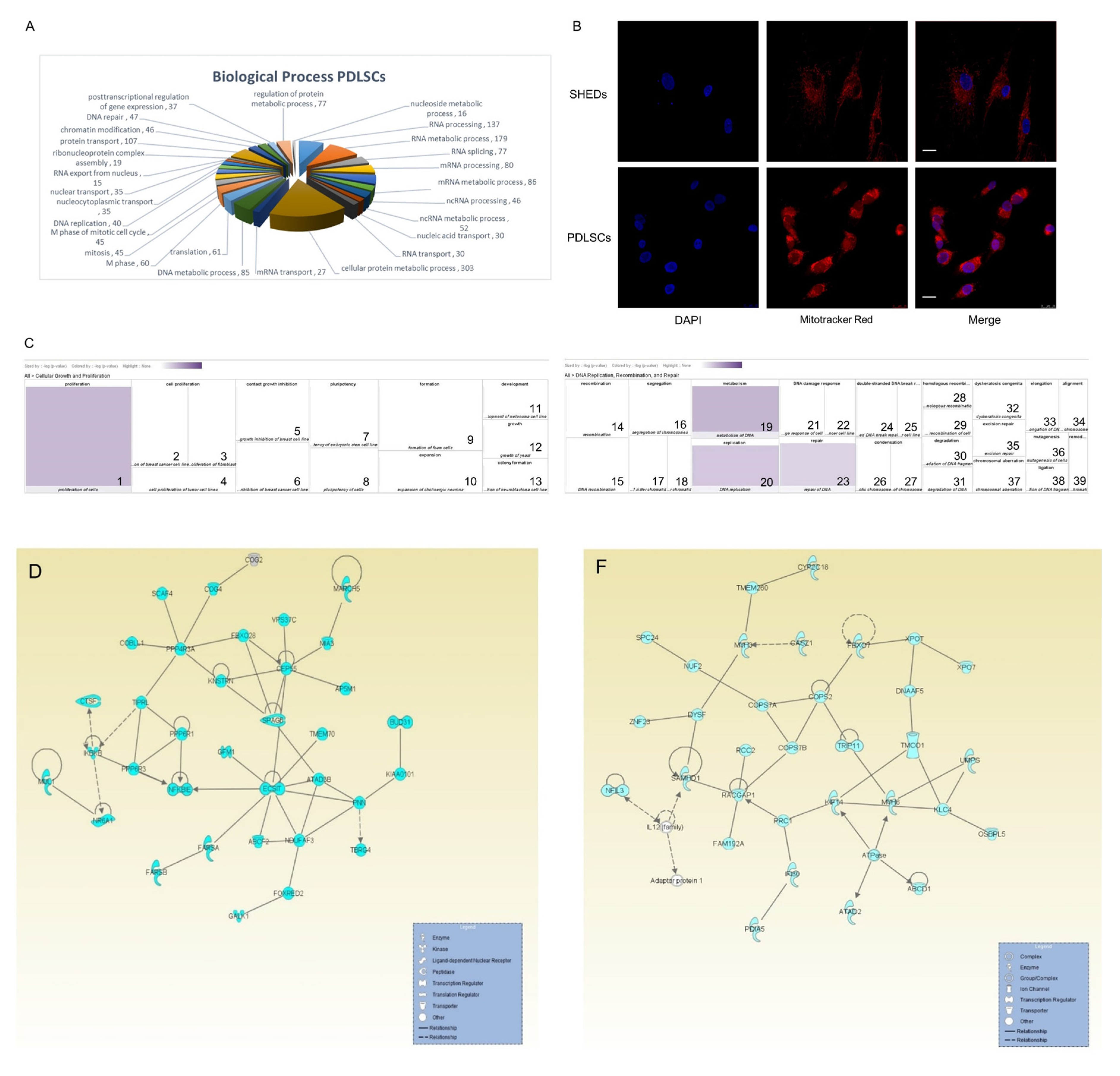
2.2. Sub-Cellular Localization of Commonly Identified Proteins in SHEDs and PDLSCs
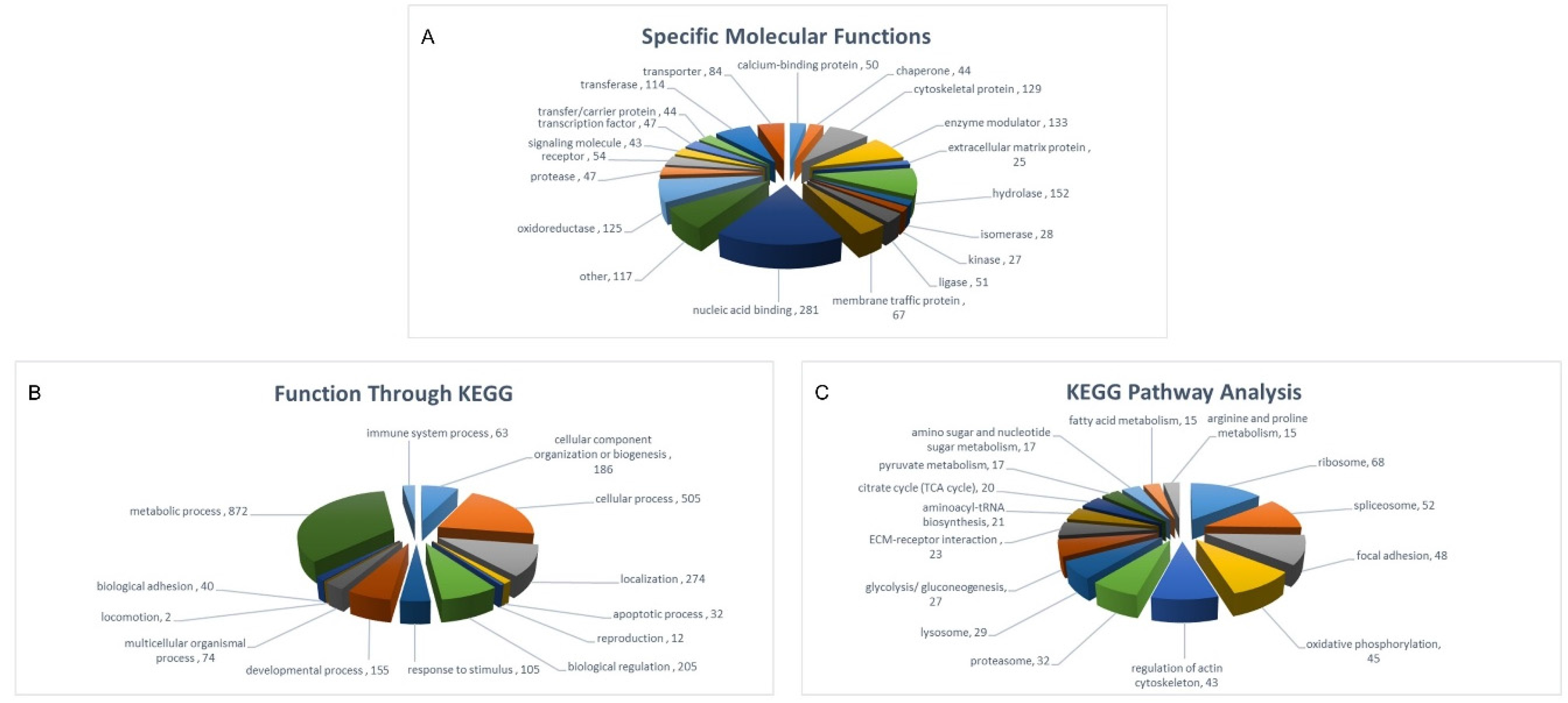
2.3. Protein Class-Function of Molecules Identified in Both SHEDs and PDLSCs
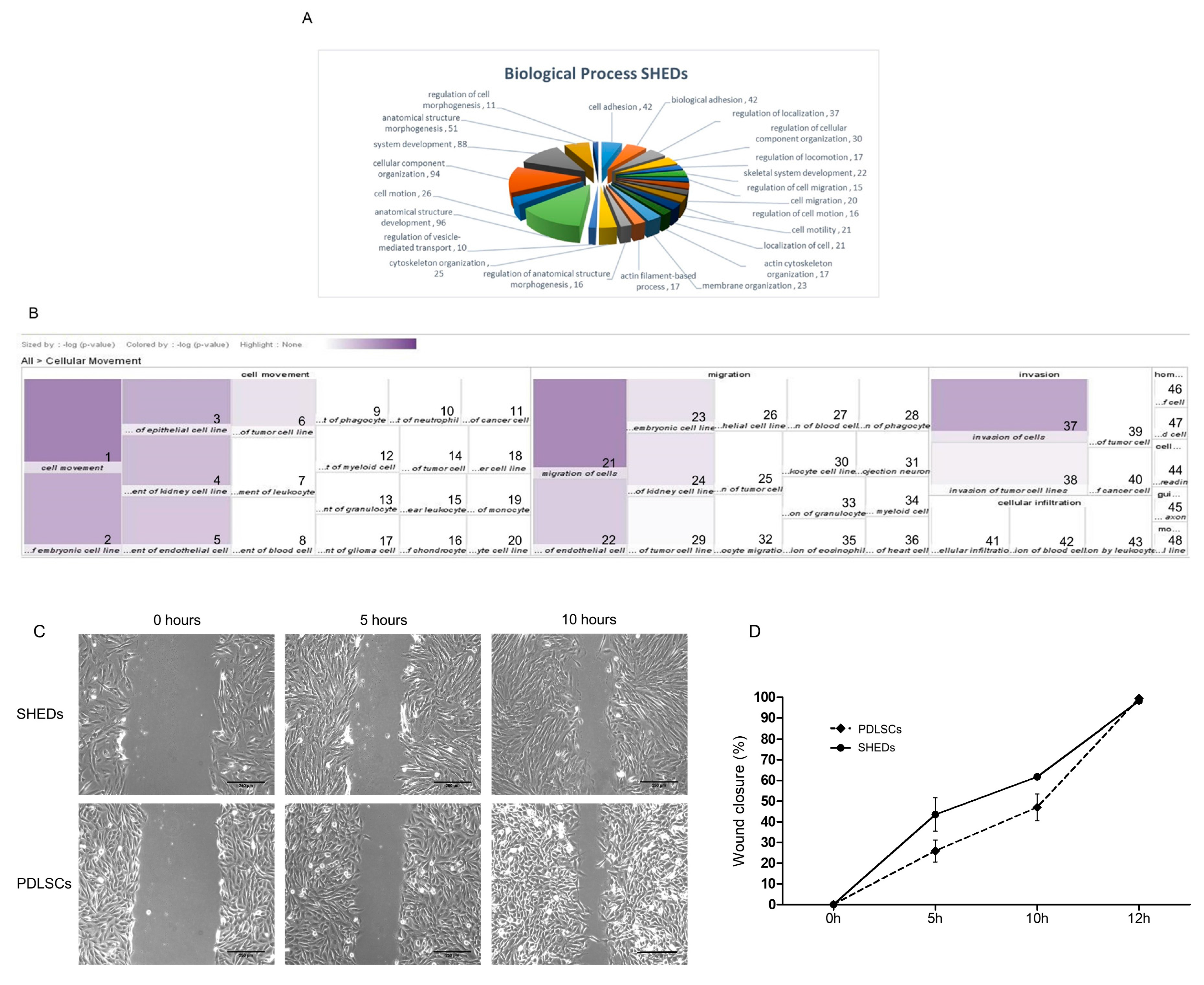
2.4. Proteins Exclusively Identified in SHEDs
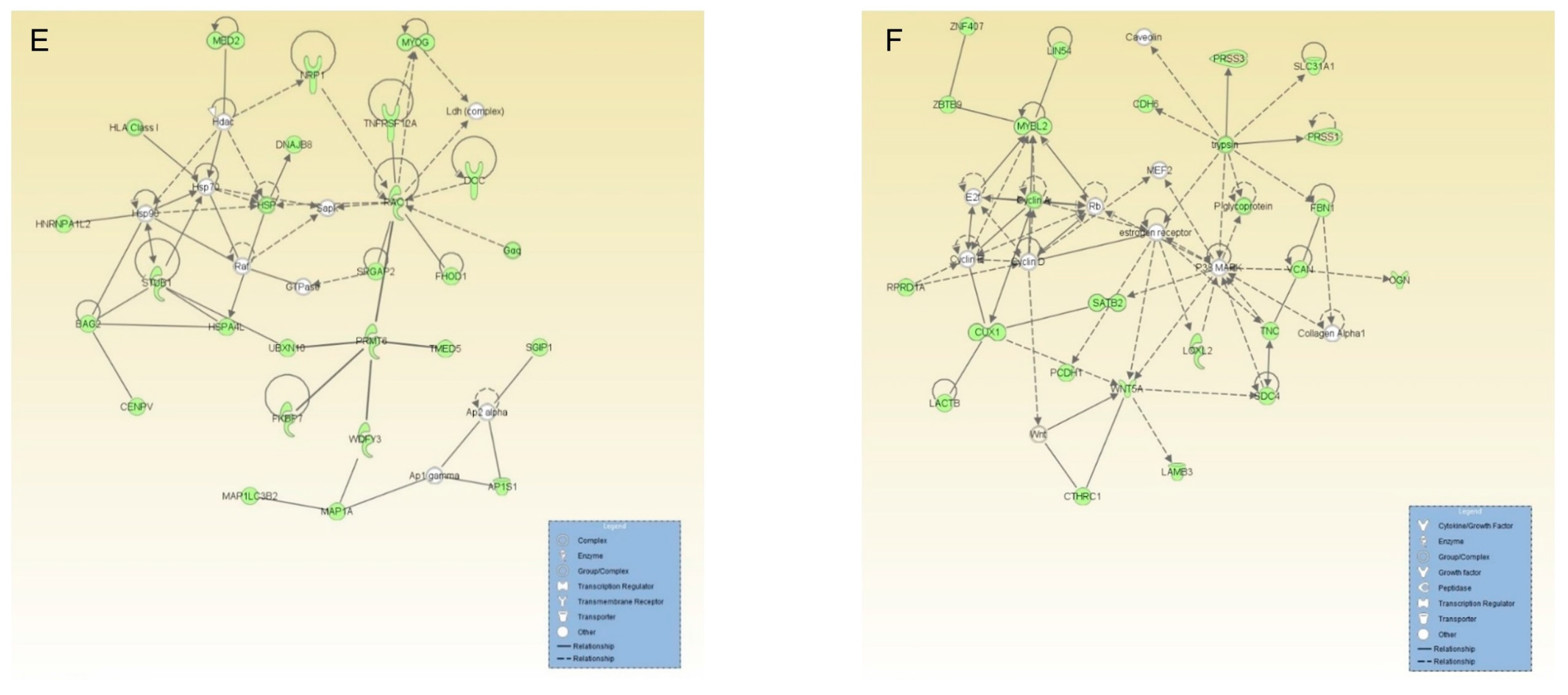
2.5. Proteins Exclusively Identified in PDLSCs
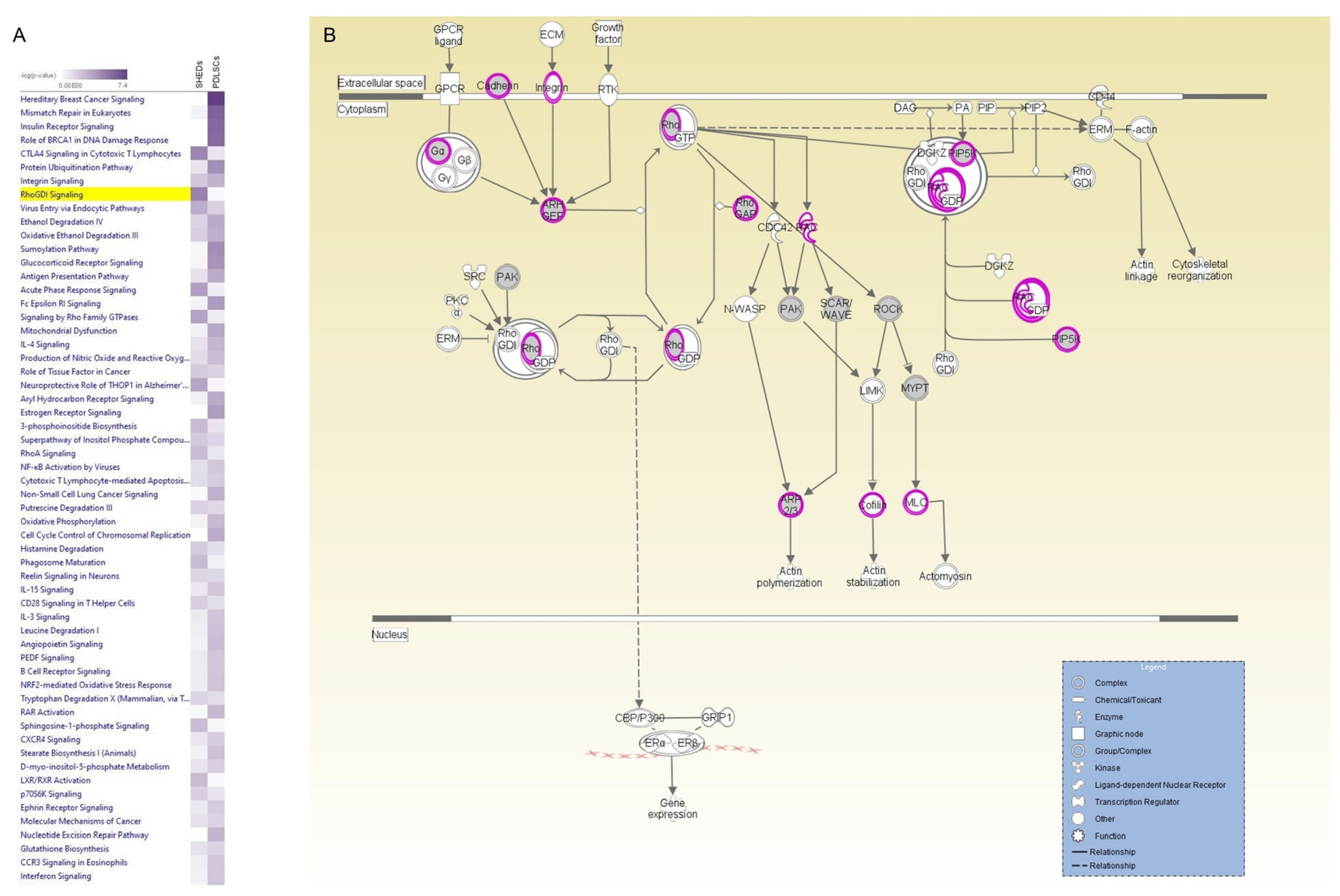
2.6. Potential Applications in the Biomarkers Field—A Paradigm of Different Markers on the Same Pathway
3. Discussion
4. Materials and Methods
4.1. Proteomics Analysis
4.1.1. LC-MS/MS-Preparation of Samples
4.1.2. LC-MS/MS and Data Bioinformatics Analysis
4.2. Immunostaining and Confocal Microscopy
4.3. Migration (Wound-Healing) Assay
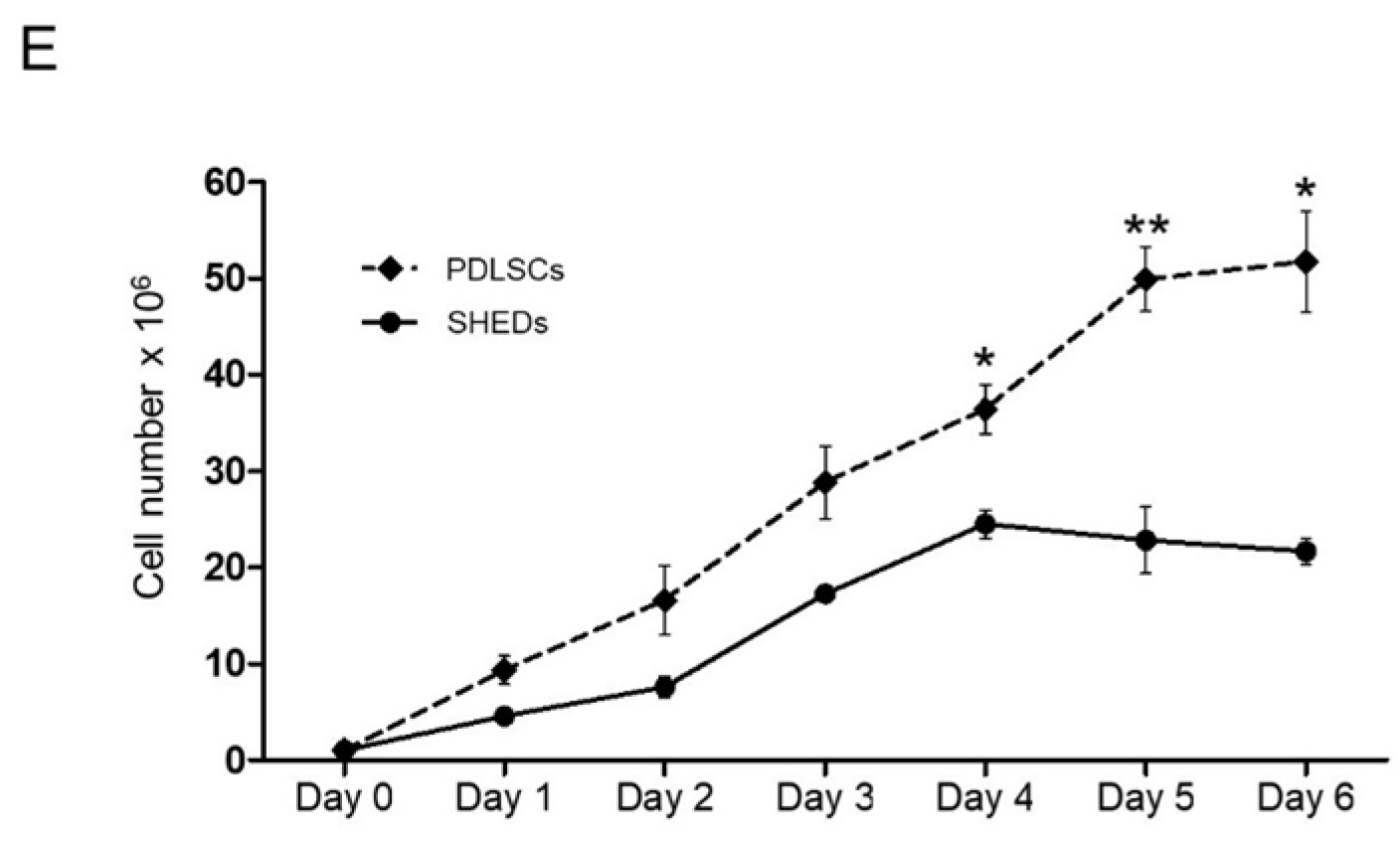
4.4. Growth Curves
4.5. Mitochondria Staining
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Karussis, D.; Karageorgiou, C.; Vaknin-Dembinsky, A.; Gowda-Kurkalli, B.; Gomori, J.M.; Kassis, I.; Bulte, J.W.; Petrou, P.; Ben-Hur, T.; Abramsky, O.; et al. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Arch. Neurol. 2010, 67, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Le Blanc, K.; Frassoni, F.; Ball, L.; Locatelli, F.; Roelofs, H.; Lewis, I.; Lanino, E.; Sundberg, B.; Bernardo, M.E.; Remberger, M.; et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: A phase II study. Lancet 2008, 371, 1579–1586. [Google Scholar] [CrossRef]
- Orozco, L.; Munar, A.; Soler, R.; Alberca, M.; Soler, F.; Huguet, M.; Sentis, J.; Sanchez, A.; Garcia-Sancho, J. Treatment of knee osteoarthritis with autologous mesenchymal stem cells: A pilot study. Transplantation 2013, 95, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, X.; Han, Z.P.; Qu, F.F.; Shao, L.; Shi, Y.F. Mesenchymal stem cells: A new trend for cell therapy. Acta Pharmacol. Sin. 2013, 34, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Lymperi, S.; Ligoudistianou, C.; Taraslia, V.; Kontakiotis, E.; Anastasiadou, E. Dental stem cells and their applications in dental tissue engineering. Open Dent. J. 2013, 7, 76–81. [Google Scholar] [PubMed]
