Gene-Transformation-Induced Changes in Chemical Functional Group Features and Molecular Structure Conformation in Alfalfa Plants Co-Expressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes: Effects of Single-Gene and Two-Gene Insertion
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Single-Gene and Double-Gene Insertion/Transformation on Changes Chemical Functional Groups and Molecular Structure Related to Protein Properties
2.2. Effect of Single-Gene and Double-Gene Insertion/Transformation on Changes in Chemical Functional Groups and Molecular Structure with Regard to Carbohydrate Properties
3. Materials and Methods
3.1. Transgenic Alfalfa Plant Material
3.2. Molecular Spectroscopy
3.3. Univariate Molecular Structure Spectral Processing and Analyses
3.4. Multivariate Molecular Spectral Analysis on Intrinsic Structure Changes by Gene Inserting
3.5. Statistical Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jonker, A.; Gruber, M.Y.; Mccaslin, M.; Wang, Y.; Coulman, B.; Mckinnon, J.J.; Christensen, D.A.; Yu, P. Nutrient composition and degradation profiles of anthocyanidin-accumulating Lc-alfalfa populations. Can. J. Anim. Sci. 2010, 90, 401–412. [Google Scholar] [CrossRef]
- Jonker, A.; Gruber, M.Y.; Wang, Y.; Coulman, B.; Azarfar, A.; McKinnon, J.J.; Christensen, D.A.; Yu, P. Modeling degradation ratios and nutrient availability of anthocyanidin-accumulating Lc-alfalfa populations in dairy cows. J. Dairy Sci. 2011, 94, 1430–1444. [Google Scholar] [CrossRef] [PubMed]
