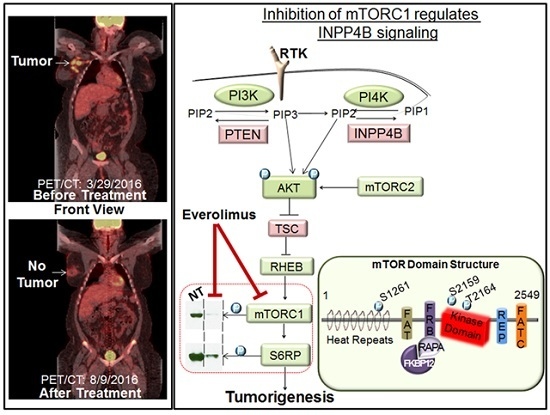
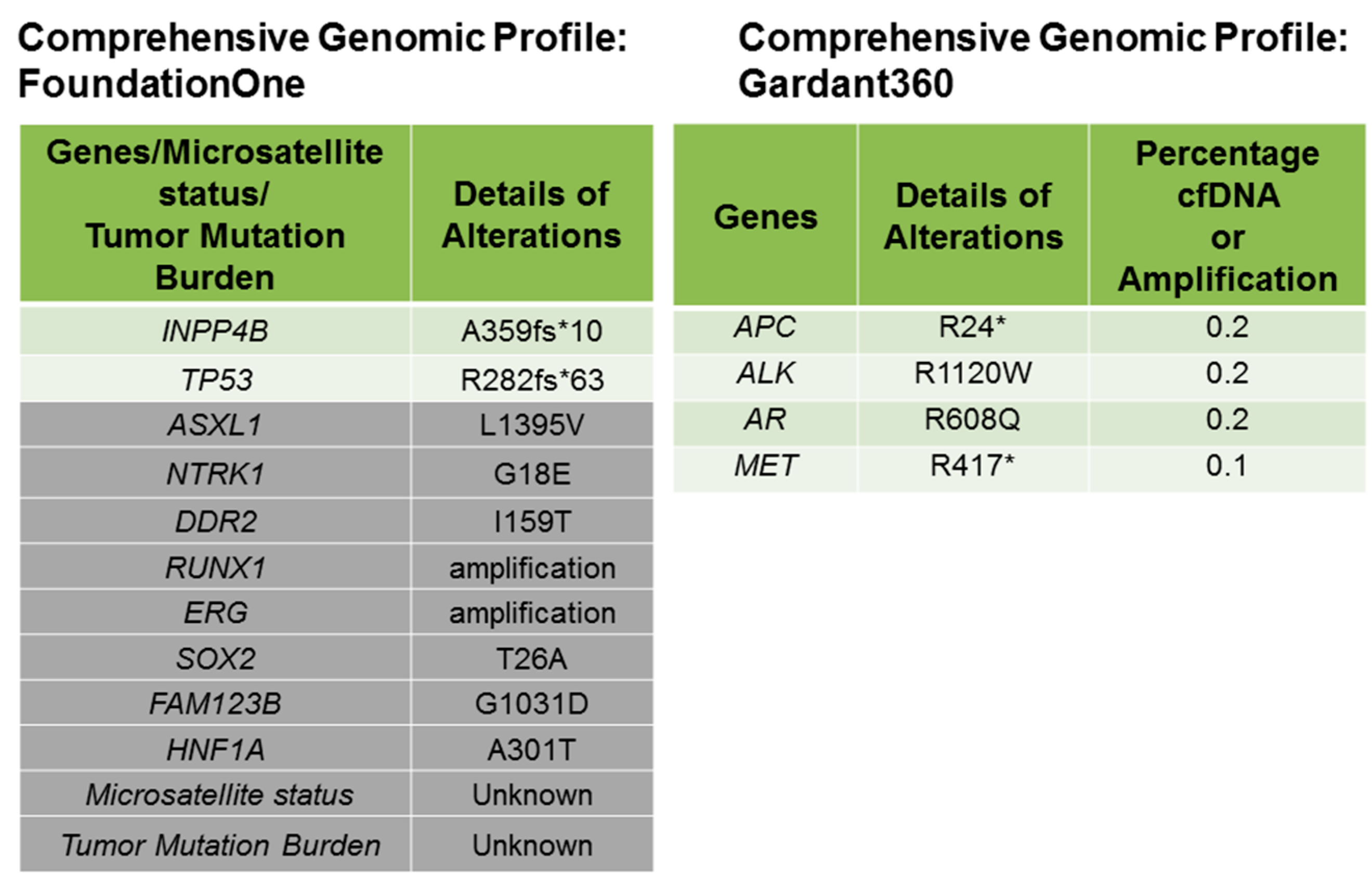
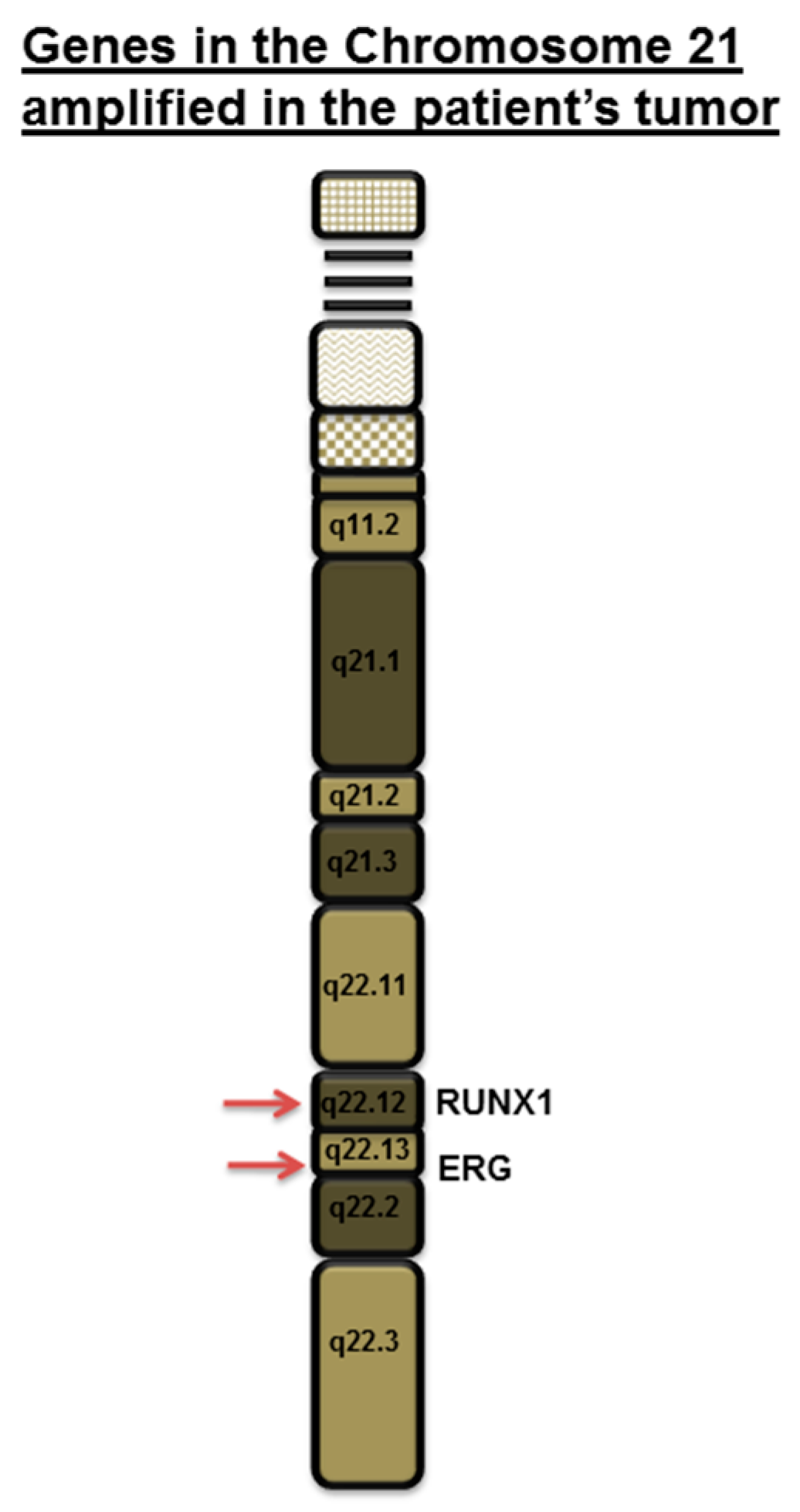
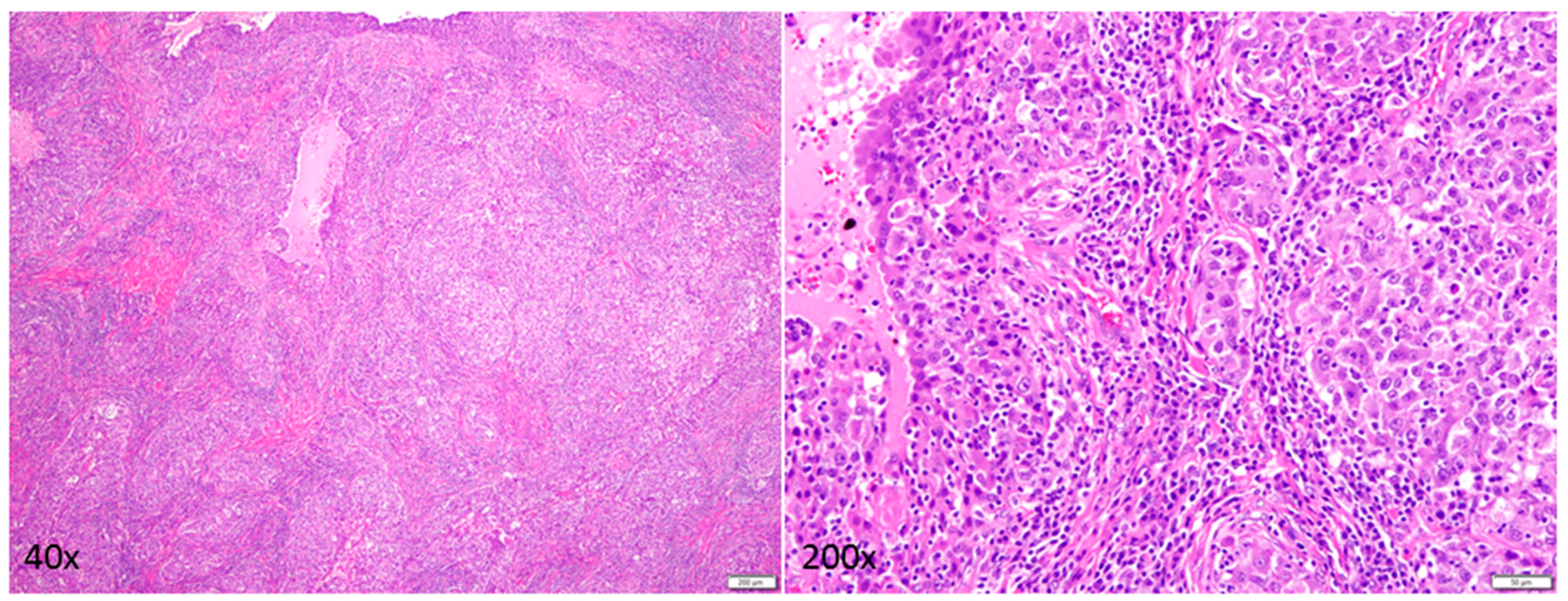
Down’s Syndrome and Triple Negative Breast Cancer: A Rare Occurrence of Distinctive Clinical Relationship
Abstract
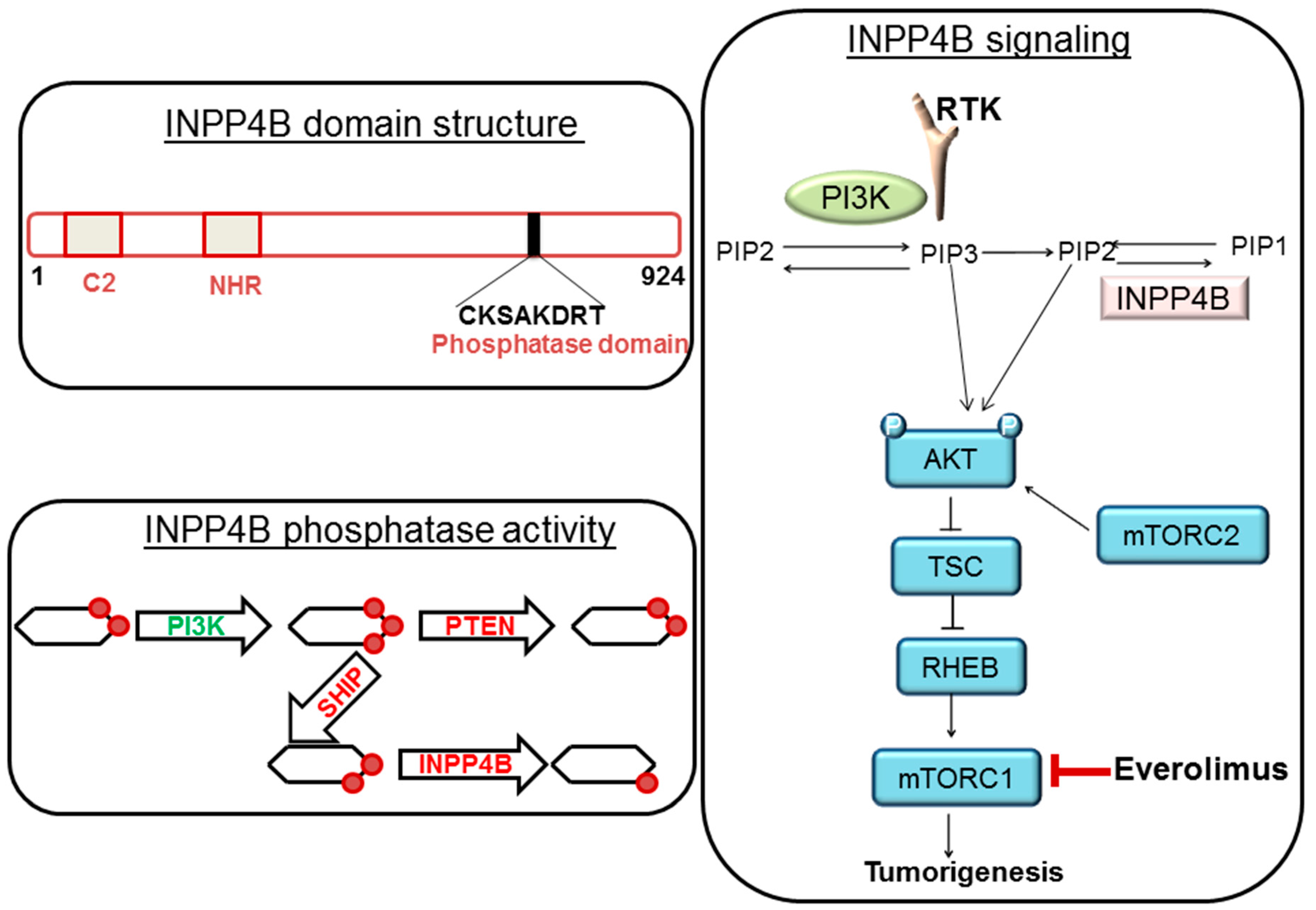
:1. Background
2. Case Description
3. Discussion
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Xavier, A.C.; Ge, Y.; Taub, J.W. Down syndrome and malignancies: A unique clinical relationship: A paper from the 2008 william beaumont hospital symposium on molecular pathology. J. Mol. Diagn. 2009, 11, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Satge, D.; Schorderet, D.F.; Balmer, A.; Beck-Popovic, M.; Addor, M.C.; Beckmann, J.S.; Munier, F.L. Association Down syndrome-retinoblastoma: A new observation. Ophthalmic Genet. 2005, 26, 151–152. [Google Scholar] [CrossRef] [PubMed]
- Hsiung Stripp, D.C.; Vaughn, D.; van Arsdalen, K.; Whittington, R. Three cases of advanced seminoma and Down’s syndrome: A possible association. Am. J. Clin. Oncol. 2003, 26, 197–199. [Google Scholar] [CrossRef] [PubMed]
- Satge, D.; Le Tourneau, A.; Verger, J.P.; Lefort, S.; Geneix, A.; Malet, P.; Diebold, J.; Vekemans, M. A case report of Down syndrome and centroblastic lymphoma. Pathol Res. Pract. 1996, 192, 1266–1269. [Google Scholar] [CrossRef]
