Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study
Abstract
:1. Introduction
2. Results
2.1. Prognostic Value of Circulating Tumor Cells (CTCs) Individual Markers
2.2. PrediCTC Prognostic Value
2.3. Efficiency of PrediCTC for Clinical Outcome Assessment
2.4. Competitive Advantage of PrediCTC versus Standard Computed Tomography (CT) Scan Monitoring
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. CTCs Immunoisolation and Gene Expression Analysis
4.3. Data evaluation and Statistical Analysis
4.4. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| mCRC | Metastatic colorectal cancer |
| CT | Computed tomography |
| CTCs | Circulating tumor cells |
| PFS | Progression-free survival |
| OS | Overall survival |
| FDA | Food and Drug Administration |
| EMT | Epithelial to mesenchymal transition |
| CEA | Carcinoembryonic antigen |
| HR | Hazard ratio |
| CI | Confidence interval |
| R | Responder |
| NR | Non-responder |
| ECOG | Eastern Cooperative Oncology Group |
| PS | Performance status |
| Mests. | metastasis |
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Cancer statistics, 2017. CA. Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D.; ESMO Guidelines Working Group. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, III1–III9. [Google Scholar] [CrossRef] [PubMed]
- Sobrero, A.F.; Maurel, J.; Fehrenbacher, L.; Scheithauer, W.; Abubakr, Y.A.; Lutz, M.P.; Vega-Villegas, M.E.; Eng, C.; Steinhauer, E.U.; Prausova, J.; et al. EPIC: Phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.; Clarke, S.; Díaz-Rubio, E.; Scheithauer, W.; Figer, A.; Wong, R.; Koski, S.; Lichinitser, M.; Yang, T.-S.; Rivera, F.; et al. Randomized phase III study of capecitabine plus oxaliplatin compared with fluorouracil/folinic acid plus oxaliplatin as first-line therapy for metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 2006–2012. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos, G.P.; Batziou, C.; Trafalis, D.; Koutantos, J.; Batzios, S.; Stathopoulos, J.; Legakis, J.; Armakolas, A. Treatment of colorectal cancer with and without bevacizumab: A phase III study. Oncology 2010, 78, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Passardi, A.; Nanni, O.; Tassinari, D.; Turci, D.; Cavanna, L.; Fontana, A.; Ruscelli, S.; Mucciarini, C.; Lorusso, V.; Ragazzini, A.; et al. Effectiveness of bevacizumab added to standard chemotherapy in metastatic colorectal cancer: Final results for first-line treatment from the ITACa randomized clinical trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumors: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Verheul, H.M.W.; Flamen, P.; Rougier, P.; Beets-Tan, R.; Glynne-Jones, R.; Seufferlein, T. Imaging in colorectal cancer: Progress and challenges for the clinicians. Cancers 2016, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; Miller, M.C.; et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009, 20, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V; Terstappen, L.W.W.M. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed]
- Hartkopf, A.D.; Wagner, P.; Wallwiener, D.; Fehm, T.; Rothmund, R. Changing levels of circulating tumor cells in monitoring chemotherapy response in patients with metastatic breast cancer. Anticancer Res. 2011, 31, 979–984. [Google Scholar] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Barbazán, J.; Muinelo-Romay, L.; Vieito, M.; Candamio, S.; Díaz-López, A.; Cano, A.; Gómez-Tato, A.; de los Ángeles Casares de Cal, M.; Abal, M.; López-López, R. A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int. J. Cancer 2014, 135, 2633–2643. [Google Scholar] [CrossRef]