- Xiao, L.; Nasu, M. From regenerative dentistry to regenerative medicine: Progress, challenges, and potential applications of oral stem cells. Stem Cells Cloning Adv. Appl. 2014, 7, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. Shed: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [PubMed]
- Sonoyama, W.; Liu, Y.; Fang, D.; Yamaza, T.; Seo, B.M.; Zhang, C.; Liu, H.; Gronthos, S.; Wang, C.Y.; Wang, S.; et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS ONE 2006, 1, e79. [Google Scholar] [CrossRef] [PubMed]
- Morsczeck, C.; Gotz, W.; Schierholz, J.; Zeilhofer, F.; Kuhn, U.; Mohl, C.; Sippel, C.; Hoffmann, K.H. Isolation of precursor cells (PCS) from human dental follicle of wisdom teeth. Matrix Biol. 2005, 24, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- La Noce, M.; Mele, L.; Tirino, V.; Paino, F.; De Rosa, A.; Naddeo, P.; Papagerakis, P.; Papaccio, G.; Desiderio, V. Neural crest stem cell population in craniomaxillofacial development and tissue repair. Eur. Cells Mater. 2014, 28, 348–357. [Google Scholar] [CrossRef]
- D’aquino, R.; De Rosa, A.; Lanza, V.; Tirino, V.; Laino, L.; Graziano, A.; Desiderio, V.; Laino, G.; Papaccio, G. Human mandible bone defect repair by the grafting of dental pulp stem/progenitor cells and collagen sponge biocomplexes. Eur. Cells Mater. 2009, 18, 75–83. [Google Scholar] [CrossRef]
- Ramamoorthi, M.; Bakkar, M.; Jordan, J.; Tran, S.D. Osteogenic potential of dental mesenchymal stem cells in preclinical studies: A systematic review using modified arrive and consort guidelines. Stem Cells Int. 2015, 2015, 378368. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Bartold, P.M.; Miura, M.; Seo, B.M.; Robey, P.G.; Gronthos, S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. Craniofac. Res. 2005, 8, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, B.G.; Serafim, R.C.; Melo, G.B.; Silva, M.C.; Lizier, N.F.; Maranduba, C.M.; Smith, R.L.; Kerkis, A.; Cerruti, H.; Gomes, J.A.; et al. Human immature dental pulp stem cells share key characteristic features with limbal stem cells. Cell Prolif. 2009, 42, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Seo, B.M.; Liu, Y.; Sonoyama, W.; Yamaza, T.; Zhang, C.; Wang, S.; Shi, S. Transplantation of mesenchymal stem cells is an optimal approach for plastic surgery. Stem Cells 2007, 25, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Chadipiralla, K.; Yochim, J.M.; Bahuleyan, B.; Huang, C.Y.; Garcia-Godoy, F.; Murray, P.E.; Stelnicki, E.J. Osteogenic