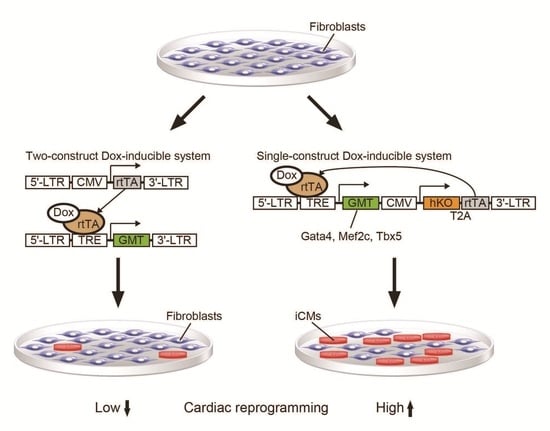
Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression
Abstract
1. Introduction
2. Results
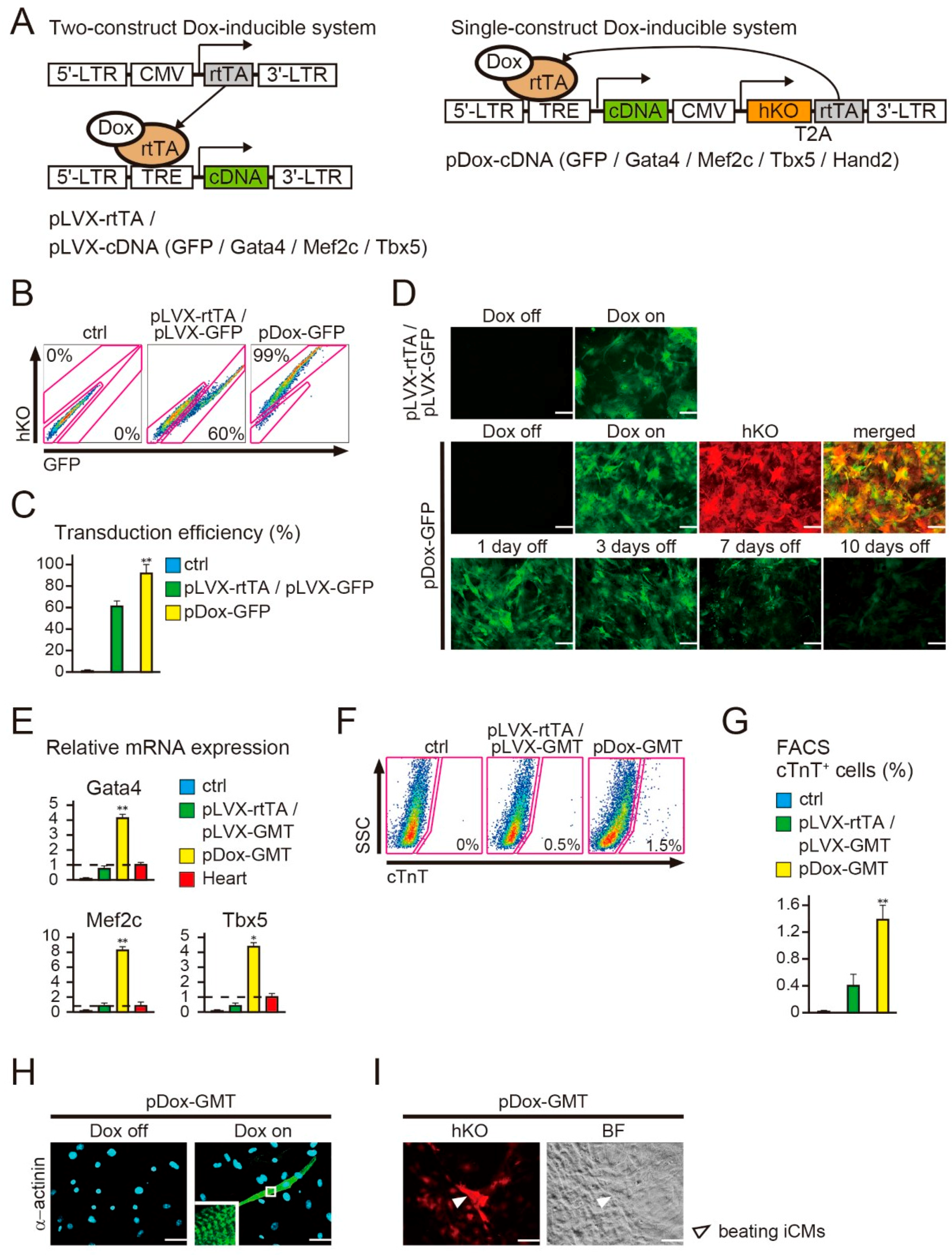
2.1. New Polycistronic Dox-Inducible Vectors Improve Direct Cardiac Reprogramming
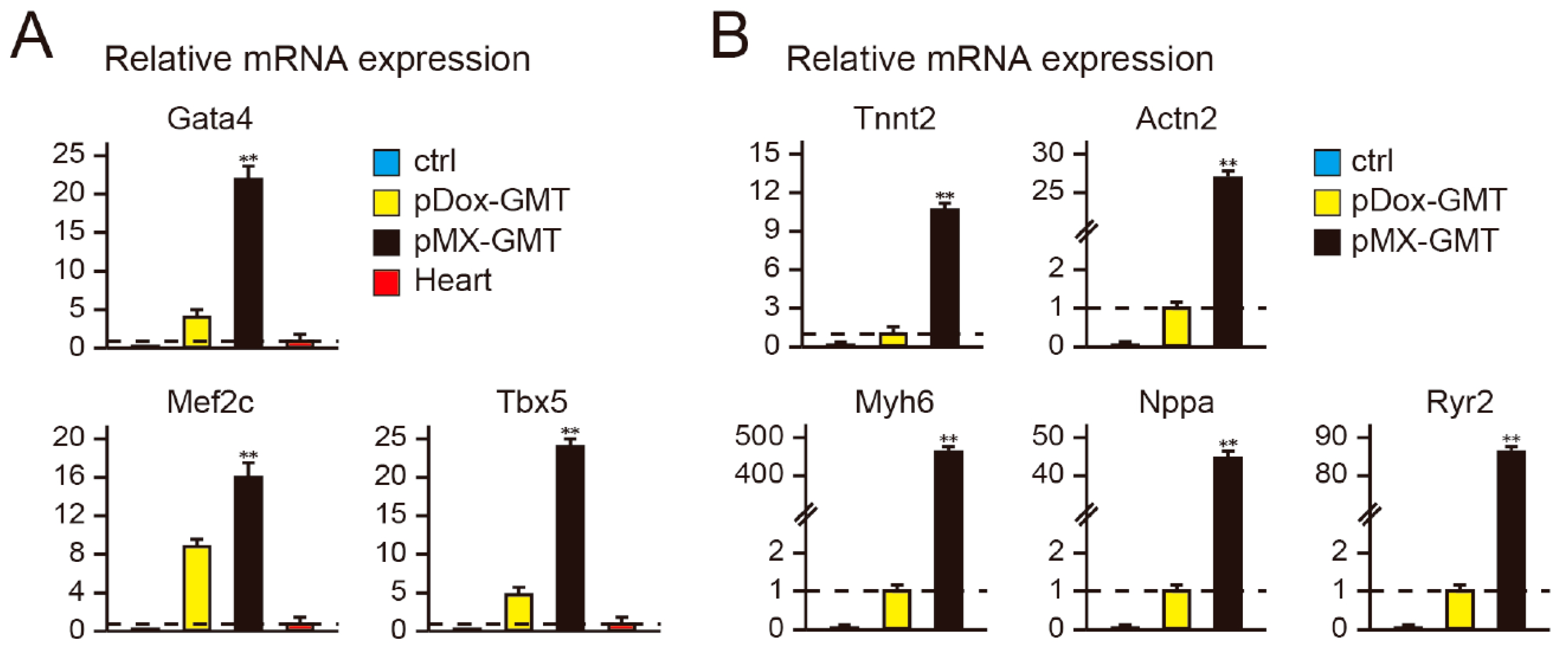
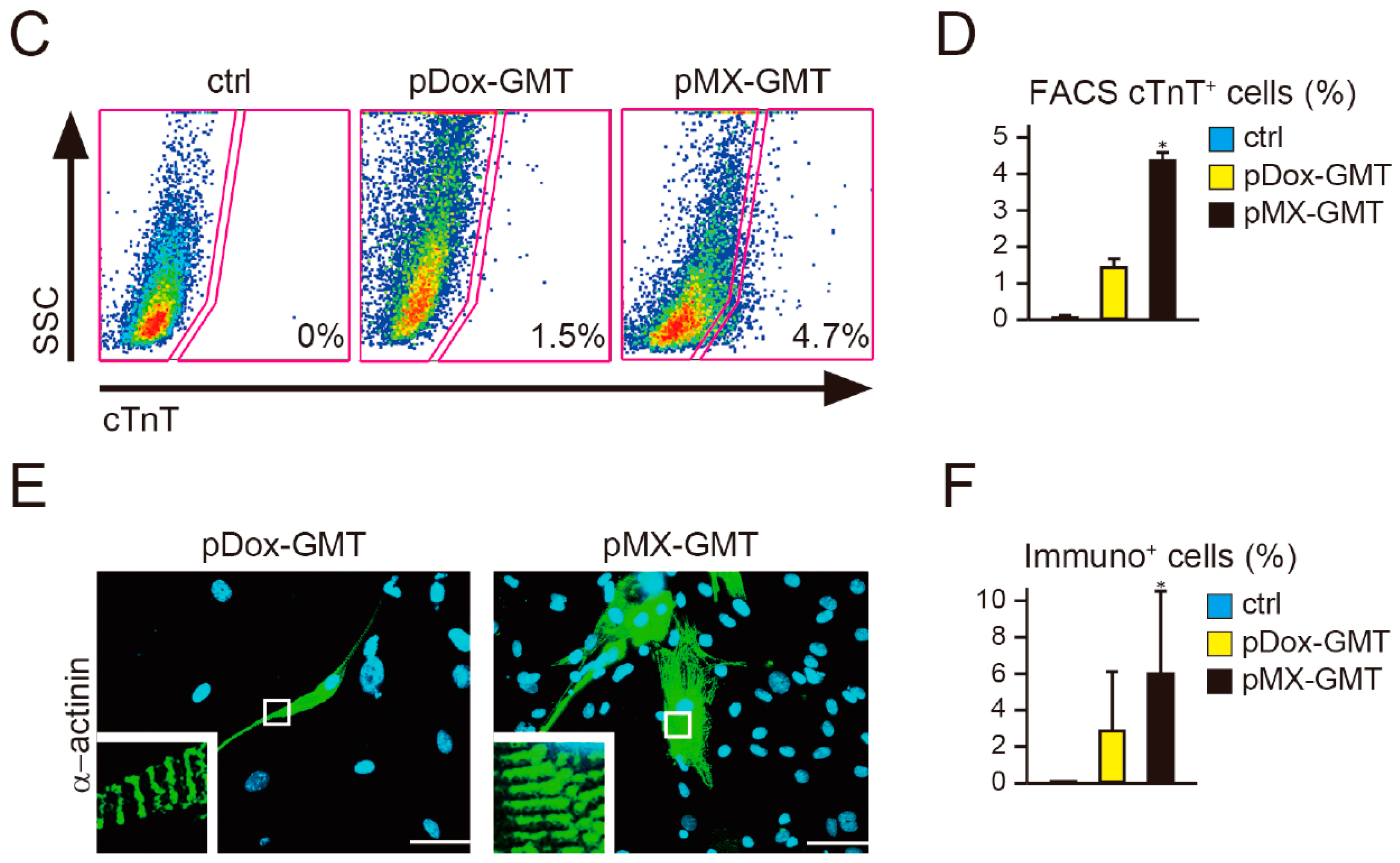
2.2. Diminished Cardiac Reprogramming with Lentiviral pDox-GMT (a Mixture of pDox-Gata4, -Mef2c, and -Tbx5) Compared to that Seen with Retroviral pMX-GMT (a Mixture of pMX-Gata4, -Mef2c, and -Tbx5)
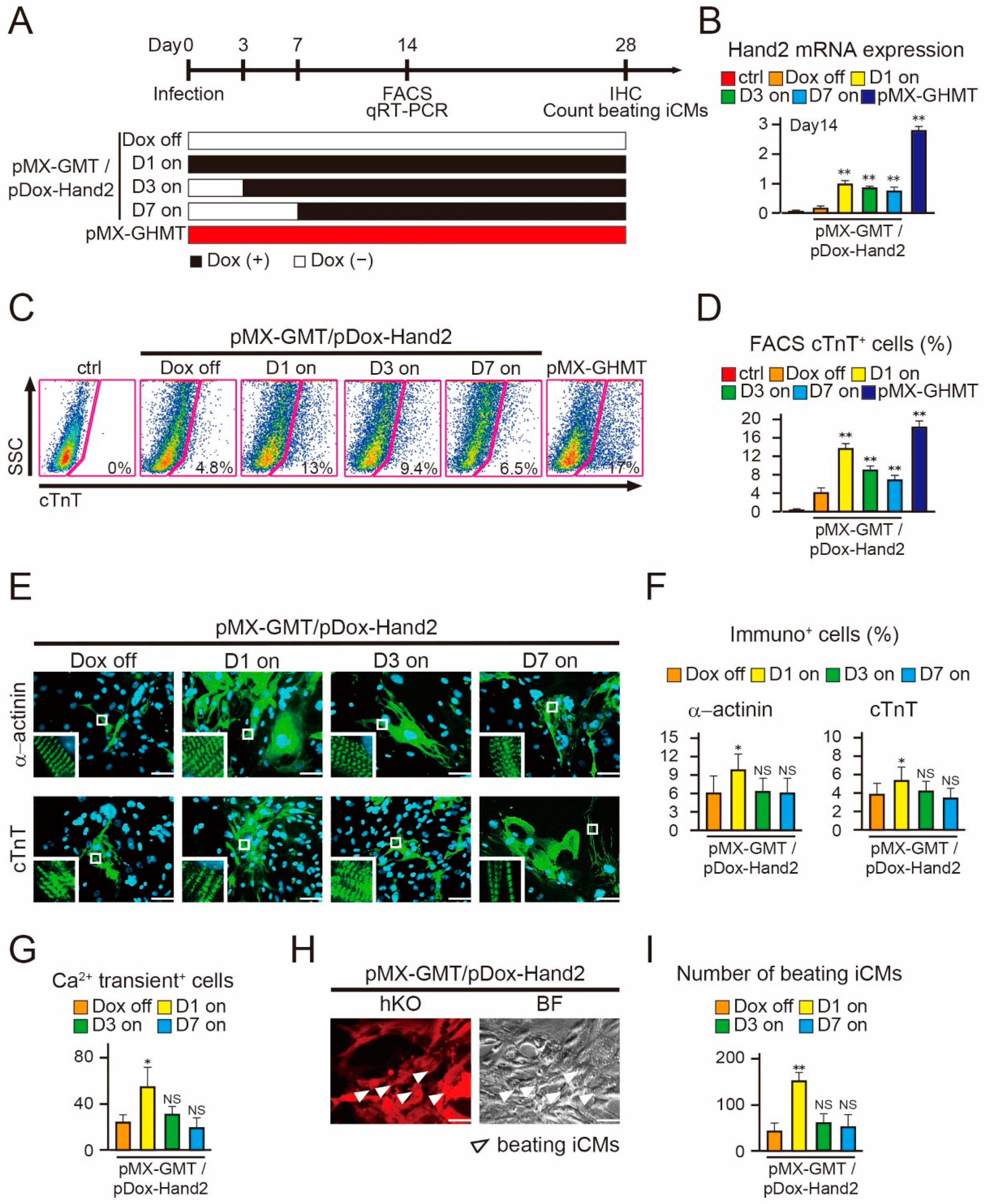
2.3. Induction of Hand2 Transgene Expression One Day after Transduction Promotes Cardiac Reprogramming
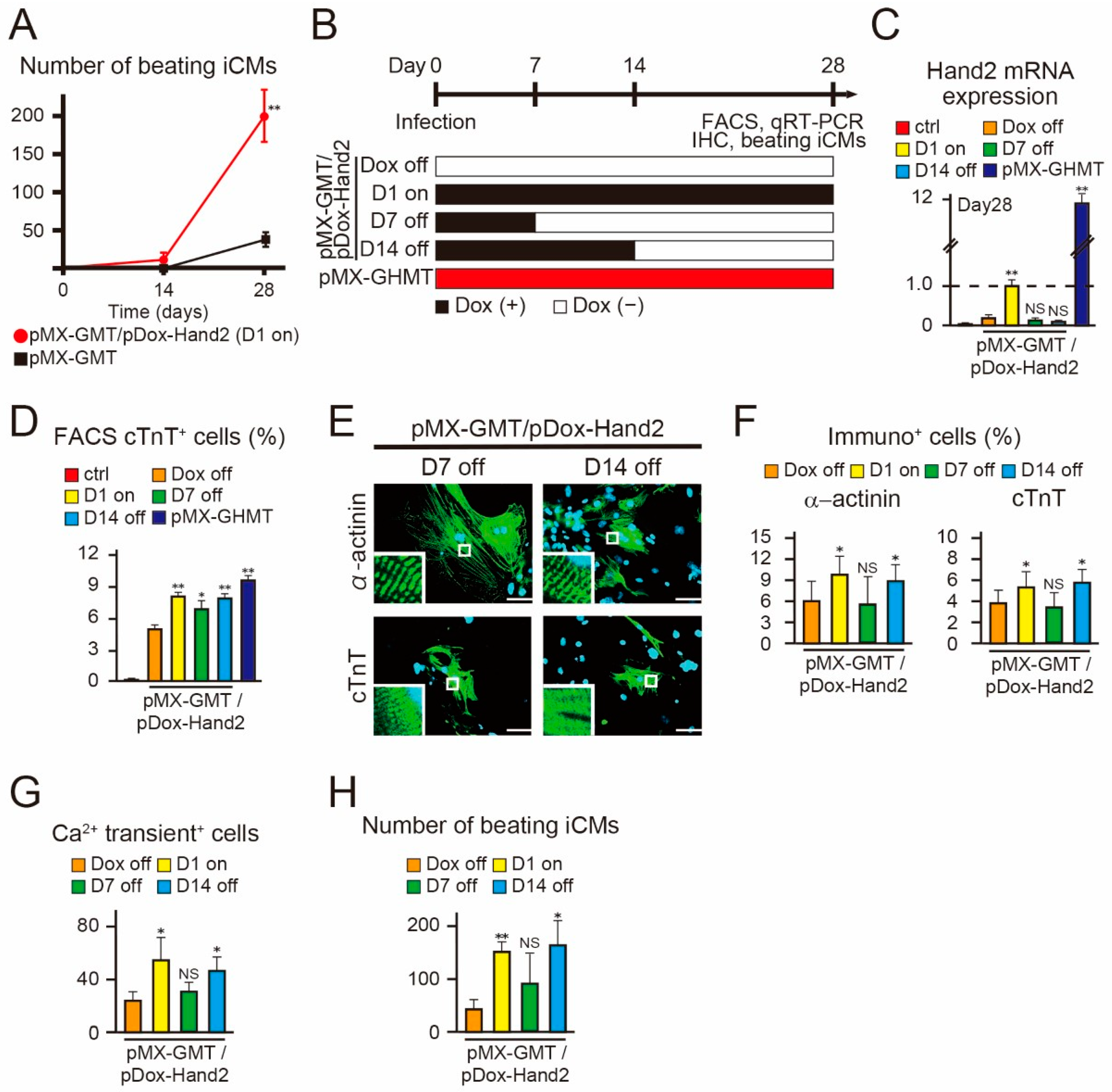