- Barbazán, J.; Alonso-Alconada, L.; Muinelo-Romay, L.; Vieito, M.; Abalo, A.; Alonso-Nocelo, M.; Candamio, S.; Gallardo, E.; Fernández, B.; Abdulkader, I.; et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PLoS ONE 2012, 7, e40476. [Google Scholar] [CrossRef] [PubMed]
- Barbazán, J.; Alonso-Alconada, L.; Elkhatib, N.; Geraldo, S.; Gurchenkov, V.; Glentis, A.; van Niel, G.; Palmulli, R.; Fernandez, B.; Viaño, P.; et al. Liver metastasis is facilitated by the adherence of circulating tumor cells to vascular fibronectin deposits. Cancer Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Barbazán, J.; Vieito, M.; Abalo, A.; Alonso-Alconada, L.; Muinelo-Romay, L.; Alonso-Nocelo, M.; León, L.; Candamio, S.; Gallardo, E.; Anido, U.; et al. A logistic model for the detection of circulating tumor cells in human metastatic colorectal cancer. J. Cell. Mol. Med. 2012, 16, 2342–2349. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, J.E.; Grover, P.; Winter, M.; Hewett, P.J.; Price, T.J.; Thierry, B. Detection and clinical significance of circulating tumor cells in colorectal cancer--20 years of progress. Mol. Med. 2015, 21, S25–S31. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Xu, H.; Wang, Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 2015, 15, 202. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kunkel, M.; Joudeh, J.; Dicker, D.T.; Scicchitano, A.; Allen, J.E.; Sarwani, N.; Yang, Z.; Kaifi, J.; Zhu, J.; et al. Clinico-pathological correlation of serial measurement of circulating tumor cells in 24 metastatic colorectal cancer patients receiving chemotherapy reveals interpatient heterogeneity correlated with CEA levels but