Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants
Abstract
:1. Introduction
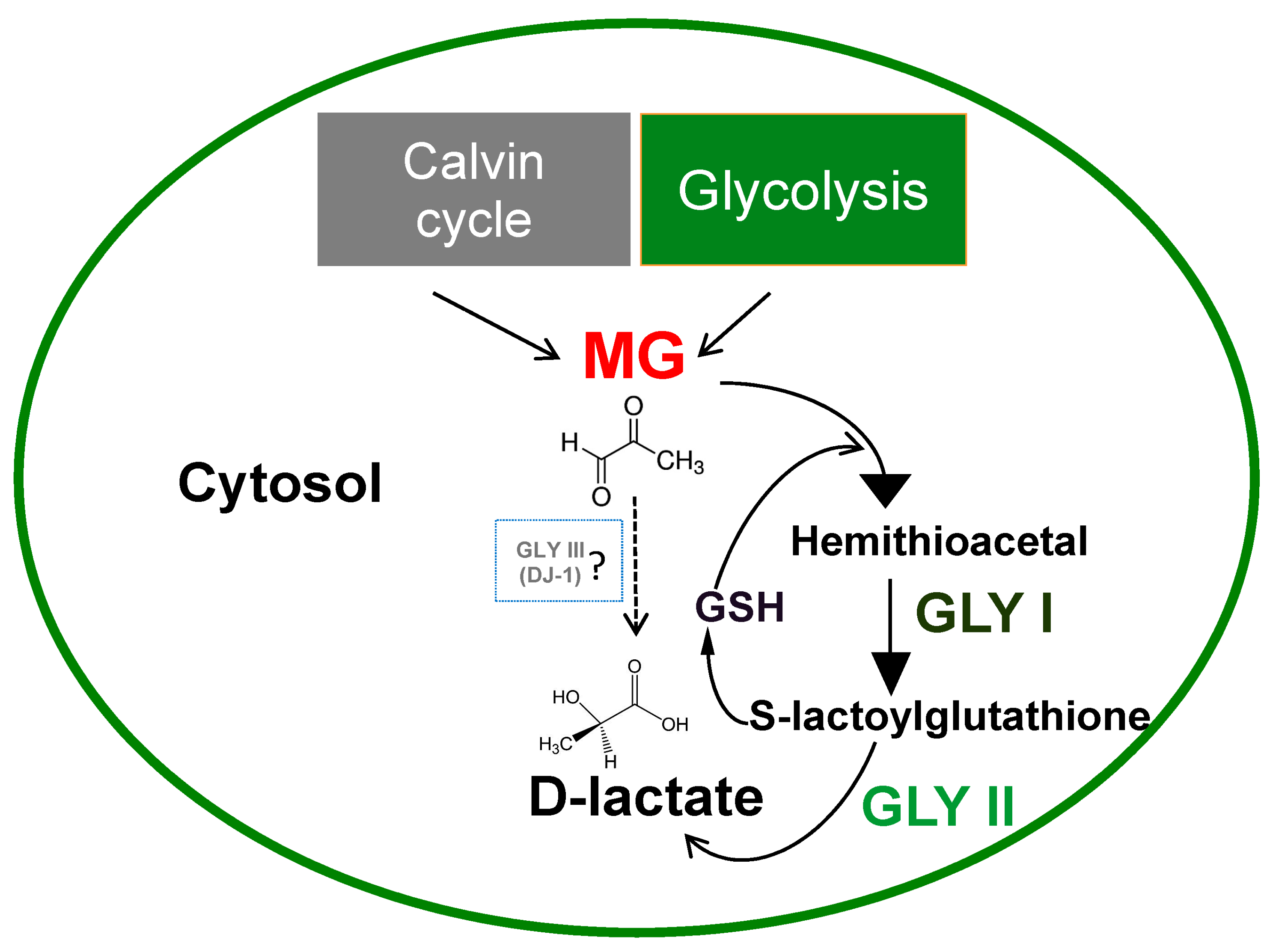
2. Methylglyoxal Detoxification System in Plants
3. Functional Roles of Glyoxalases in Plants: From the Past to the Present
3.1. Glyoxalase as a Marker for Cell Division
3.2. Glyoxalase as a Mitigator of Abiotic Stress
3.3. Glyoxalases and Biotic Stress
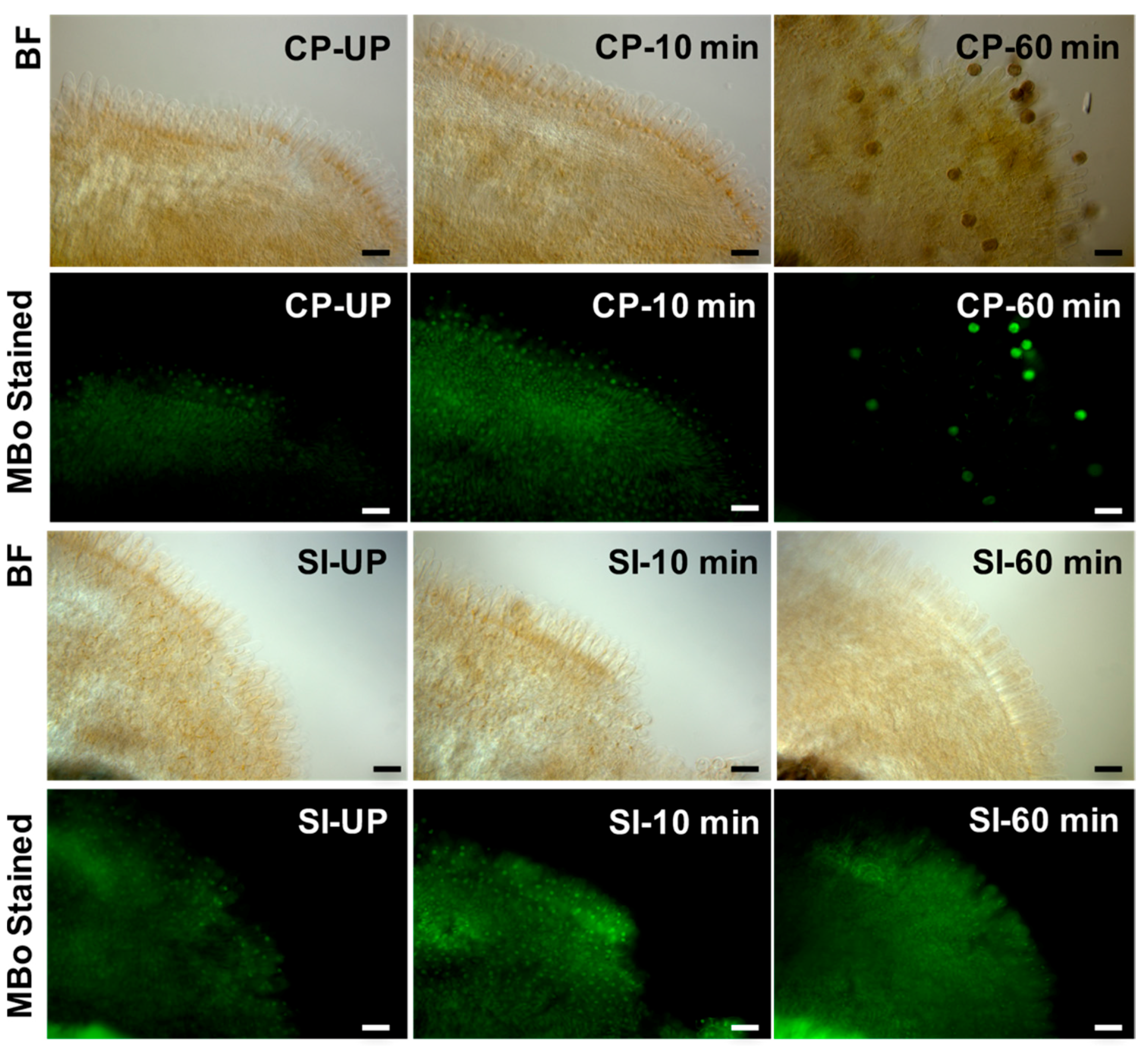
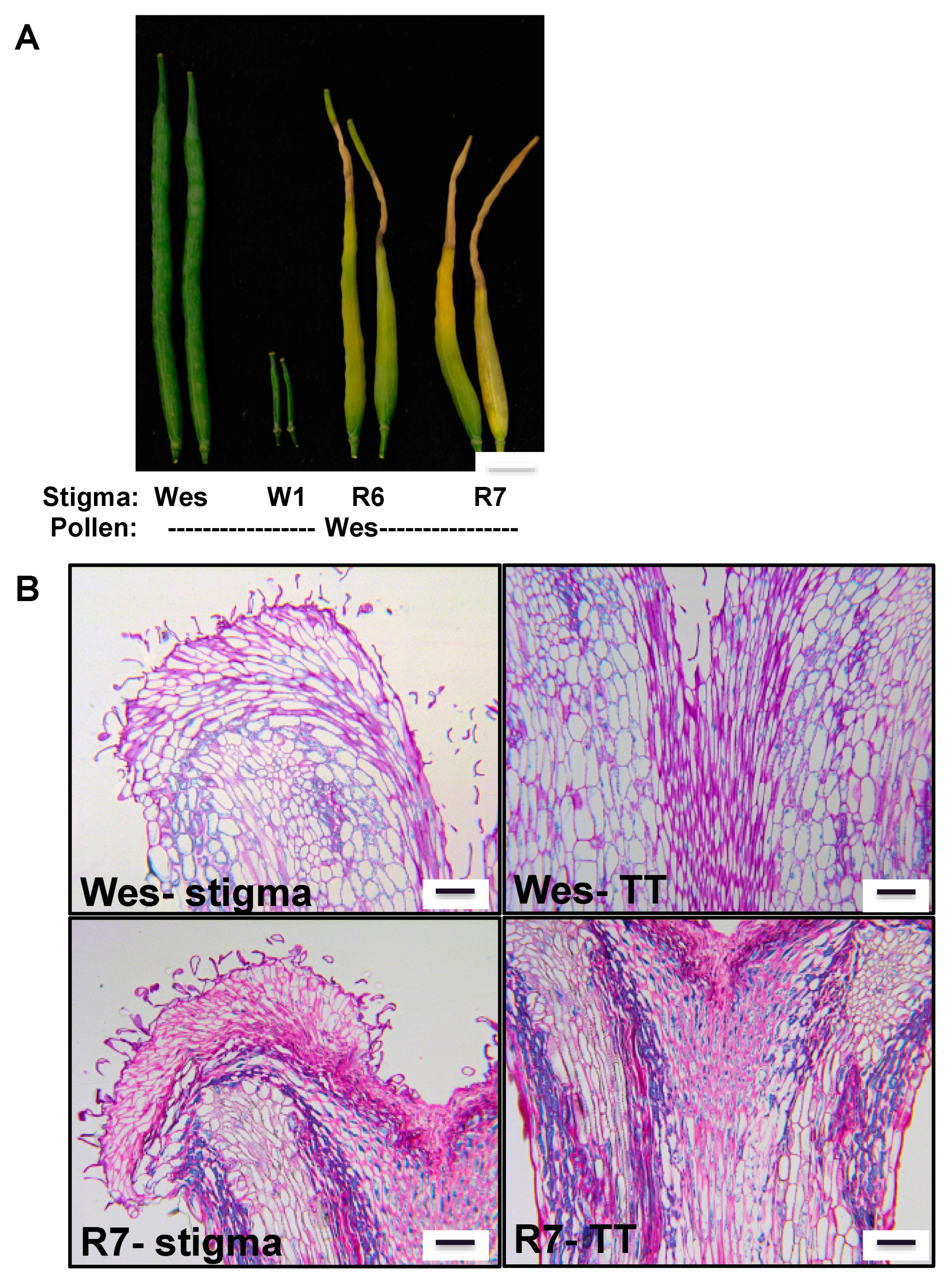
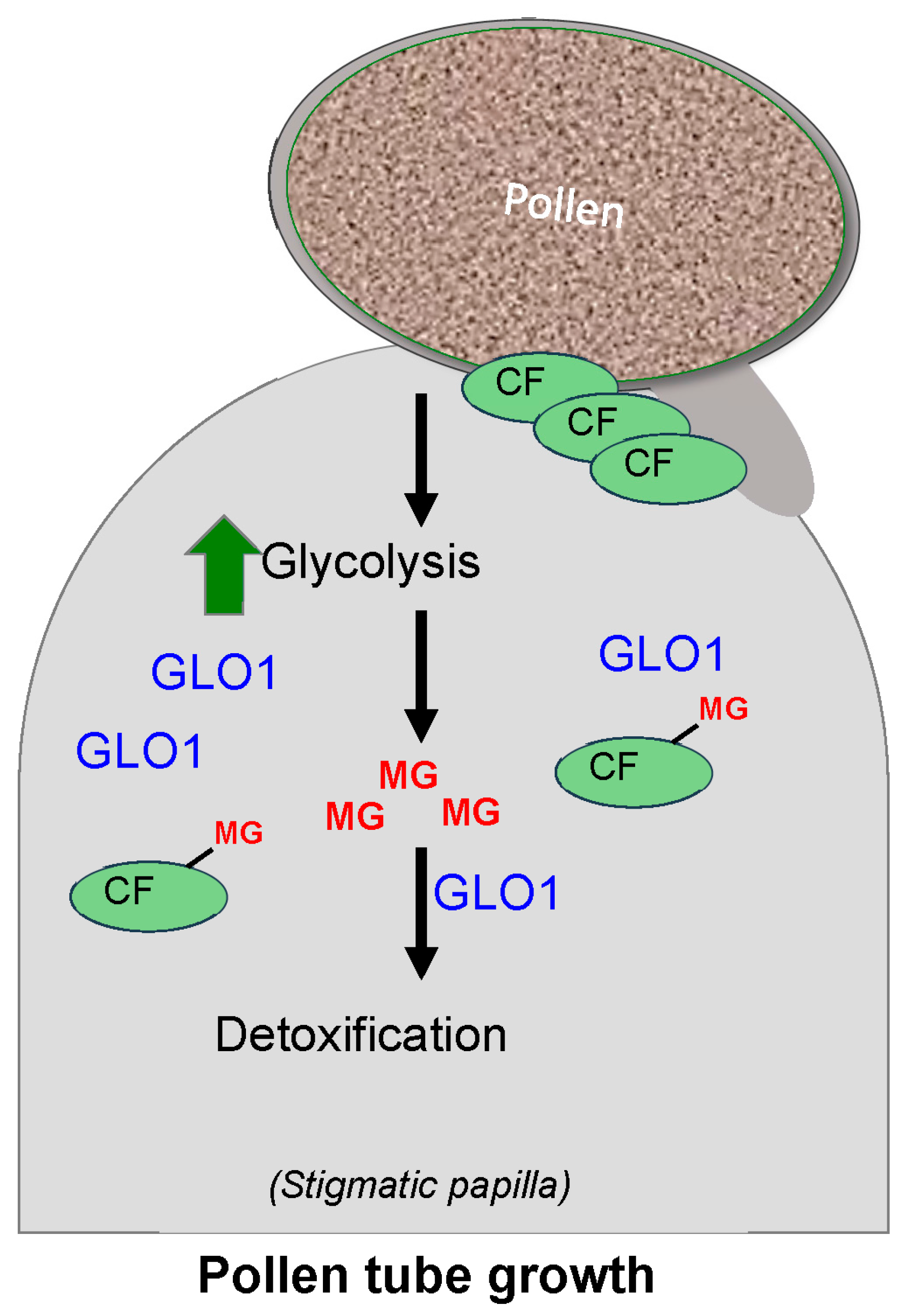
3.4. Glyoxalase as a Regulator of Plant Reproduction
3.5. Roles of MG in Cellular Signaling
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Racker, E. The mechanism of action of Glyoxalase. J. Biol. Chem. 1951, 190, 685–696. [Google Scholar] [PubMed]
- Thornalley, P.J. The glyoxalase system: New developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990, 269, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Subedi, K.P.; Choi, D.; Kim, I.; Min, B.; Park, C. Hsp31 of Escherichia coli k-12 is Glyoxalase III. Mol. Microbiol. 2011, 81, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Presence of unique Gyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Xue, M.; Thornalley, P.J. Activity, regulation, copy number and function in the glyoxalase system. Biochem. Soc. Trans. 2014, 42, 419–424. [Google Scholar] [CrossRef] [PubMed]
- McLellan, A.C.; Thornalley, P.J. Glyoxalase activity in human red blood cells fractioned by age. Mech. Ageing Dev. 1989, 48, 63–71. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Glyoxalase in diabetes, obesity and related disorders. Semin. Cell Dev. Biol. 2011, 22, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. The critical role of methylglyoxal and Glyoxalase I in diabetic nephropathy. Diabetes 2014, 63, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.L.; Tsai, M.M.; Tsai, C.Y.; Huang, Y.H.; Chen, C.Y.; Chi, H.C.; Tseng, Y.H.; Chao, I.W.; Lin, W.C.; Wu, S.M.; et al. Glyoxalase I is a novel prognosis factor associated with gastric cancer progression. PLoS ONE 2012, 7, e34352. [Google Scholar] [CrossRef]