differentiation of stem cells derived from human periodontal ligaments and pulp of human exfoliated deciduous teeth. Cell Tissue Res. 2010, 340, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Ponnaiyan, D.; Bhat, K.M.; Bhat, G.S. Comparison of immuno-phenotypes of stem cells from human dental pulp and periodontal ligament. Int. J. Immunopathol. Pharmacol. 2012, 25, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Li, K.; Li, B.; Gao, L.N.; Chen, F.M.; Jin, Y. Mesenchymal stem cell characteristics of dental pulp and periodontal ligament stem cells after in vivo transplantation. Biomaterials 2014, 35, 6332–6343. [Google Scholar] [CrossRef] [PubMed]
- Hakki, S.S.; Kayis, S.A.; Hakki, E.E.; Bozkurt, S.B.; Duruksu, G.; Unal, Z.S.; Turac, G.; Karaoz, E. Comparison of mesenchymal stem cells isolated from pulp and periodontal ligament. J. Periodontol. 2015, 86, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, V.R.; Nadig, R.R.; Nadig, R.; Prasanna, J.S.; Karthik, J.; Pai, V.S. Differentiation of isolated and characterized human dental pulp stem cells and stem cells from human exfoliated deciduous teeth: An in vitro study. J. Conserv. Dent. 2013, 16, 423–428. [Google Scholar] [PubMed]
- Shekar, R.; Ranganathan, K. Phenotypic and growth characterization of human mesenchymal stem cells cultured from permanent and deciduous teeth. Indian J. Dent. Res. 2012, 23, 838–839. [Google Scholar] [PubMed]
- Suchanek, J.; Visek, B.; Soukup, T.; El-Din Mohamed, S.K.; Ivancakova, R.; Mokry, J.; Aboul-Ezz, E.H.; Omran, A. Stem cells from human exfoliated deciduous teeth—Isolation, long term cultivation and phenotypical analysis. Acta Medica 2010, 53, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sha, X.J.; Li, G.H.; Yang, F.S.; Ji, K.; Wen, L.Y.; Liu, S.Y.; Chen, L.; Ding, Y.; Xuan, K. Comparative characterization of stem cells from human exfoliated deciduous teeth and dental pulp stem cells. Arch. Oral Biol. 2012, 57, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.C.; Zhu, S.; Xu, J.; Yuan, C.; Gong, T.; Zhang, C. Effects of decellularized matrices derived from periodontal ligament stem cells and shed on the adhesion, proliferation and osteogenic differentiation of human dental pulp stem cells in vitro. Tissue Cell 2016, 48, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Eleuterio, E.; Trubiani, O.; Sulpizio, M.; di Giuseppe, F.; Pierdomenico, L.; Marchisio, M.; Giancola, R.; Giammaria, G.; Miscia, S.; Caputi, S.; et al. Proteome of human stem cells from periodontal ligament and dental pulp. PLoS ONE 2013, 8, e71101. [Google Scholar] [CrossRef] [PubMed]
