BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction
Abstract
:1. Introduction
2. Results
2.1. BDNF Binds Its Pro-Peptide with High Affinity
2.2. The BDNF Val66Met Polymorphism Stabilizes Interaction with Its Pro-Peptide
2.3. The Met-Containing BDNF Pro-Peptide Binds Stably to BDNF over A Wider pH Range
2.4. Structural Analysis of BDNF/Pro-Peptide Interaction and the Influence of the Val66Met Polymorphism
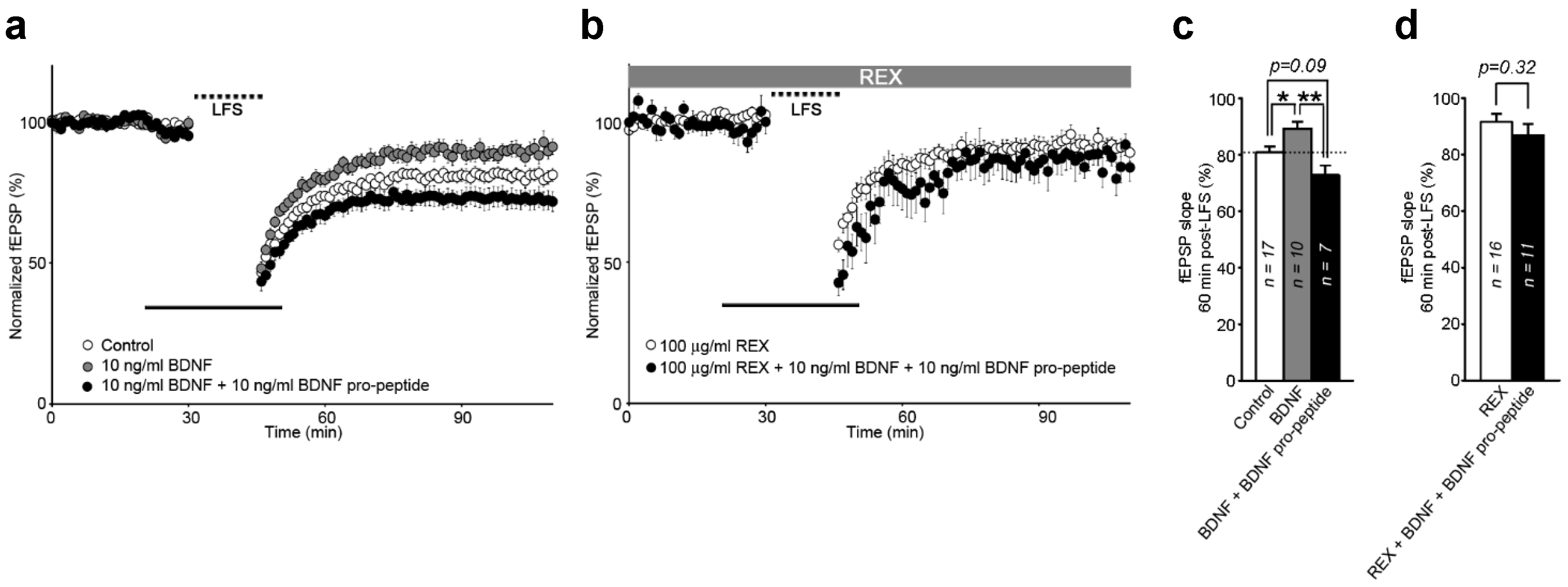
2.5. Interaction of BDNF and Its Pro-Peptide May Affect the Biological Function of BDNF
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Production of Recombinant Proteins
4.3. BIAcore Assay
4.4. In Vitro Binding of BDNF and Its Pro-Peptide
4.5. Computational Modeling
4.6. Hippocampal Slice Preparation
4.7. Electrophysiology
4.8. Statistics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bibel, M.; Barde, Y.A. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000, 14, 2919–2937. [Google Scholar] [CrossRef] [PubMed]
- Leibrock, J.; Lottspeich, F.; Hohn, A.; Hofer, M.; Hengerer, B.; Masiakowski, P.; Thoenen, H.; Barde, Y.A. Molecular cloning and expression of brain-derived neurotrophic factor. Nature 1989, 341, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.F. Emerging themes in structural biology of neurotrophic factors. Trends Neurosci. 1998, 21, 438–444. [Google Scholar] [CrossRef]
- Bothwell, M. Functional interactions of neurotrophins and neurotrophin receptors. Annu. Rev. Neurosci. 1995, 18, 223–253. [Google Scholar] [CrossRef] [PubMed]
- West, A.E.; Pruunsild, P.; Timmusk, T. Neurotrophins: Transcription and translation. Handb. Exp. Pharmacol. 2014, 220, 67–100. [Google Scholar] [PubMed]