independent of KRAS and BRAF mutation. Cancer Biol. Ther. 2015, 16, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Stein, A.; Quidde, J.; Hauch, S.; Röck, K.; Riethdorf, S.; Joosse, S.A.; Pantel, K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the CellSearch® System and the AdnaTest®. PLoS ONE 2016, 11, e0155126. [Google Scholar] [CrossRef] [PubMed]
- Vojtechova, G.; Benesova, L.; Belsanova, B.; Minarikova, P.; Levy, M.; Lipska, L.; Suchanek, S.; Zavoral, M.; Minarik, M. Monitoring of circulating tumor cells by a combination of immunomagnetic enrichment and RT-PCR in colorectal cancer patients undergoing surgery. Adv. Clin. Exp. Med. 2016, 25, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumor cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Hinz, S.; Hendricks, A.; Witting, A.; Schafmayer, C.; Tepel, J.; Kalthoff, A.; Becker, T.; Röder, C. Detection of circulating tumor cells with CK20 RT-PCR is an independent negative prognostic marker in colon cancer patients—A prospective study. BMC Cancer 2017, 17, 53. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Zhang, K.; Yang, X.; Ding, J.; Wang, Z.; Li, J. Prognostic value of circulating tumor DNA in patients with colon cancer: Systematic review. PLoS ONE 2017, 12, e0171991. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.R.C.; Zhou, L.; El-Deiry, W.S. Circulating tumor cells versus circulating tumor DNA in colorectal cancer: Pros and cons. Curr. Colorectal Cancer Rep. 2016, 12, 151–161. [Google Scholar] [CrossRef] [PubMed]
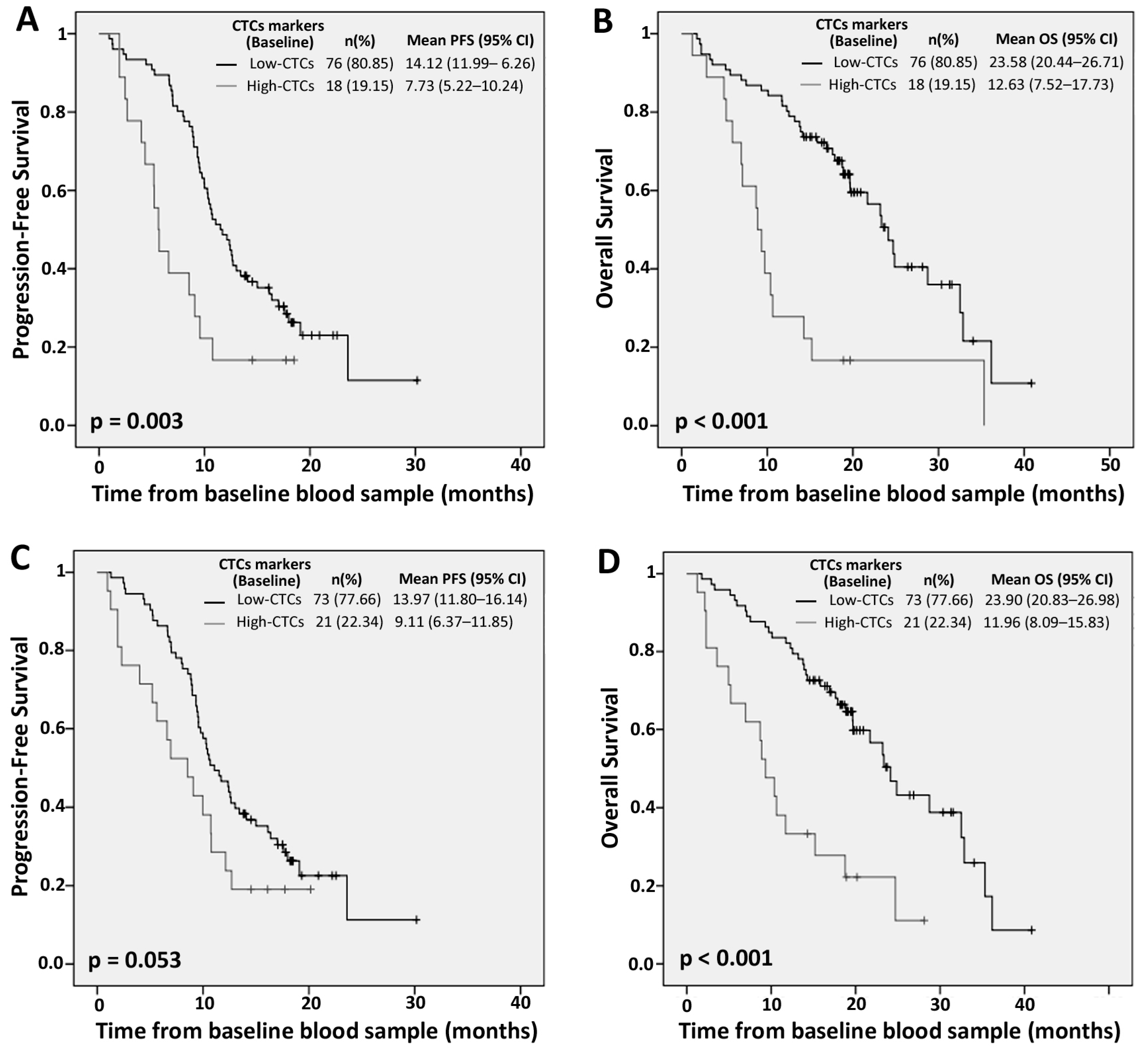
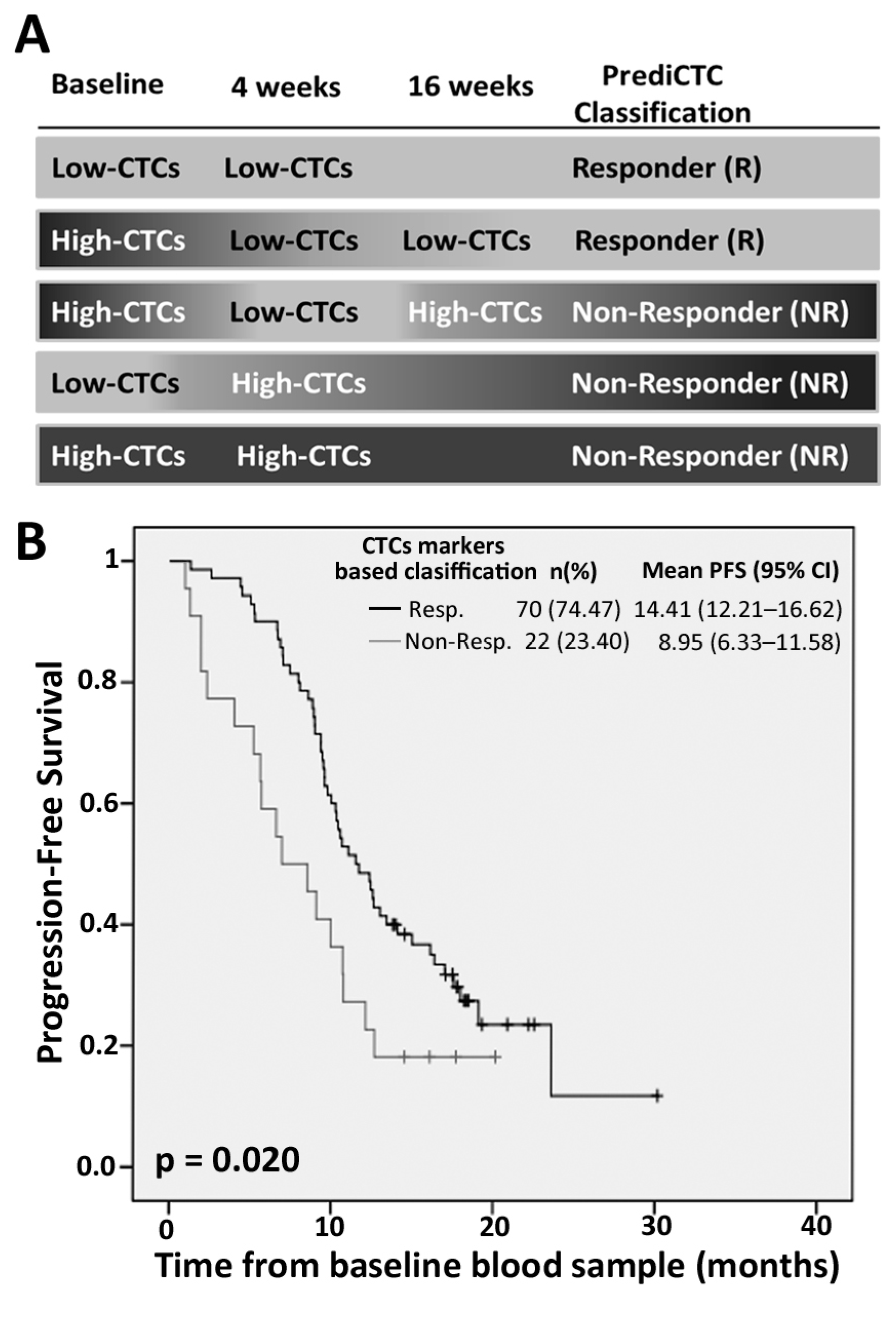
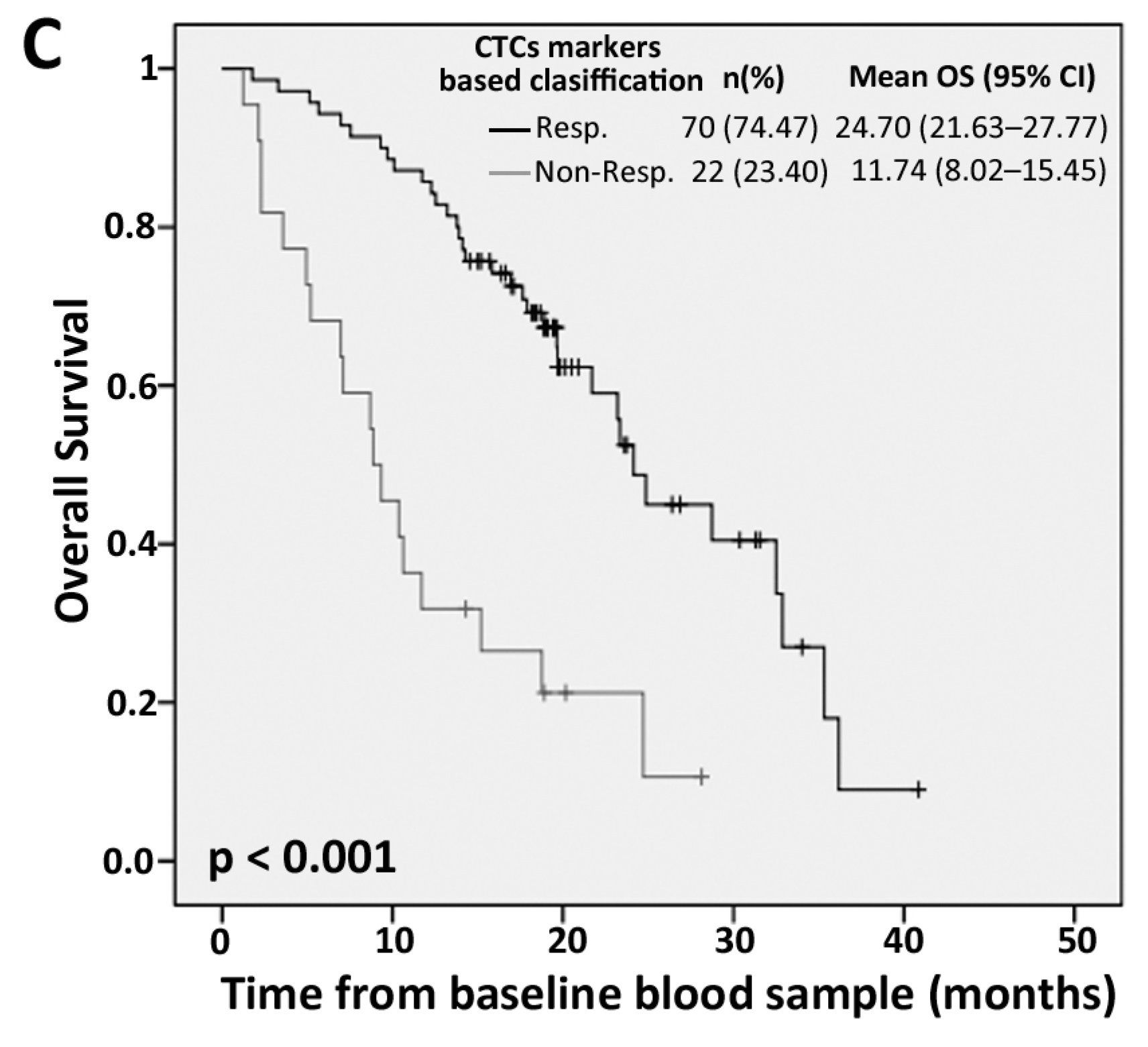
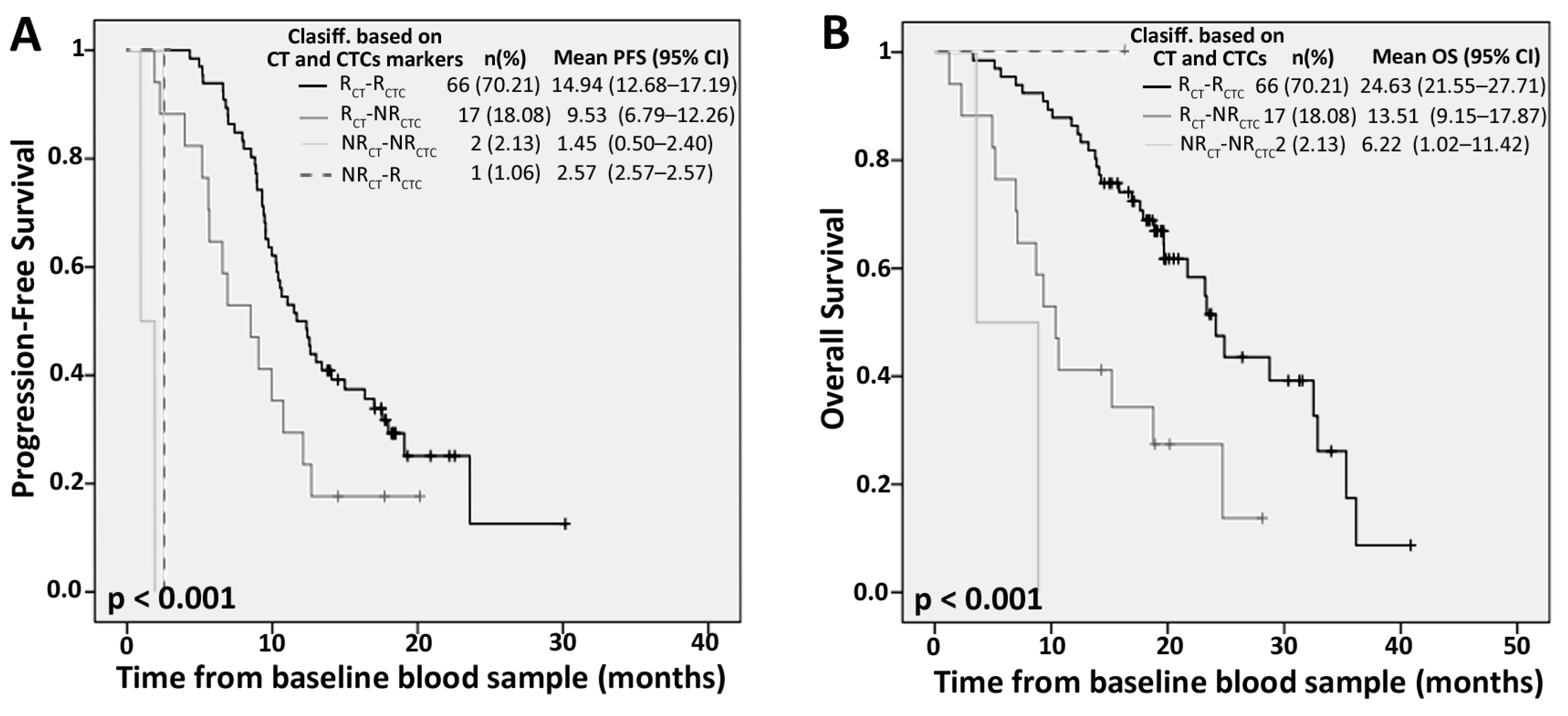
| Marker Levels | PFS | OS | ||||
|---|---|---|---|---|---|---|