- Wang, Y.F.; Kuramitsu, Y.; Ueno, T.; Suzuki, N.; Yoshino, S.; Iizuka, N.; Akada, J.; Kitagawa, T.; Oka, M.; Nakamura, K. Glyoxalase I (GLO1) is up-regulated in pancreatic cancerous tissues compared with related non-cancerous tissues. Anticancer Res. 2012, 32, 3219–3222. [Google Scholar] [PubMed]
- Takagi, D.; Inoue, H.; Odawara, M.; Shimakawa, G.; Miyake, C. The calvin cycle inevitably produces sugar-derived reactive carbonyl methylglyoxal during photosynthesis: A potential cause of plant diabetes. Plant Cell Physiol. 2014, 55, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.M.; Johnson, M.A. Methylglyoxal—Enzyme distributions relative to its presence in Douglas-fir needles and absence in Douglas-fir needle callus. Arch. Biochem. Biophys. 1981, 208, 431–439. [Google Scholar] [CrossRef]
- Talesa, V.; Rosi, G.; Contenti, S.; Mangiabene, C.; Lupattelli, M.; Norton, S.J.; Giovannini, E.; Principato, G.B. Presence of Glyoxalase II in mitochondria from spinach leaves: Comparison with the enzyme from cytosol. Biochem. Int. 1990, 22, 1115–1120. [Google Scholar] [PubMed]
- Norton, S.J.; Talesa, V.; Yuan, W.J.; Principato, G.B. Glyoxalase I and Glyoxalase II from Aloe vera: Purification, characterization and comparison with animal glyoxalases. Biochem. Int. 1990, 22, 411–418. [Google Scholar] [PubMed]
- Deswal, R.; Chakaravarty, T.N.; Sopory, S.K. The glyoxalase system in higher plants: Regulation in growth and differentiation. Biochem. Soc. Trans. 1993, 21, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Kollner, B.; Jacobsen, H.J. Physiological and biochemical characterization of glyoxalase I, a general marker for cell proliferation, from a soybean cell suspension. Planta 1993, 189, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, T.N.; Sopory, S.K. Blue light stimulation of cell proliferation and Glyoxalase I activity in callus cultures of Amaranthus paniculatus. Plant Sci. 1998, 132, 63–69. [Google Scholar] [CrossRef]
- Kaur, C.; Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Glyoxalases and stress tolerance in plants. Biochem. Soc. Trans. 2014, 42, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Shimakawa, G.; Suzuki, M.; Yamamoto, E.; Saito, R.; Iwamoto, T.; Nishi, A.; Miyake, C. Why don’t plants have diabetes? Systems for scavenging reactive carbonyls in photosynthetic organisms. Biochem. Soc. Trans. 