2.4. Hand2 Transgene Expression for Two Weeks is Sufficient to Promote Cardiac Reprogramming
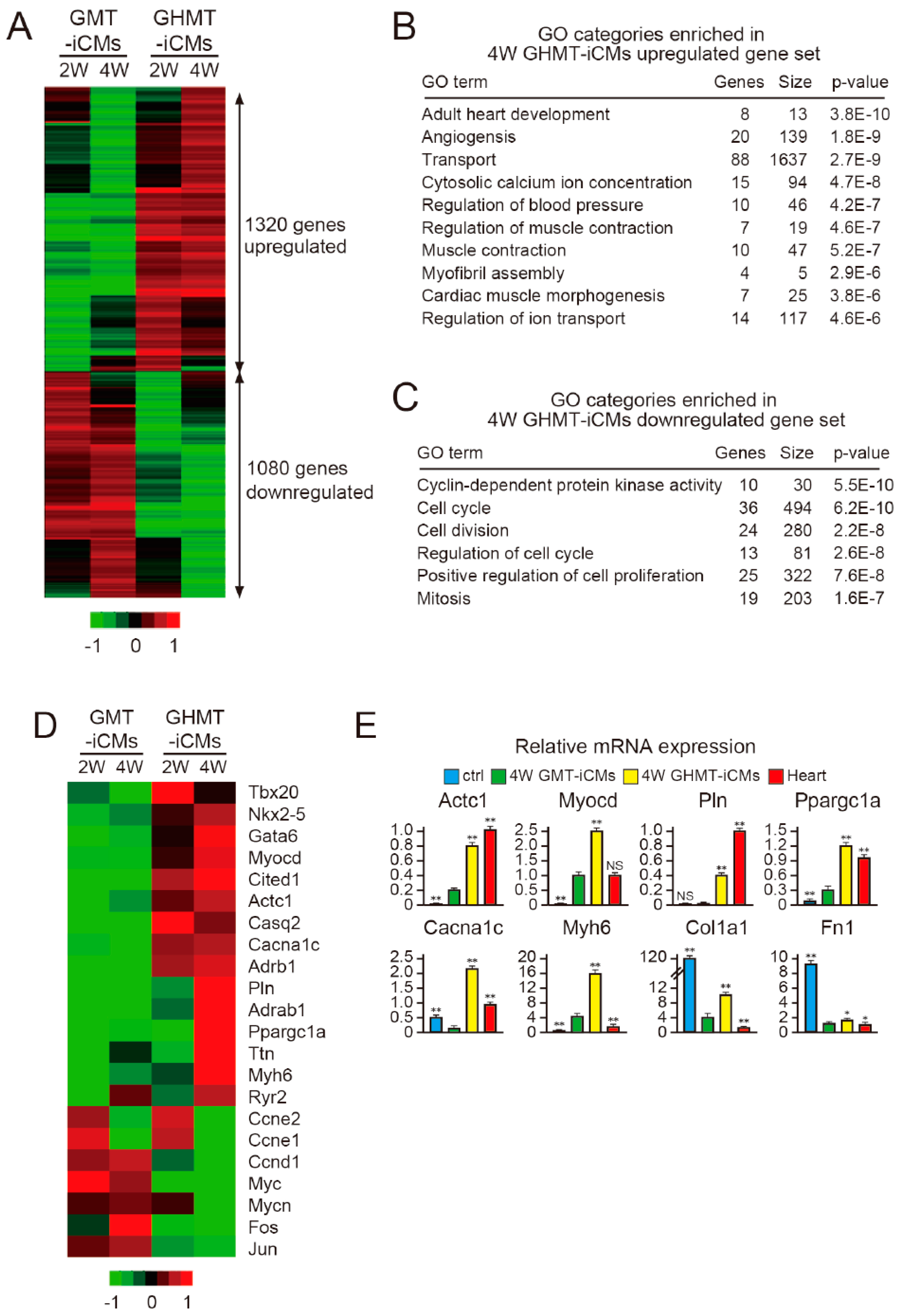
2.5. Hand2 Activates a Cardiac Program and Represses Cell Cycle-Promoting Genes During Cardiac Reprogramming
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Generation of α Myosin Heavy Chain (αMHC)-Green Fluorescent Protein (GFP) Mice
4.3. Mouse Embryonic Fibroblast (MEF) Isolation
4.4. Lentivirus Production
4.5. Retroviral and Lentiviral Infection
4.6. Fluorescence Activated Cell Sorting (FACS) Analyses and Sorting
4.7. Quantitative RT-PCR
4.8. Immunocytochemistry
4.9. Ca2+ Imaging and Counting Beating Cells
4.10. Microarray Analyses