| Mean | 95% CI | p Value | Mean | 95% CI | p Value | |
| Baseline | ||||||
| GAPDH | ||||||
| Low | 14.05 | 11.83–16.26 | 0.052 | 22.93 | 19.61–26.26 | 0.041 |
| High | 9.40 | 6.84–11.96 | 17.30 | 11.79–22.81 | ||
| VIL1 | ||||||
| Low | 14.15 | 11.94–16.63 | 0.009 | 23.60 | 20.41–26.79 | 0.001 |
| High | 8.69 | 6.43–10.95 | 13.90 | 10.11–17.69 | ||
| CLU | ||||||
| Low | 14.49 | 12.24–16.75 | 0.001 | 23.24 | 19.92–26.56 | 0.014 |
| High | 7.97 | 5.93–10.00 | 16.01 | 10.96–21.05 | ||
| TIMP1 | ||||||
| Low | 14.24 | 12.09–16.39 | 0.008 | 24.32 | 21.07–27.58 | <0.001 |
| High | 8.34 | 5.92–10.77 | 12.74 | 8.37–17.11 | ||
| TLN1 | ||||||
| Low | 13.66 | 11.49–15.83 | 0.210 | 23.21 | 20.03–26.39 | 0.004 |
| High | 9.95 | 7.33–12.56 | 15.67 | 10.74–20.61 | ||
| LOXL3 | ||||||
| Low | 14.30 | 11.97–16.63 | 0.012 | 23.10 | 19.78–26.43 | 0.019 |
| High | 8.94 | 6.77–11.11 | 16.76 | 11.45–22.06 | ||
| ZEB2 | ||||||
| Low | 14.08 | 11.78–16.37 | 0.064 | 23.53 | 19.93–27.13 | 0.009 |
| High | 9.39 | 7.05–11.74 | 16.32 | 11.24–21.38 | ||
| Four-week follow-up | ||||||
| GAPDH | ||||||
| Low | 14.03 | 11.91–16.15 | 0.046 | 24.05 | 21.02–27.07 | <0.001 |
| High | 9.01 | 6.16–11.87 | 11.85 | 7.83–15.87 | ||
| VIL1 | ||||||
| Low | 13.64 | 11.48–15.80 | 0.277 | 23.06 | 20.17–25.95 | 0.006 |
| High | 10.31 | 7.62–13.01 | 15.165 | 9.46–20.86 | ||
| CLU | ||||||
| Low | 13.66 | 11.47–15.84 | 0.241 | 22.80 | 19.90–25.70 | 0.021 |
| High | 10.63 | 7.69–13.57 | 16.38 | 10.45–22.31 | ||
| TIMP1 | ||||||
| Low | 13.81 | 11.65–15.96 | 0.144 | 23.89 | 20.76–27.02 | <0.001 |
| High | 9.62 | 6.96–12.28 | 12.83 | 9.07–16.59 | ||
| TLN1 | ||||||
| Low | 13.66 | 11.57–15.75 | 0.268 | 23.74 | 20.66–26.82 | <0.001 |
| High | 9.83 | 7.00–12.66 | 13.06 | 9.10–17.02 | ||
| LOXL3 | ||||||