2014, 42, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on Glyoxalase I and Glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett. 2005, 579, 6265–6271. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 2014, 9, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Rabbani, N.; Mullineaux, P.M.; Thornalley, P.J. Quantitative measurement of specific biomarkers for protein oxidation, nitration and glycation in Arabidopsis leaves. Plant J. 2009, 59, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Crook, E.M.; Law, K. Glyoxalase: The role of the components. Biochem. J. 1952, 52, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Ghosh, A.; Tripathi, A.K.; Kaur, C.; Ganguly, A.K.; Bhavesh, N.S.; Tripathi, J.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A unique Ni2+-dependent and methylglyoxal-inducible rice Glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 2014, 78, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Tripathi, A.K.; Nutan, K.K.; Sharma, S.; Ghosh, A.; Tripathi, J.K.; Pareek, A.; Singla-Pareek, S.L.; Sopory, S.K. A nucleus-localized rice Glyoxalase I enzyme, OsGlyi-8 functions in the detoxification of methylglyoxal in the nucleus. Plant J. 2016, 89, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Schilling, O.; Wenzel, N.; Naylor, M.; Vogel, A.; Crowder, M.; Makaroff, C.; Meyer-Klaucke, W. Flexible metal binding of the metallo-beta-lactamase domain: Glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry 2003, 42, 11777–11786. [Google Scholar] [CrossRef] [PubMed]
- Limphong, P.; McKinney, R.M.; Adams, N.E.; Makaroff, C.A.; Bennett, B.; Crowder, M.W. The metal ion requirements of Arabidopsis thaliana Glx2-2 for catalytic activity. J. Biol. Inorg. Chem. 2010, 15, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Misra, K.; Banerjee, A.B.; Ray, S.; Ray, M. Glyoxalase III from Escherichia coli: A single novel enzyme for the conversion of methylglyoxal into d-lactate without reduced glutathione. Biochem. J. 1995, 305 Pt 3, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.; Choi, D.; Hyun, J.K.; Jung, H.S.; Baek, K.; Park, C. Novel glyoxalases from Arabidopsis thaliana. FEBS J. 2013, 280, 3328–3339. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, D.H.; Fleming, T.; Nawroth, P.; Teleman, A.A. Evidence against a role for the parkinsonism-associated protein Dj-1 in methylglyoxal detoxification. J. Biol. Chem. 2017, 292, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Oberschall, A.; Deak, M.; Torok, K.; Sass, L.; Vass, I.; Kovacs, I.; Feher, A.; Dudits, D.; Horvath, G.V. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 2000, 24, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, A.; Erdei, S.; Janda, T.; Toth, E.; Horvath, G.; Dudits, D. Transgenic tobacco plants overproducing alfalfa aldose/aldehyde reductase show higher tolerance to low temperature and cadmium stress. Plant Sci. 2004, 166, 1329–1333. [Google Scholar] [CrossRef]
- Turoczy, Z.; Kis, P.; Torok, K.; Cserhati, M.; Lendvai, A.; Dudits, D.; Horvath, G.V. Overproduction of a rice Aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol. Biol. 2011, 75, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Ku, M.; Kang, S.O. NAD+-linked Alcohol dehydrogenase 1 regulates methylglyoxal concentration in Candida albicans. FEBS Lett. 2014, 588, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, O.; Pal, S.; Guhamukherjee, S.; Sopory, S.K. Presence of Glyoxalase I in pea. Biochem. Int. 1983, 7, 307–318. [Google Scholar]
- Ramaswamy, O.; Pal, S.; Guha-Mukherjee, S.; Sopory, S.K. Correlation of Glyoxalase I activity with cell proliferation in Datura callus culture. Plant Cell Rep. 1984, 3, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Espartero, J.; Sanchez-Aguayo, I.; Pardo, J.M. Molecular characterization of Glyoxalase I from a higher plant: Upregulation by stress. Plant Mol. Biol. 1995, 29, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Veena; Reddy, V.S.; Sopory, S.K. Glyoxalase I from Brassica juncea: Molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 1999, 17, 385–395. [Google Scholar] [PubMed]
- Hossain, M.A.; Hossain, M.Z.; Fujita, M. Stress-induced changes of Methylglyoxal level and Glyoxalase I activity in pumpkin seedlings and cDNA cloning of Glyoxalase I gene. Aust. J. Crop Sci. 2009, 3, 53–64. [Google Scholar]