- Ray, H.; Yu, M.; Auser, P.; Blahut-Beatty, L.; McKersie, B.; Bowley, S.; Westcott, N.; Coulman, B.; Lloyd, A.; Gruber, M.Y. Expression of anthocyanins and proanthocyanidins after transformation of alfalfa with maize Lc. Plant Physiol. 2003, 132, 1448–1463. [Google Scholar] [CrossRef] [PubMed]
- Peel, G.J.; Pang, Y.; Modolo, L.V.; Dixon, R.A. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 2009, 59, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Hancock, K.R.; Collette, V.; Fraser, K.; Greig, M.; Xue, H.; Richardson, K.; Jones, C.; Rasmussen, S. Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol. 2012, 159, 1204–1220. [Google Scholar] [CrossRef] [PubMed]
- Doiron, K.; Yu, P.; McKinnon, J.J.; Christensen, D.A. Heat-induced protein structure and subfractions in relation to protein degradation kinetics and intestinal availability in dairy cattle. J. Dairy Sci. 2009, 92, 3319–3330. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Zhang, X.; Yu, P. Using synchrotron radiation-based infrared microspectroscopy to reveal microchemical structure characterization: Frost damaged wheat vs. normal wheat. Int. J. Mol. Sci. 2013, 14, 16706–16718. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Christensen, C.R.; Christensen, D.A.; McKinnon, J.J. Ultrastructural-chemical make-up of yellow-seeded (Brassica rapa) and brown-seeded (Brassica napus) canola within cellular dimensions, explored with synchrotron reflection FTIR microspectroscopy. Can. J. Plant Sci. 2005, 85, 533–541. [Google Scholar] [CrossRef]