| Low | 14.24 | 12.05–16.43 | 0.013 | 23.75 | 20.64–26.86 | 0.001 |
| High | 8.53 | 6.01–11.05 | 14.31 | 9.06–19.57 | ||
| ZEB2 | ||||||
| Low | 14.02 | 11.87–16.17 | 0.049 | 22.96 | 19.87–26.06 | 0.022 |
| High | 9.31 | 6.22–12.39 | 16.35 | 10.78–21.91 | ||
| Covariate | n | PFS | p Value | OS | p Value |
|---|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | ||||
| Age (≥66 vs. <66 years) | 94 | 0.76 (0.48–1.22) | 0.255 | 0.99 (0.58–1.69) | 0.966 |
| Sex (female vs. Male) | 94 | 0.69 (0.42–1.14) | 0.148 | 0.45 (0.24–0.87) | 0.017 |
| T stage (4 vs. ≤3) | 64 | 0.86 (0.44–1.69) | 0.667 | 0.84 (0.36–1.93) | 0.678 |
| N stage (2 vs. ≤1) | 59 | 1.50 (0.85–2.66) | 0.161 | 0.90 (0.44–1.87) | 0.786 |
| Hepatic mets. (yes vs. no) | 91 | 0.76 (0.36–1.60) | 0.474 | 0.98 (0.42–2.30) | 0.964 |
| Lung mets. (yes vs. no) | 91 | 2.60 (1.54–4.38) | <0.001 | 2.45 (1.39–4.34) | 0.002 |
| Peritoneal mets. (yes vs. no) | 91 | 1.27 (0.71–2.30) | 0.422 | 1.53 (0.81–2.92) | 0.192 |
| Ganglionar mets. (yes vs. no) | 91 | 1.26 (0.75–2.12) | 0.391 | 1.00 (0.54–1.85) | 0.996 |
| No. of metastatic sites (≥2 vs. 1) | 91 | 1.41 (0.88–2.26) | 0.151 | 1.40 (0.80–2.44) | 0.237 |
| KRAS (mut. vs. WT) | 91 | 0.92 (0.56–1.52) | 0.754 | 1.07 (0.60–1.91) | 0.815 |
| Baseline CEA (≥10 vs. <10ng/mL) | 91 | 1.27 (0.75–2.14) | 0.379 | 1.03 (0.56–1.89) | 0.918 |
| ECOG PS (2 vs. ≤1) | 94 | 1.51 (0.81–2.82) | 0.199 | 1.91 (0.98–3.74) | 0.058 |
| Antibodies therapy (yes vs. no) | 94 | 1.02 (0.64–1.63) | 0.932 | 1.11 (0.65–1.90) | 0.711 |
| Baseline CTCs model (high-CTCs vs. low-CTCs) | 94 | 2.33 (1.30–4.15) | 0.004 | 3.37 (1.84–6.18) | <0.001 |
| 4 weeks CTCs model (high-CTCs vs. low-CTCs) | 94 | 1.71 (0.99–2.95) | 0.056 | 3.30 (1.82–5.99) | <0.001 |