- Lin, F.; Xu, J.; Shi, J.; Li, H.; Li, B. Molecular cloning and characterization of a novel glyoxalase i gene TaGly I in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2010, 37, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, D.; Trivedi, P.K.; Misra, P.; Tiwari, M.; Shri, M.; Shukla, D.; Kumar, S.; Rai, A.; Pandey, A.; Nigam, D.; et al. Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 2009, 74, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Bisht, R.; Roy, S.D.; Sopory, S.K.; Bhalla-Sarin, N. Cloning and characterization of a mitochondrial Glyoxalase II from Brassica juncea that is upregulated by NaCl, Zn, and ABA. Biochem. Biophys. Res. Commun. 2005, 336, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Kumar, M.; Pareek, A.; Saxena, M.; Sarin, N.B.; Sopory, S.K. Characterization and functional validation of Glyoxalase II from rice. Protein Expr. Purif. 2007, 51, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ekman, D.R.; Lorenz, W.W.; Przybyla, A.E.; Wolfe, N.L.; Dean, J.F. Sage analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol. 2003, 133, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Devanathan, S.; Erban, A.; Perez-Torres, R.; Kopka, J.; Makaroff, C.A. Arabidopsis thaliana Glyoxalase 2-1 is required during abiotic stress but is not essential under normal plant growth. PLoS ONE 2014, 9, e95971. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xiong, F.; Yu, X.; Gong, X.; Luo, J.; Jiang, Y.; Kuang, H.; Gao, B.; Niu, X.; Liu, Y. Overexpression of a glyoxalase gene, OsGly I, improves abiotic stress tolerance and grain yield in rice (Oryza sativa l.). Plant Physiol. Biochem. 2016, 109, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Reddy, M.K.; Sopory, S.K. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Enhancing salt tolerance in a crop plant by overexpression of Glyoxalase II. Transgenic Res. 2008, 17, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Gosal, S.S. Introduction of OsGlyII gene into Oryza sativa for increasing salinity tolerance. Biol. Plant. 2011, 55, 536–540. [Google Scholar] [CrossRef]
- Saxena, M.; Roy, S.B.; Singla-Pareek, S.L.; Sopory, S.K.; Bhalla-Sarin, N. Overexpression of the Glyoxalase II gene leads to enhanced salinity tolerance in Brassica juncea. Open Plant Sci. J. 2011, 5, 23–28. [Google Scholar] [CrossRef]