- Arancio, O.; Chao, M.V. Neurotrophins, synaptic plasticity and dementia. Curr. Opin. Neurobiol. 2007, 17, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Cory, S.; Kidane, A.H.; Shirkey, N.J.; Marshak, S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Dev. Neurobiol. 2010, 70, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Poo, M.M. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 2013, 14, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Lessmann, V.; Brigadski, T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: An update. Neurosci. Res. 2009, 65, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Nagappan, G.; Lu, Y. BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb. Exp. Pharmacol. 2014, 220, 223–250. [Google Scholar] [PubMed]
- Kolbeck, R.; Jungbluth, S.; Barde, Y.A. Characterisation of neurotrophin dimers and monomers. Eur. J. Biochem. 1994, 225, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Dieni, S.; Matsumoto, T.; Dekkers, M.; Rauskolb, S.; Ionescu, M.S.; Deogracias, R.; Gundelfinger, E.D.; Kojima, M.; Nestel, S.; Frotscher, M.; et al. BDNF and its pro-peptide are stored in presynaptic dense core vesicles in brain neurons. J. Cell. Biol. 2012, 196, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Anastasia, A.; Deinhardt, K.; Chao, M.V.; Will, N.E.; Irmady, K.; Lee, F.S.; Hempstead, B.L.; Bracken, C. Val66Met polymorphism of BDNF alters prodomain structure to induce neuronal growth cone retraction. Nat. Commun. 2013, 4, 2490. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ji, Y.; Ding, Y.; Jiang, W.; Sun, Y.; Lu, B.; Nagappan, G. BDNF pro-peptide regulates dendritic spines via caspase-3. Cell Death Dis. 2016, 7, e2264. [Google Scholar] [CrossRef] [PubMed]