- Patja, K.; Eero, P.; Iivanainen, M. Cancer incidence among people with intellectual disability. J. Intellect. Disabil. Res. 2001, 45, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Satge, D.; Sasco, A.J.; Pujol, H.; Rethore, M.O. Breast cancer in women with trisomy 21. Bull. Acad Natl Med. 2001, 185, 1239–1252, discussion 1252–1254. [Google Scholar] [PubMed]
- Scholl, T.; Stein, Z.; Hansen, H. Leukemia and other cancers, anomalies and infections as causes of death in Down’s syndrome in the United States during 1976. Dev. Med. Child. Neurol. 1982, 24, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Chicoine, B.; Roth, M.; Chicoine, L.; Sulo, S. Breast cancer screening for women with Down syndrome: Lessons learned. Intellect Dev. Disabil. 2015, 53, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Martel-Billard, C.; Cordier, C.; Tomasetto, C.; Jegu, J.; Mathelin, C. Trisomy 21 and breast cancer: A genetic abnormality which protects against breast cancer? Gynecol. Obstet Fertil. 2016, 44, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Korbel, J.O.; Tirosh-Wagner, T.; Urban, A.E.; Chen, X.N.; Kasowski, M.; Dai, L.; Grubert, F.; Erdman, C.; Gao, M.C.; Lange, K.; et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA 2009, 106, 12031–12036. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Bursts of chromosome changes drive TNBC. Cancer Discov. 2016, 6, 1075. [Google Scholar]