- Viveros, M.F.A.; Inostroza-Blancheteau, C.; Timmermann, T.; Gonzalez, M.; Arce-Johnson, P. Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum mill.) plants confers salt tolerance by decreasing oxidative stress. Mol. Biol. Rep. 2013, 40, 3281–3290. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Phys. 2006, 140, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Peng, K.; Zhaohui, C.; Wang, S.; Zhang, Q. The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci. China C Life Sci. 2002, 45, 449–467. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Srivastava, S.; Rahman, M.H.; Strelkov, S.E.; Kav, N.N. Proteome changes in leaves of Brassica napus L. As a result of sclerotinia sclerotiorum challenge. J. Agric. Food. Chem. 2008, 56, 1963–1976. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Manrique, S.; Garcia-Martinez, J.; Segundo, B.S. Production of cecropin a in transgenic rice plants has an impact on host gene expression. Plant Biotechnol. J. 2008, 6, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of Glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15185. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shiba, H.; Iwano, M.; Shimosato, H.; Che, F.S.; Kai, N.; Watanabe, M.; Suzuki, G.; Hinata, K.; Isogai, A. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 2000, 97, 1920–1925. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001, 293, 1824–1826. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Chong, Y.T.; Haasen, K.E.; Aldea-Brydges, M.G.; Stone, S.L.; Goring, D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70a1, a putative component of the exocyst complex. Plant Cell 2009, 21, 2655–2671. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen acceptance or rejection: A tale of two pathways. Trends Plant Sci. 2016, 21, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.A.; Goring, D.R. Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J. Exp. Bot. 2010, 61, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Deb, S.; Chatfield-Reed, K.; Kwon, E.J.; Chua, G.; Samuel, M.A. Deciphering the stigmatic transcriptional landscape of compatible and self-incompatible pollinations in Brassica napus reveals a rapid stigma senescence response following compatible pollination. Mol. Plant 2013, 6, 1988–1991. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Tang, W.; Jamshed, M.; Northey, J.; Patel, D.; Smith, D.; Siu, K.W.; Muench, D.G.; Wang, Z.Y.; Goring, D.R. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role for microtubule dynamics during pollen responses. Mol. Cell. Proteom. 2011, 10, M111.011338. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Proteomics approaches advance our understanding of plant self-incompatibility response. J. Proteome Res. 2013, 12, 4717–4726. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Douglass, E.F., Jr.; Fitzgerald, K.J.; Spiegel, D.A. A “turn-on” fluorescent sensor for methylglyoxal. J. Am. Chem. Soc. 2013, 135, 12429–12433. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Kushwaha, H.R.; Mustafiz, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Analysis of global gene expression profile of rice in response to methylglyoxal indicates its possible role as a stress signal molecule. Front. Plant Sci. 2015, 6, 682. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; De, S.; Ray, M.; Ray, S. Methylglyoxal can completely replace the requirement of kinetin to induce differentiation of plantlets from some plant calluses. Plant Growth Regul. 2004, 44, 33–45. [Google Scholar] [CrossRef]
- Ray, A.; Ray, S.; Mukhopadhyay, S.; Ray, M. Methylglyoxal with glycine or succinate enhances differentiation and shoot morphogenesis in Nicotiana tabacum callus. Biol. Plant. 2013, 57, 219–223. [Google Scholar] [CrossRef]
- Hoque, T.S.; Uraji, M.; Tuya, A.; Nakamura, Y.; Murata, Y. Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in Arabidopsis. Plant Biol. (Stuttg.) 2012, 14, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Hoque, T.S.; Okuma, E.; Uraji, M.; Furuichi, T.; Sasaki, T.; Hoque, M.A.; Nakamura, Y.; Murata, Y. Inhibitory effects of methylglyoxal on light-induced stomatal opening and inward K+ channel activity in Arabidopsis. Biosci. Biotechnol. Biochem. 2012, 76, 617–619. [Google Scholar] [CrossRef] [PubMed]