- Mizui, T.; Ishikawa, Y.; Kumanogoh, H.; Lume, M.; Matsumoto, T.; Hara, T.; Yamawaki, S.; Takahashi, M.; Shiosaka, S.; Itami, C.; et al. BDNF pro-peptide actions facilitate hippocampal ltd and are altered by the common BDNF polymorphism val66met. Proc. Natl. Acad. Sci. USA 2015, 112, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.F.; Kojima, M.; Callicott, J.H.; Goldberg, T.E.; Kolachana, B.S.; Bertolino, A.; Zaitsev, E.; Gold, B.; Goldman, D.; Dean, M.; et al. The BDNF Val66Met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003, 112, 257–269. [Google Scholar] [CrossRef]
- Chen, J.X.; Li, W.; Zhao, X.; Yang, J.X. Effects of the chinese traditional prescription xiaoyaosan decoction on chronic immobilization stress-induced changes in behavior and brain BDNF, TRKB, and NT-3 in rats. Cell. Mol. Neurobiol. 2008, 28, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Dincheva, I.; Glatt, C.E.; Lee, F.S. Impact of the BDNF Val66Met polymorphism on cognition: Implications for behavioral genetics. Neuroscientist 2012, 18, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Notaras, M.; Hill, R.; van den Buuse, M. The BDNF gene Val66Met polymorphism as a modifier of psychiatric disorder susceptibility: Progress and controversy. Mol. Psychiatry 2015, 20, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.V.; Bothwell, M. Neurotrophins: To cleave or not to cleave. Neuron 2002, 33, 9–12. [Google Scholar] [CrossRef]
- Lu, B. Pro-region of neurotrophins: Role in synaptic modulation. Neuron 2003, 39, 735–738. [Google Scholar] [CrossRef]