- Yu, P. Plant-based food and feed protein structure changes induced by gene-transformation, heating and bio-ethanol processing: A synchrotron-based molecular structure and nutrition research program. Food Mol. Nutr. 2010, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Nuez-Ortín, W.G. Relationship of protein molecular structure to metabolisable proteins in different types of dried distillers grains with solubles: A novel approach. Br. J. Nutr. 2010, 104, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Mantsch, H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Jonker, A.; Gruber, M. Molecular basis of protein structure in proanthocyanidin and anthocyanin-enhanced Lc-transgenic alfalfa in relation to nutritive value using synchrotron-radiation FTIR microspectroscopy: A novel approach. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2009, 73, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, D.L.; Eilert, A.J.; Pietrzak, L.N.; Miller, S.S.; Sweat, J.A. Ultraspatially resolved synchrotron infrared microspectroscopyof plant tissue in situ. Cell. Mol. Biol. 1998, 44, 145–168. [Google Scholar] [PubMed]
- Yu, P. Application of advanced synchrotron radiation-based Fourier transform infrared (SR-FTIR) microspectroscopy to animal nutrition and feed science: A novel approach. Br. J. Nutr. 2004, 92, 869. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Block, H.; Niu, Z.; Doiron, K. Rapid characterization of molecular chemistry, nutrient make-up and microlocation of internal seed tissue. J. Synchrotron Radiat. 2007, 14, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Fick, G.W.; Mueller, S.C. Alfalfa-quality, maturity and mean stage of development. Inf. Bull. 1989, 217, 1–16. [Google Scholar]