- Akpinar, G.; Kasap, M.; Aksoy, A.; Duruksu, G.; Gacar, G.; Karaoz, E. Phenotypic and proteomic characteristics of human dental pulp derived mesenchymal stem cells from a natal, an exfoliated deciduous, and an impacted third molar tooth. Stem Cells Int. 2014, 2014, 457059. [Google Scholar] [CrossRef] [PubMed]
- Issaq, H.; Veenstra, T. Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE): Advances and perspectives. Biotechniques 2008, 44, 697–698, 700. [Google Scholar] [CrossRef] [PubMed]
- Tirino, V.; Paino, F.; de Rosa, A.; Papaccio, G. Identification, isolation, characterization, and banking of human dental pulp stem cells. Methods Mol. Biol. 2012, 879, 443–463. [Google Scholar] [PubMed]
- Tirino, V.; Paino, F.; d’Aquino, R.; Desiderio, V.; de Rosa, A.; Papaccio, G. Methods for the identification, characterization and banking of human dpscs: Current strategies and perspectives. Stem Cell Rev. 2011, 7, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.H.; Chang, C.C.; Kiupel, M.; Webster, J.D.; Olson, L.K.; Trosko, J.E. Oct4 expression in adult human stem cells: Evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 2005, 26, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; Seo, K.W.; So, A.Y.; Seo, M.S.; Yu, K.R.; Kang, S.K.; Kang, K.S. Sox2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012, 19, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Hergt, M.; Grund, C. Rearrangement of the vimentin cytoskeleton during adipose conversion: Formation of an intermediate filament cage around lipid globules. Cell 1987, 49, 131–141. [Google Scholar] [CrossRef]
- Moustakas, A.; Heldin, C.H. Ecsit-ement on the crossroads of toll and bmp signal transduction. Genes Dev. 2003, 17, 2855–2859. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.S.; Lee, M.W.; Ryu, D.; Kim, J.H.; Ma, H.; Seo, W.Y.; Kim, Y.N.; Kim, S.S.; Lee, C.H.; Hunter, T.; et al. Suppressor of mek null (SMEK)/protein phosphatase 4 catalytic subunit (PP4C) is a key regulator of hepatic gluconeogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17704–17709. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, A.; Glotzer, M.; Mishima, M. Clustering of centralspindlin is essential for its accumulation to the central spindle and the midbody. Curr. Biol. 2009, 19, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Cui, L.; Gao, J.; Yan, W.; Liu, Y.; Xu, S.; Wu, B. Proteomic analysis of mesenchymal stem cells from normal and deep carious dental pulp. PLoS ONE 2014, 9, e97026. [Google Scholar] [CrossRef] [PubMed]
- Ivaska, J.; Vuoriluoto, K.; Huovinen, T.; Izawa, I.; Inagaki, M.; Parker, P.J. Pkcepsilon-mediated phosphorylation of vimentin controls integrin recycling and motility. EMBO J. 2005, 24, 3834–3845. [Google Scholar] [CrossRef] [PubMed]
- Kadri, T.; Lataillade, J.J.; Doucet, C.; Marie, A.; Ernou, I.; Bourin, P.; Joubert-Caron, R.; Caron, M.; Lutomski, D. Proteomic study of galectin-1 expression in human mesenchymal stem cells. Stem Cells Dev. 2005, 14, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Shin, H.I.; Cha, S.S.; Lee, C.S.; Hong, B.S.; Lim, S.; Jang, H.J.; Kim, J.; Yang, Y.R.; Kim, Y.H.; et al. DJ-1 promotes angiogenesis and osteogenesis by activating FGF receptor-1 signaling. Nat. Commun. 2012, 3, 1296. [Google Scholar] [CrossRef] [PubMed]
- Park, H.W.; Shin, J.S.; Kim, C.W. Proteome of mesenchymal stem cells. Proteomics 2007, 7, 2881–2894. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Lee, H.M.; Jang, Y.J.; Kim, C.H.; Ryu, C.J. Heterogeneous nuclear ribonucleoprotein A2/B1 regulates the self-renewal and pluripotency of human embryonic stem cells via the control of the G1/S transition. Stem Cells 2013, 31, 2647–2658. [Google Scholar] [CrossRef] [PubMed]
- Roche, S.; Delorme, B.; Oostendorp, R.A.; Barbet, R.; Caton, D.; Noel, D.; Boumediene, K.; Papadaki, H.A.; Cousin, B.; Crozet, C.; et al. Comparative proteomic analysis of human mesenchymal and embryonic stem cells: Towards the definition of a mesenchymal stem cell proteomic signature. Proteomics 2009, 9, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.A., Jr.; Covas, D.T.; Panepucci, R.A.; Proto-Siqueira, R.; Siufi, J.L.; Zanette, D.L.; Santos, A.R.; Zago, M.A. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 2003, 21, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Oser, M.; Condeelis, J. The cofilin activity cycle in lamellipodia and invadopodia. J. Cell. Biochem. 2009, 108, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Kurita, S.; Gunji, E.; Ohashi, K.; Mizuno, K. Actin filaments-stabilizing and -bundling activities of cofilin-phosphatase slingshot-1. Genes Cells 2007, 12, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta 2004, 1692, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Rho GTpases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar] [PubMed]
- Ridley, A.J. Rho GTpases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Gardberg, M.; Kaipio, K.; Lehtinen, L.; Mikkonen, P.; Heuser, V.D.; Talvinen, K.; Iljin, K.; Kampf, C.; Uhlen, M.; Grenman, R.; et al. Fhod1, a formin upregulated in epithelial-mesenchymal transition, participates in cancer cell migration and invasion. PLoS ONE 2013, 8, e74923. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.K.; Cain, S.A.; Lennon, R.; Godwin, A.; Merry, C.L.; Kielty, C.M. Epithelial-mesenchymal status influences how cells deposit fibrillin microfibrils. J. Cell Sci. 2014, 127, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Jossin, Y.; Cooper, J.A. Reelin, rap1 and n-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 2011, 14, 697–703. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Wang, K.C.; Koprivica, V.; Ming, G.; Song, H.J. Knowing how to navigate: Mechanisms of semaphorin signaling in the nervous system. Sci. STKE 2002, 2002, re1. [Google Scholar] [CrossRef] [PubMed]
- Hirata, T.; Nakazawa, M.; Yoshihara, S.; Miyachi, H.; Kitamura, K.; Yoshihara, Y.; Hibi, M. Zinc-finger gene fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development 2006, 133, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Nagayasu, Y.; Kato, K.; Sato, R.; Yokoyama, S. Apolipoprotein A-I induces translocation of cholesterol, phospholipid, and caveolin-1 to cytosol in rat astrocytes. J. Biol. Chem. 2002, 277, 7929–7935. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.Y.; Dong, H.; Wan, Y.; Li, J.; Yee, A.; Yang, B.B.; Lu, W.Y. Versican G3 domain regulates neurite growth and synaptic transmission of hippocampal neurons by activation of epidermal growth factor receptor. J. Biol. Chem. 2006, 281, 19358–19368. [Google Scholar] [CrossRef] [PubMed]