- Chao, M.V. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003, 4, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Jing, D.; Bath, K.G.; Ieraci, A.; Khan, T.; Siao, C.J.; Herrera, D.G.; Toth, M.; Yang, C.; McEwen, B.S.; et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science 2006, 314, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Seet, B.T.; Dikic, I.; Zhou, M.M.; Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006, 7, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.R.; Reichardt, L.F. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc. Natl. Acad. Sci. USA 1990, 87, 8060–8064. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.K.; Felice, S.; Kim, T.; Hempstead, B.L. Understanding proneurotrophin actions: Recent advances and challenges. Dev. Neurobiol. 2010, 70, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Mizui, T.; Ishikawa, Y.; Kumanogoh, H.; Kojima, M. Neurobiological actions by three distinct subtypes of brain-derived neurotrophic factor: Multi-ligand model of growth factor signaling. Pharmacol. Res. 2016, 105, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Kliemannel, M.; Rattenholl, A.; Golbik, R.; Balbach, J.; Lilie, H.; Rudolph, R.; Schwarz, E. The mature part of prongf induces the structure of its pro-peptide. FEBS Lett. 2004, 566, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Ieraci, A.; Teng, H.; Dall, H.; Meng, C.X.; Herrera, D.G.; Nykjaer, A.; Hempstead, B.L.; Lee, F.S. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J. Neurosci. 2005, 25, 6156–6166. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.N. Intermolecular and Surface Forces: With Applications to Colloidal and Biological Systems; Academic Press: London, UK, 1985. [Google Scholar]