- Pfefferle, A.D.; Agrawal, Y.N.; Koboldt, D.C.; Kanchi, K.L.; Herschkowitz, J.I.; Mardis, E.R.; Rosen, J.M.; Perou, C.M. Genomic profiling of murine mammary tumors identifies potential personalized drug targets for p53-deficient mammary cancers. Dis. Model. Mech. 2016, 9, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Smith, B.R.; Leyland-Jones, B. Targeting basal-like breast cancers. Curr. Drug Targets 2012, 13, 1510–1524. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Ellis, M.J.; Perou, C.M. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 2013, 3, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Fedele, C.G.; Ooms, L.M.; Ho, M.; Vieusseux, J.; O’Toole, S.A.; Millar, E.K.; Lopez-Knowles, E.; Sriratana, A.; Gurung, R.; Baglietto, L.; et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2010, 107, 22231–22236. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Carlson, J.H.; Leyland-Jones, B.; Dey, N. PI3K-AKT-mTOR Pathway Cooperates with the DNA Damage Repair Pathway: Carcinogenesis in Triple-Negative Breast Cancers and Beyond; Teicher, B.A., Ed.; Springer: Heidelberg, Germany, 2016. [Google Scholar]
- Dey, N.; De, P.; Leyland-Jones, B. PI3K–AKT–mTOR inhibitors in breast cancers: From tumor cell signaling to clinical trials. Pharmacol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Dey, N.; Leyland-Jones, B. Growth factor and signaling networks. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 365–369. [Google Scholar]
- Hattori, M.; Fujiyama, A.; Taylor, T.D.; Watanabe, H.; Yada, T.; Park, H.S.; Park, A.; Toyoda, K.; Ishii, Y.; Totoki, D.-K.; et al. The DNA sequence of human chromosome 21. Nature 2000, 405, 311–319. [Google Scholar] [PubMed]
- Janes, K.A. RUNX1 and its understudied role in breast cancer. Cell Cycle 2011, 10, 3461–3465. [Google Scholar] [CrossRef] [PubMed]
- Browne, G.; Taipaleenmaki, H.; Bishop, N.M.; Madasu, S.C.; Shaw, L.M.; van Wijnen, A.J.; Stein, G.S.; Lian, J.B. RUNX1 is associated with breast cancer progression in MMTV–PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J. Cell. Physiol. 2015, 230, 2522–2532. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, N.; Mohammed, Z.M.; Nixon, C.; Mason, S.M.; Mallon, E.; McMillan, D.C.; Morris, J.S.; Cameron, E.R.; Edwards, J.; Blyth, K. Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS ONE 2014, 9, e100759. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Hong, D.; Lajoie, B.R.; McCord, R.P.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Dekker, J.; Imbalzano, A.N.; Stein, G.S.; et al. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim. Biophys. Acta 2016, 1859, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.O.; Ahmed-Alnassar, S.; Frenkel, B. Relationship between RUNX1 and AXIN1 in ER-negative versus ER-positive breast cancer. Cell Cycle 2017, 16, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Barwick, B.G.; Moreno, C.S.; Ordanic-Kodani, M.; Chen, Z.; Oprea-Ilies, G.; Tang, W.; Catzavelos, C.; Kerstann, K.F.; Sledge, G.W., Jr.; et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer 2013, 13, 537. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Young, B.; Abramovitz, M.; Bouzyk, M.; Barwick, B.; De, P. Differential activation of Wnt-β-catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PLoS ONE 2013, 8, e77425. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Carlson, J.H.; Jepperson, T.; Willis, S.; Leyland-Jones, B.; Dey, N. RAC1 GTP-ase signals Wnt-β-catenin pathway mediated integrin-directed metastasis-associated tumor cell phenotypes in triple negative breast cancers. Oncotarget 2017, 8, 3072–3103. [Google Scholar] [PubMed]
- De, P.; Carlson, J.H.; Wu, H.; Marcus, A.; Leyland-Jones, B.; Dey, N. Wnt-β-catenin pathway signals metastasis-associated tumor cell phenotypes in triple negative breast cancers. Oncotarget 2016, 7, 43124–43149. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, N.; Krie, A.; Klein, J.; Williams, K.; McMillan, A.; Elsey, R.; Sun, Y.; Williams, C.; De, P.; Leyland-Jones, B. Down’s Syndrome and Triple Negative Breast Cancer: A Rare Occurrence of Distinctive Clinical Relationship. Int. J. Mol. Sci. 2017, 18, 1218. https://doi.org/10.3390/ijms18061218
Dey N, Krie A, Klein J, Williams K, McMillan A, Elsey R, Sun Y, Williams C, De P, Leyland-Jones B. Down’s Syndrome and Triple Negative Breast Cancer: A Rare Occurrence of Distinctive Clinical Relationship. International Journal of Molecular Sciences. 2017; 18(6):1218. https://doi.org/10.3390/ijms18061218
Chicago/Turabian StyleDey, Nandini, Amy Krie, Jessica Klein, Kirstin Williams, Amanda McMillan, Rachel Elsey, Yuliang Sun, Casey Williams, Pradip De, and Brian Leyland-Jones. 2017. "Down’s Syndrome and Triple Negative Breast Cancer: A Rare Occurrence of Distinctive Clinical Relationship" International Journal of Molecular Sciences 18, no. 6: 1218. https://doi.org/10.3390/ijms18061218
APA StyleDey, N., Krie, A., Klein, J., Williams, K., McMillan, A., Elsey, R., Sun, Y., Williams, C., De, P., & Leyland-Jones, B. (2017). Down’s Syndrome and Triple Negative Breast Cancer: A Rare Occurrence of Distinctive Clinical Relationship. International Journal of Molecular Sciences, 18(6), 1218. https://doi.org/10.3390/ijms18061218