4.11. Statistical Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Srivastava, D.; DeWitt, N. In vivo cellular reprogramming: The next generation. Cell 2016, 166, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Sadahiro, T.; Yamanaka, S.; Ieda, M. Direct cardiac reprogramming: Progress and challenges in basic biology and clinical applications. Circ. Res. 2015, 116, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Yamanaka, S. Induced pluripotent stem cells 10 years later: For cardiac applications. Circ. Res. 2017, 120, 1958–1968. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.; Delgado-Oguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Qian, L. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Nam, Y.J.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Inagawa, K.; Miyamoto, K.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Umei, T.; Wada, R.; Katsumata, Y.; Kaneda, R.; Nakada, K.; et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Brambrink, T.; Foreman, R.; Welstead, G.G.; Lengner, C.J.; Wernig, M.; Suh, H.; Jaenisch, R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008, 2, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2014, 116, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Ieda, M. Critical factors for cardiac reprogramming. Circ. Res. 2012, 111, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Krane, M.; Deutsch, M.A.; Wang, L.; Rav-Acha, M.; Gregoire, S.; Engels, M.C.; Rajarajan, K.; Karra, R.; Abel, E.D.; et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Ieda, M. Stoichiometry of transcription factors is critical for cardiac reprogramming. Circ. Res. 2015, 116, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Vaseghi, H.R.; Yin, C.; Zhou, Y.; Wang, L.; Liu, J.; Qian, L. Generation of an inducible fibroblast cell line for studying direct cardiac reprogramming. Genesis. 2016, 54, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D. Making or breaking the heart: from lineage determination to morphogenesis. Cell 2006, 126, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.S.; Cserjesi, P.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 2002, 277, 24390–24398. [Google Scholar] [CrossRef] [PubMed]
- Zang, M.X.; Li, Y.; Xue, L.X.; Jia, H.T.; Jing, H. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J. Cell. Biochem. 