- Koshimizu, H.; Kiyosue, K.; Hara, T.; Hazama, S.; Suzuki, S.; Uegaki, K.; Nagappan, G.; Zaitsev, E.; Hirokawa, T.; Tatsu, Y.; et al. Multiple functions of precursor BDNF to CNS neurons: Negative regulation of neurite growth, spine formation and cell survival. Mol. Brain 2009, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Uegaki, K.; Otomo, T.; Sakahira, H.; Shimizu, M.; Yumoto, N.; Kyogoku, Y.; Nagata, S.; Yamazaki, T. Structure of the CAD domain of caspase-activated DNase and interaction with the CAD domain of its inhibitor. J. Mol. Biol. 2000, 297, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
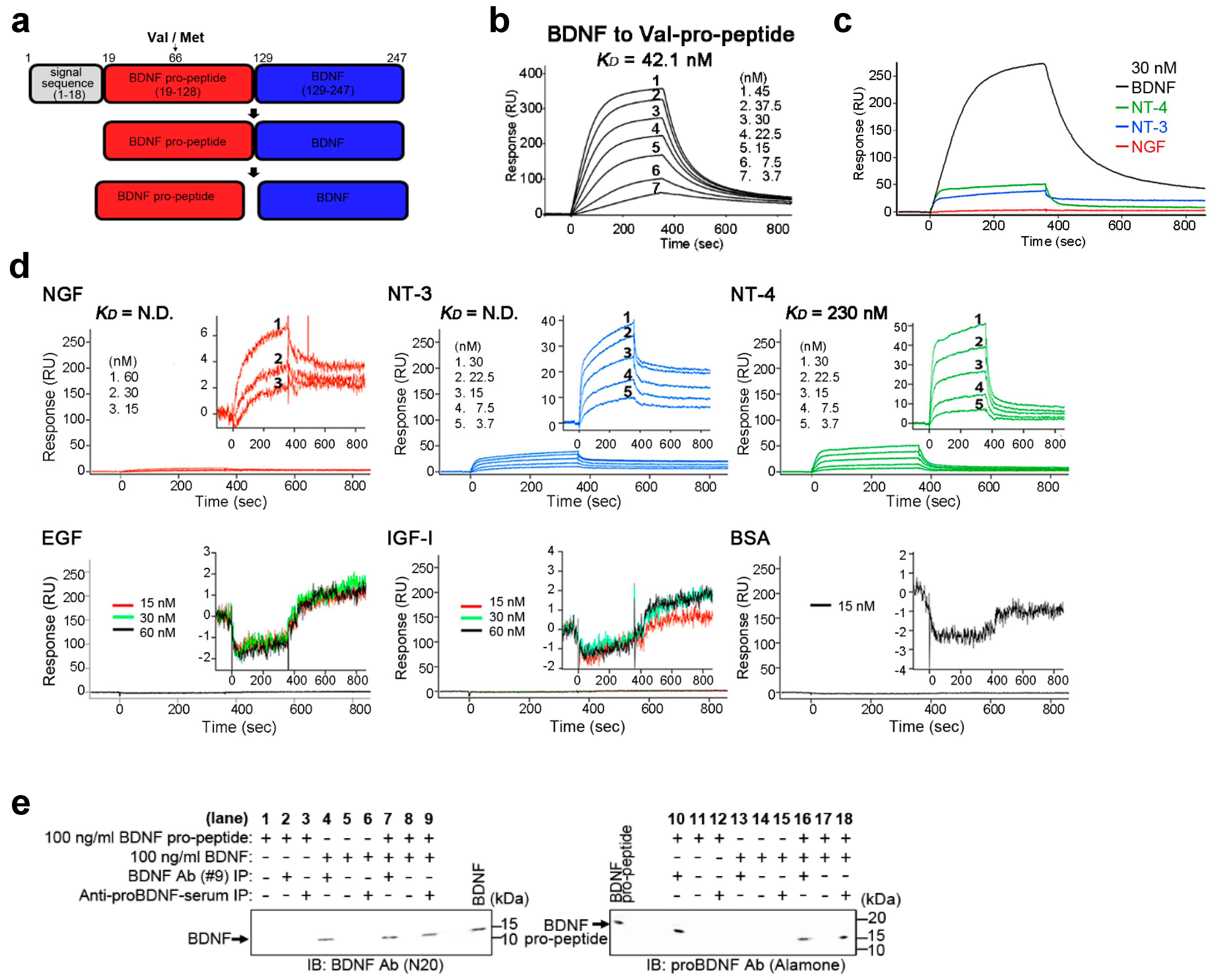
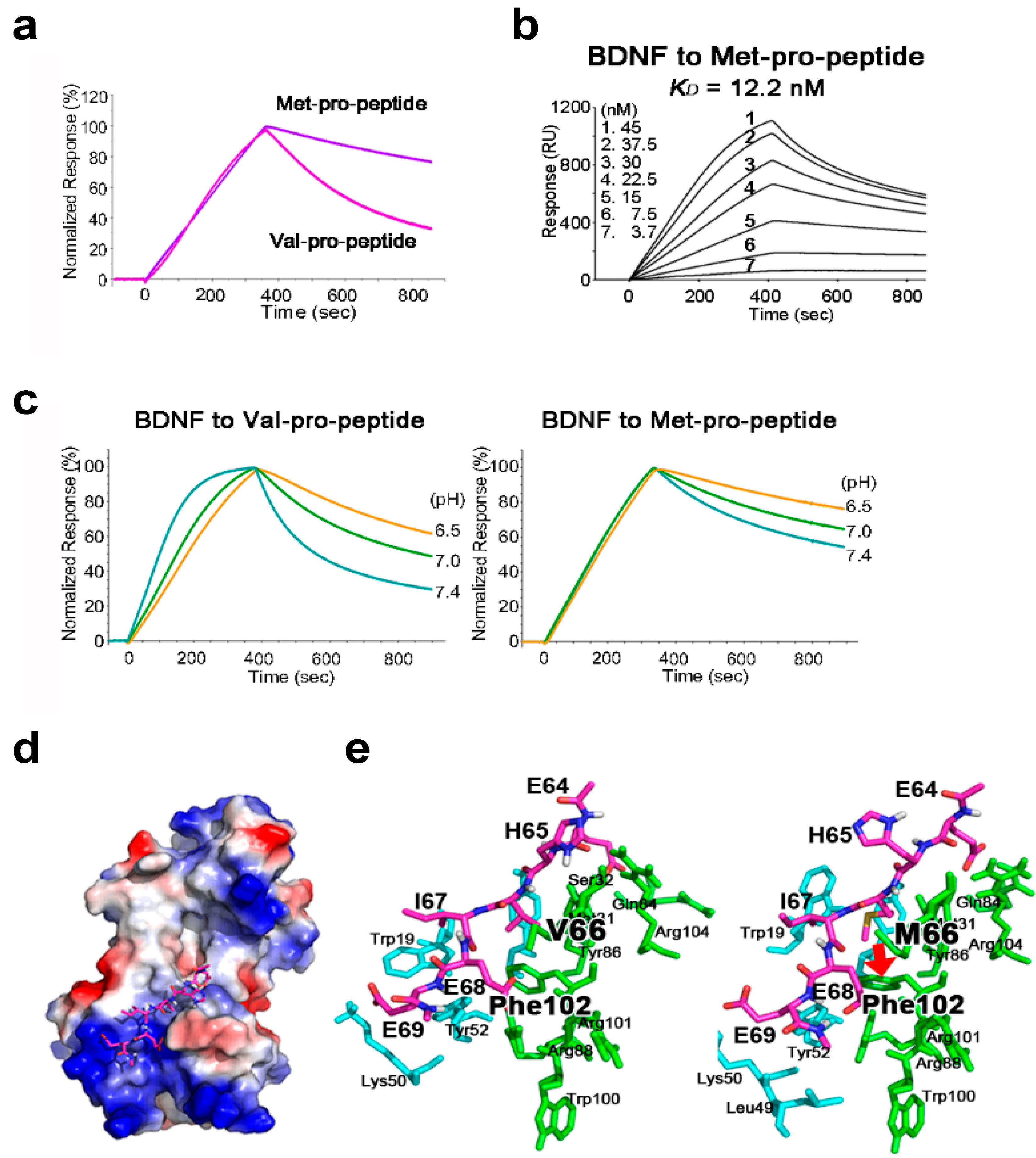
| Type of Pro-Peptide | ka (1/Ms) | kd (1/s) | KD (M) |
|---|---|---|---|
| Val-pro-peptide | 3.78 × 105 | 1.59 × 10−2 | 4.21 × 10−8 |
| Met-pro-peptide | 3.63 × 104 | 4.44 × 10−4 | 1.22 × 10−8 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uegaki, K.; Kumanogoh, H.; Mizui, T.; Hirokawa, T.; Ishikawa, Y.; Kojima, M. BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction. Int. J. Mol. Sci. 2017, 18, 1042. https://doi.org/10.3390/ijms18051042
Uegaki K, Kumanogoh H, Mizui T, Hirokawa T, Ishikawa Y, Kojima M. BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction. International Journal of Molecular Sciences. 2017; 18(5):1042. https://doi.org/10.3390/ijms18051042
Chicago/Turabian StyleUegaki, Koichi, Haruko Kumanogoh, Toshiyuki Mizui, Takatsugu Hirokawa, Yasuyuki Ishikawa, and Masami Kojima. 2017. "BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction" International Journal of Molecular Sciences 18, no. 5: 1042. https://doi.org/10.3390/ijms18051042
APA StyleUegaki, K., Kumanogoh, H., Mizui, T., Hirokawa, T., Ishikawa, Y., & Kojima, M. (2017). BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction. International Journal of Molecular Sciences, 18(5), 1042. https://doi.org/10.3390/ijms18051042