| CTCs model (NR vs. R) | 92 | 1.88 (1.09–3.24) | 0.022 | 3.83 (2.10–6.94) | <0.001 |
| Covariates | n | PFS | p Value | χ2 (1) | OS | p Value | χ2 (1) |
|---|---|---|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | ||||||
| Lung metastasis (yes vs. no) | 91 | 2.82 (1.66–4.78) | <0.001 | 18.63 | 2.75 (1.54–4.90) | 0.001 | 22.29 |
| Baseline CTCs model (high-CTCs vs. low-CTCs) | 2.42 (1.33–4.43) | 0.004 | 3.66 (1.93–6.92) | <0.001 | |||
| Lung metastasis (yes vs. no) | 91 | 2.81 (1.65–4.77) | <0.001 | 15.31 | 2.49 (1.41–4.15) | 0.002 | 20.85 |
| 4 weeks CTCs model (high-CTCs vs. low-CTCs) | 1.80 (1.02–3.19) | 0.043 | 3.22 (1.74–5.95) | <0.001 | |||
| Lung metastasis (yes vs. no) | 89 | 3.09 (1.80–5.30) | <0.001 | 18.50 | 2.76 (1.54–4.95) | 0.001 | 25.80 |
| CTCs model (NR vs. R) | 2.06 (1.17–3.62) | 0.012 | 3.82 (2.06–7.09) | <0.001 |
| Characteristic | n (%) | Characteristic | n (%) |
|---|---|---|---|
| Gender | Metastasis location | ||
| Male | 59 (62.77) | Liver | 38 (40.43) |
| Female | 35 (37.23) | Liver and other | 44 (46.81) |
| Primary tumor location | Nonliver | 9 (9.57) | |
| Colon | 63 (67.02) | Unknown | 3 (3.19) |
| Rectum | 29 (30.85) | ECOG PS grade | |
| Both | 2 (2.13) | 0 | 10 (10.64) |
| KRAS status | 1 | 70 (74.47) | |
| Wild type | 55 (58.51) | 2 | 14 (14.89) |
| Mutated | 36 (38.30) | First-line chemotherapy | |
| Unknown | 3 (3.19) | Folfox | 70 (74.47) |
| T | Folfiri | 11 (11.70) | |
| T1–T2 | 3 (3.19) | Capecitabine | 5 (5.32) |
| T3 | 46 (48.94) | Capecitabine-Oxaliplatin | 6 (6.38) |
| T4 | 15 (15.96) | Irinotecan | 2 (2.13) |
| Tx | 30 (31.91) | First-line combined biological therapy | |
| N | Anti-EGFR | 35 (37.23) | |
| N0 | 10 (10.63) | Anti-VEGF | 12 (12.76) |
| N1 | 24 (25.53) | None | 47 (50) |
| N2 | 25 (26.59) | ||