2004, 93, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Tanos, R.; El-Hachem, N.; Kurban, M.; Bouvagnet, P.; Bitar, F.; Nemer, G. A HAND to TBX5 explains the link between thalidomide and cardiac diseases. Sci. Rep. 2017, 7, 1416. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umei, T.C.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Kurotsu, S.; Tamura, F.; Osakabe, R.; et al. Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. Int. J. Mol. Sci. 2017, 18, 1805. https://doi.org/10.3390/ijms18081805
Umei TC, Yamakawa H, Muraoka N, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Kurotsu S, Tamura F, Osakabe R, et al. Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. International Journal of Molecular Sciences. 2017; 18(8):1805. https://doi.org/10.3390/ijms18081805
Chicago/Turabian StyleUmei, Tomohiko C., Hiroyuki Yamakawa, Naoto Muraoka, Taketaro Sadahiro, Mari Isomi, Sho Haginiwa, Hidenori Kojima, Shota Kurotsu, Fumiya Tamura, Rina Osakabe, and et al. 2017. "Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression" International Journal of Molecular Sciences 18, no. 8: 1805. https://doi.org/10.3390/ijms18081805
APA StyleUmei, T. C., Yamakawa, H., Muraoka, N., Sadahiro, T., Isomi, M., Haginiwa, S., Kojima, H., Kurotsu, S., Tamura, F., Osakabe, R., Tani, H., Nara, K., Miyoshi, H., Fukuda, K., & Ieda, M. (2017). Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. International Journal of Molecular Sciences, 18(8), 1805. https://doi.org/10.3390/ijms18081805