- Swiercz, J.M.; Kuner, R.; Offermanns, S. Plexin-B1/RHoGEF-mediated RhoA activation involves the receptor tyrosine kinase ErbB-2. J. Cell Biol. 2004, 165, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Janebodin, K.; Horst, O.V.; Ieronimakis, N.; Balasundaram, G.; Reesukumal, K.; Pratumvinit, B.; Reyes, M. Isolation and characterization of neural crest-derived stem cells from dental pulp of neonatal mice. PLoS ONE 2011, 6, e27526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Niu, C.; He, W.; Hou, T.; Sun, X.; Xu, L.; Zhang, Y. Upregulation of centrosomal protein 55 is associated with unfavorable prognosis and tumor invasion in epithelial ovarian carcinoma. Tumour Biol. 2016, 37, 6239–6254. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, J.R.; Toyoda, Y.; Hegemann, B.; Poser, I.; Heriche, J.K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Seki, A.; Fang, G. Skap associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle 2009, 8, 2819–2827. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Kubota, Y.; Arata, T.; Shindo, Y.; Hanaoka, F. Structure of the human gins complex and its assembly and functional interface in replication initiation. Nat. Struct. Mol. Biol. 2007, 14, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, R.; Qi, W.; Yu, H. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 2004, 279, 13076–13085. [Google Scholar] [CrossRef] [PubMed]
- Vergnolle, M.A.; Taylor, S.S. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 2007, 17, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, J.B.; Wueseke, O.; Hyman, A.A. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130459. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, C.B.; Khodjakov, A.L. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 2007, 120, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Doxsey, S.; Zimmerman, W.; Mikule, K. Centrosome control of the cell cycle. Trends Cell Biol. 2005, 15, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.; Le Peuch, C.; Labbe, J.C.; Meyer, A.N.; Donoghue, D.J.; Doree, M. Initial activation of cyclin-B1-CDC2 kinase requires phosphorylation of cyclin b1. EMBO Rep. 2002, 3, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Ma, W.; Stafford, L.J.; Marcus, S.; Xiong, W.C.; Liu, M. Regulation of the P21-activated kinase (PAK) by a human gβ -like wd-repeat protein, hpip1. Proc. Natl. Acad. Sci. USA 2001, 98, 6174–6179. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, V.; Piel, M.; Collomb, N.; Saoudi, Y.; Frank, R.; Paintrand, M.; Narumiya, S.; Bornens, M.; Job, D. The rho-associated protein kinase p160ROCK is required for centrosome positioning. J. Cell Biol. 2002, 157, 807–817. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. Uniprot: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the panther classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. Panther in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taraslia, V.; Lymperi, S.; Pantazopoulou, V.; Anagnostopoulos, A.K.; Papassideri, I.S.; Basdra, E.K.; Bei, M.; Kontakiotis, E.G.; Tsangaris, G.T.; Stravopodis, D.J.; et al. A High-Resolution Proteomic Landscaping of Primary Human Dental Stem Cells: Identification of SHED- and PDLSC-Specific Biomarkers. Int. J. Mol. Sci. 2018, 19, 158. https://doi.org/10.3390/ijms19010158
Taraslia V, Lymperi S, Pantazopoulou V, Anagnostopoulos AK, Papassideri IS, Basdra EK, Bei M, Kontakiotis EG, Tsangaris GT, Stravopodis DJ, et al. A High-Resolution Proteomic Landscaping of Primary Human Dental Stem Cells: Identification of SHED- and PDLSC-Specific Biomarkers. International Journal of Molecular Sciences. 2018; 19(1):158. https://doi.org/10.3390/ijms19010158
Chicago/Turabian StyleTaraslia, Vasiliki, Stefania Lymperi, Vasiliki Pantazopoulou, Athanasios K. Anagnostopoulos, Issidora S. Papassideri, Efthimia K. Basdra, Marianna Bei, Evangelos G. Kontakiotis, George Th. Tsangaris, Dimitrios J. Stravopodis, and et al. 2018. "A High-Resolution Proteomic Landscaping of Primary Human Dental Stem Cells: Identification of SHED- and PDLSC-Specific Biomarkers" International Journal of Molecular Sciences 19, no. 1: 158. https://doi.org/10.3390/ijms19010158
APA StyleTaraslia, V., Lymperi, S., Pantazopoulou, V., Anagnostopoulos, A. K., Papassideri, I. S., Basdra, E. K., Bei, M., Kontakiotis, E. G., Tsangaris, G. T., Stravopodis, D. J., & Anastasiadou, E. (2018). A High-Resolution Proteomic Landscaping of Primary Human Dental Stem Cells: Identification of SHED- and PDLSC-Specific Biomarkers. International Journal of Molecular Sciences, 19(1), 158. https://doi.org/10.3390/ijms19010158