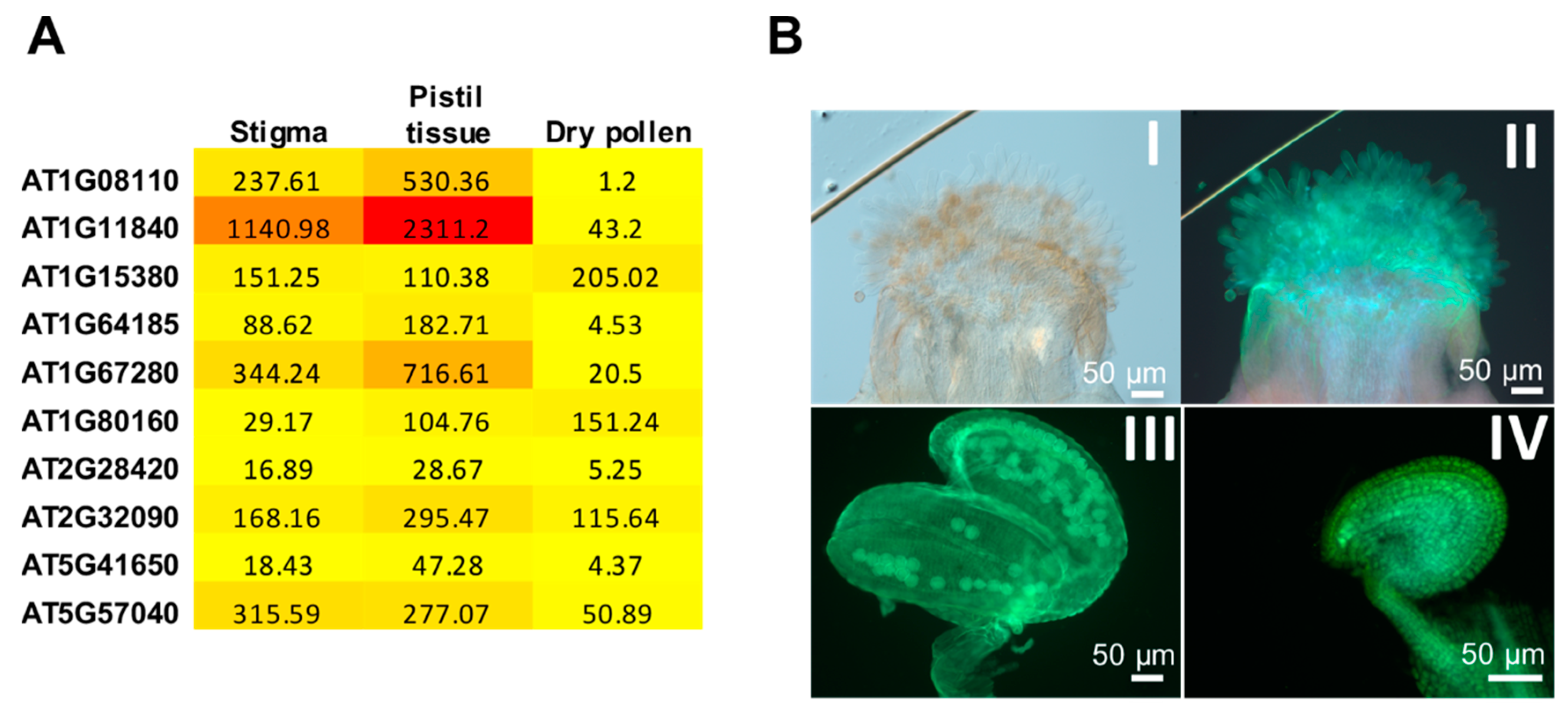
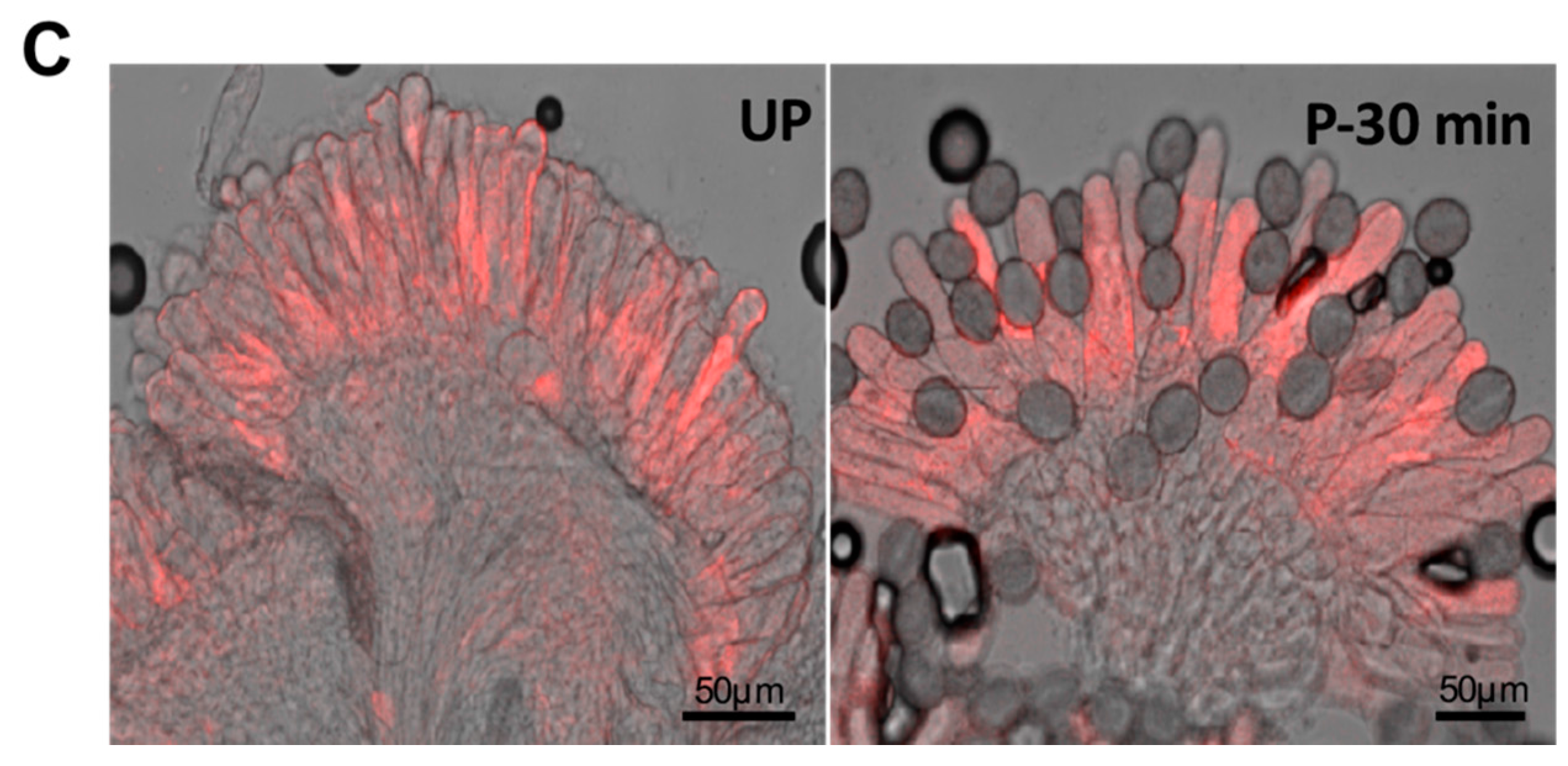
| Biological role | Plant species | References |
|---|---|---|
| Marker for cell division | Pea (Pisum sativum), Datura, Soybean (Glycine max L.) | [16,39,40] |
| Mitigator of abiotic stress (Salinity, Drought, Mannitol, ABA, extreme temperatures and heavy metal stress) | Tomato (Solanum lycopersicum), Indian Mustard (Brassica juncea), Rice (Oryza sativa), Soybean (Glycine max), Tobacco (Nicotiana tabacum) , Pumpkin and Arabidopsis | [30,31,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57] |
| Biotic stress tolerance (Evidence obtained mainly from expression analysis of fungal infected plants and promoter analysis of the genes of Glyoxalase family) | Rice (Oryza sativa), Wheat (Triticum aestivum), Canola (Brassica napus), Soybean (Glycine max) | [31,57,58,59] |
| Involvement in Plant reproduction | Canola (Brassica napus), Arabidopsis | [60], this paper |
| Regulation of protein turn-over (Indirectly by regulating MG levels) | Canola (Brassica napus) | [60] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankaranarayanan, S.; Jamshed, M.; Kumar, A.; Skori, L.; Scandola, S.; Wang, T.; Spiegel, D.; Samuel, M.A. Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants. Int. J. Mol. Sci. 2017, 18, 898. https://doi.org/10.3390/ijms18040898
Sankaranarayanan S, Jamshed M, Kumar A, Skori L, Scandola S, Wang T, Spiegel D, Samuel MA. Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants. International Journal of Molecular Sciences. 2017; 18(4):898. https://doi.org/10.3390/ijms18040898
Chicago/Turabian StyleSankaranarayanan, Subramanian, Muhammad Jamshed, Abhinandan Kumar, Logan Skori, Sabine Scandola, Tina Wang, David Spiegel, and Marcus A. Samuel. 2017. "Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants" International Journal of Molecular Sciences 18, no. 4: 898. https://doi.org/10.3390/ijms18040898
APA StyleSankaranarayanan, S., Jamshed, M., Kumar, A., Skori, L., Scandola, S., Wang, T., Spiegel, D., & Samuel, M. A. (2017). Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants. International Journal of Molecular Sciences, 18(4), 898. https://doi.org/10.3390/ijms18040898