| N3 | 1 (1.06) | ||
| Nx | 34 (36.17) | ||
| Number of metastatic sites | |||
| 1 | 45 (47.88) | ||
| ≥2 | 46 (48.93) | ||
| Unknown | 3 (3.19) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidal Insua, Y.; De la Cámara, J.; Brozos Vázquez, E.; Fernández, A.; Vázquez Rivera, F.; Villanueva Silva, M.J.; Barbazán, J.; Muinelo-Romay, L.; Candamio Folgar, S.; Abalo, A.; et al. Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study. Int. J. Mol. Sci. 2017, 18, 1265. https://doi.org/10.3390/ijms18061265
Vidal Insua Y, De la Cámara J, Brozos Vázquez E, Fernández A, Vázquez Rivera F, Villanueva Silva MJ, Barbazán J, Muinelo-Romay L, Candamio Folgar S, Abalo A, et al. Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study. International Journal of Molecular Sciences. 2017; 18(6):1265. https://doi.org/10.3390/ijms18061265
Chicago/Turabian StyleVidal Insua, Yolanda, Juan De la Cámara, Elena Brozos Vázquez, Ana Fernández, Francisca Vázquez Rivera, Mª José Villanueva Silva, Jorge Barbazán, Laura Muinelo-Romay, Sonia Candamio Folgar, Alicia Abalo, and et al. 2017. "Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study" International Journal of Molecular Sciences 18, no. 6: 1265. https://doi.org/10.3390/ijms18061265
APA StyleVidal Insua, Y., De la Cámara, J., Brozos Vázquez, E., Fernández, A., Vázquez Rivera, F., Villanueva Silva, M. J., Barbazán, J., Muinelo-Romay, L., Candamio Folgar, S., Abalo, A., López-López, R., Abal, M., & Alonso-Alconada, L. (2017). Predicting Outcome and Therapy Response in mCRC Patients Using an Indirect Method for CTCs Detection by a Multigene Expression Panel: A Multicentric Prospective Validation Study. International Journal of Molecular Sciences, 18(6), 1265. https://doi.org/10.3390/ijms18061265