- Yari, M.; Valizadeh, R.; Naserian, A.A.; Jonker, A.; Yu, P. Protein molecular structures in alfalfa hay cut at three stages of maturity and in the afternoon and morning and relationship with nutrient availability in ruminants. J. Sci. Food Agric. 2013, 93, 3072–3080. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Short communication: Relationship of carbohydrate molecular spectroscopic features to carbohydrate nutrient profiles in co-products from bioethanol production. J. Dairy Sci. 2012, 95, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Applications of hierarchical cluster analysis (CLA) and principal component analysis (PCA) in feed structure and feed molecular chemistry research, using synchrotron-based fourier transform infrared (FTIR) microspectroscopy. J. Agric. Food Chem. 2005, 53, 7115–7127. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Protein secondary structures (α-helix and β-sheet) at a cellular level and protein fractions in relation to rumen degradation behaviours of protein: A new approach. Br. J. Nutr. 2005, 94, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Yu, P. Synchrotron-based microspectroscopic analysis of molecular and biopolymer structures using multivariate techniques and advanced multi-components modeling. Can. J. Anal. Sci. Spectrosc. 2008, 53, 220–231. [Google Scholar]
- Saxton, A.M. A macro for converting mean separation output to letter groupings in PROC MIXED. In Proceedings of the 23rd SAS Users Group International, Nashville, Tennessee, 22–25 March 1998; SAS Institute: Cary, NC, USA, 1998; pp. 1243–1246. [Google Scholar]
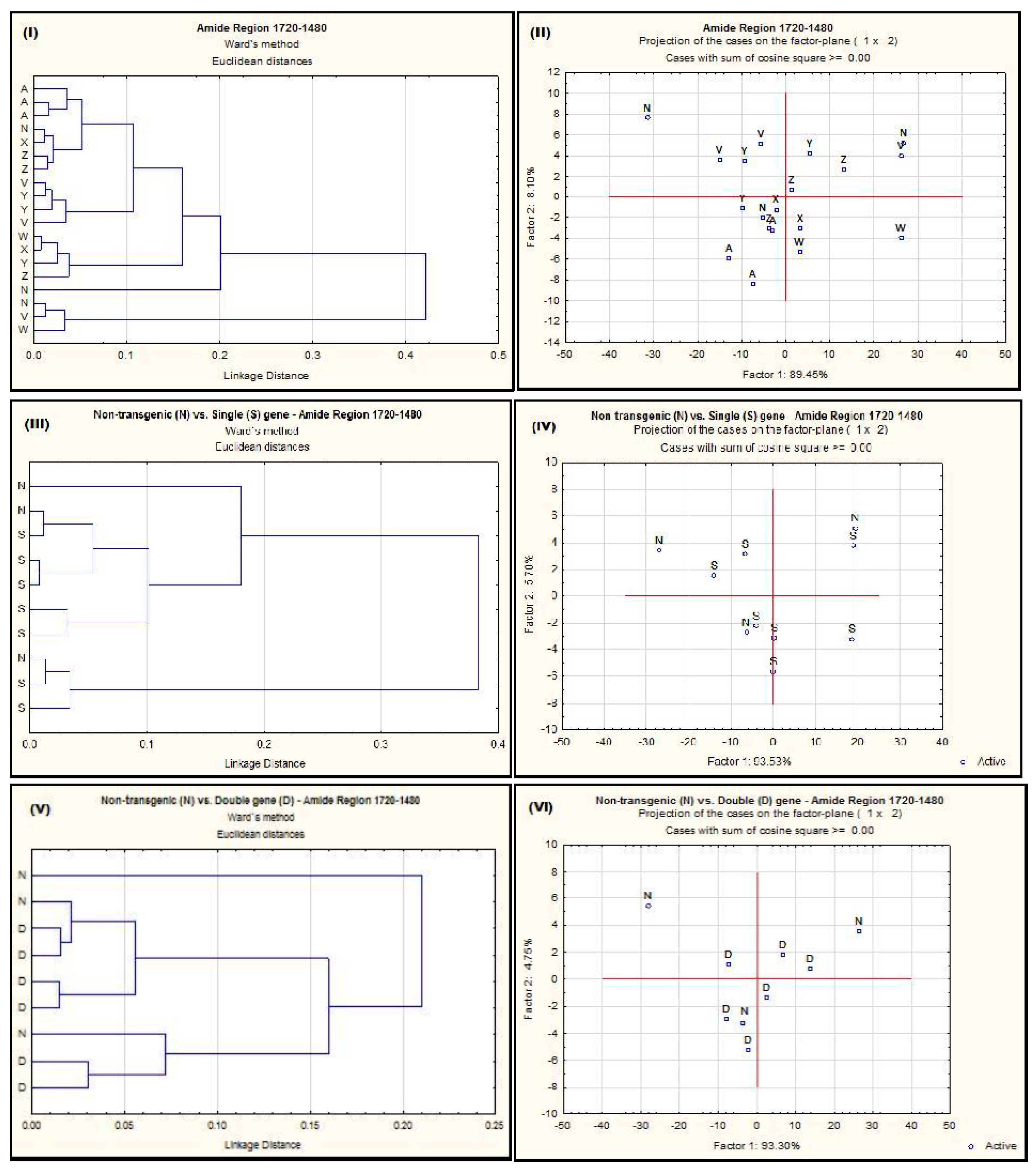
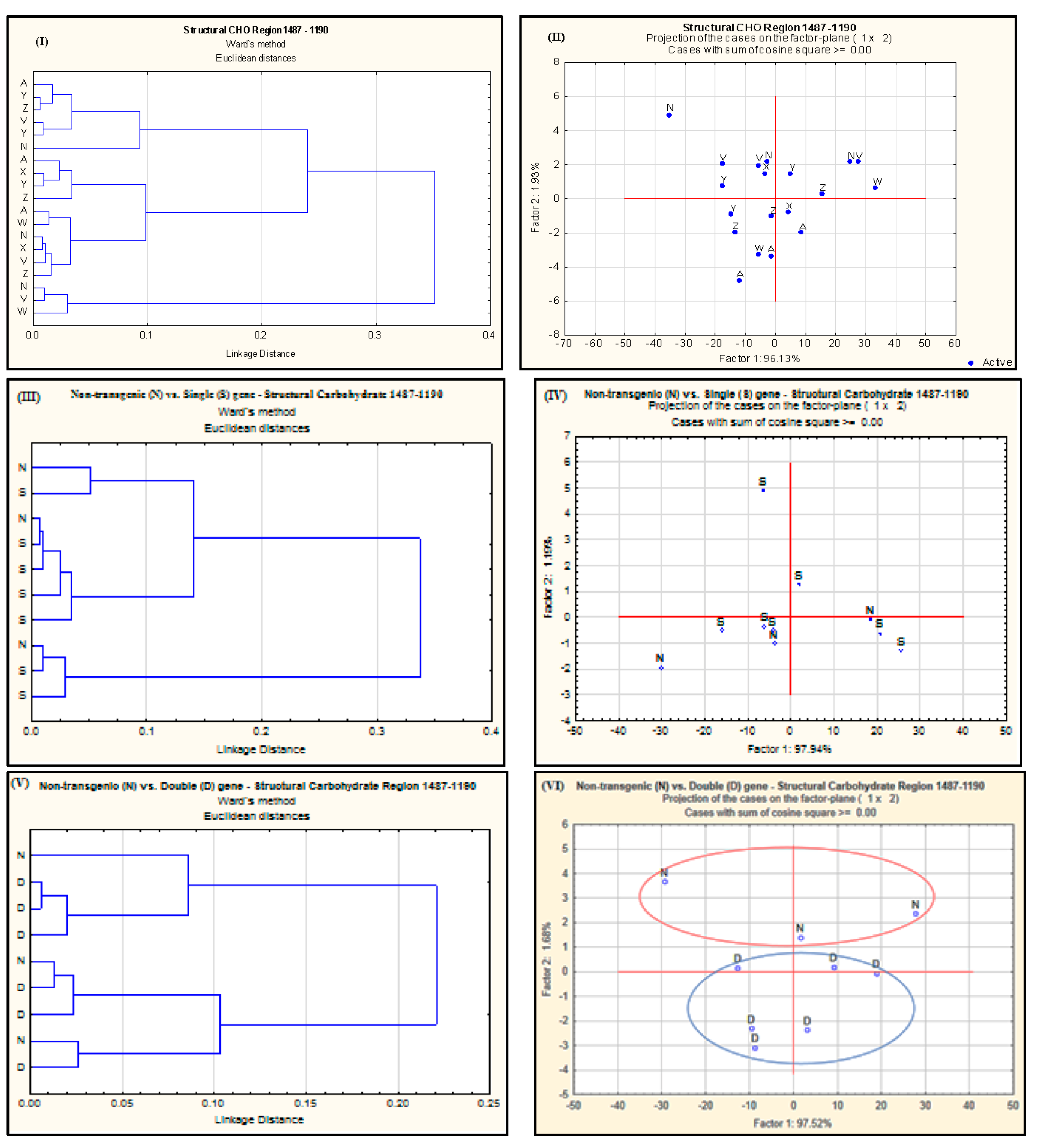
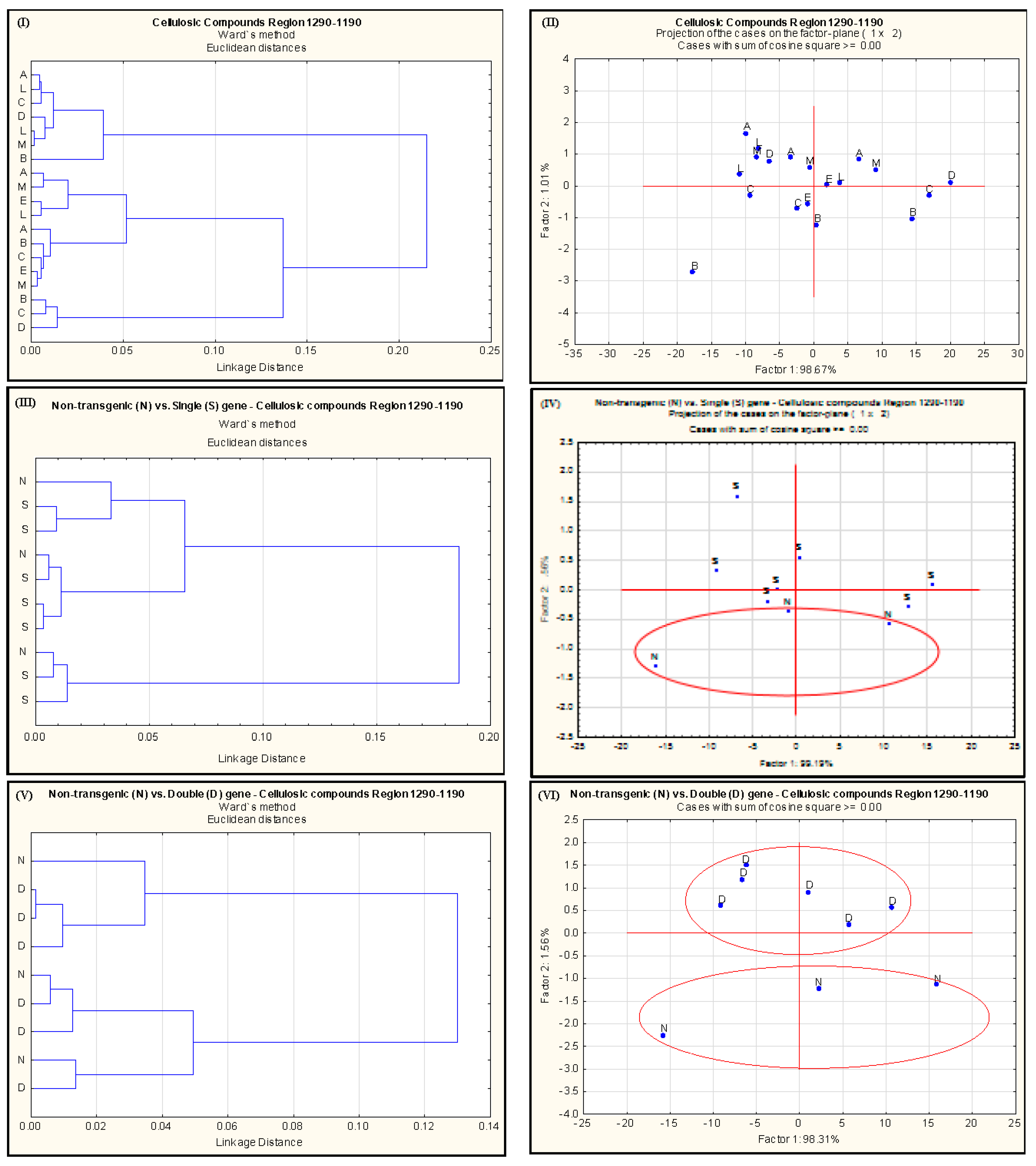
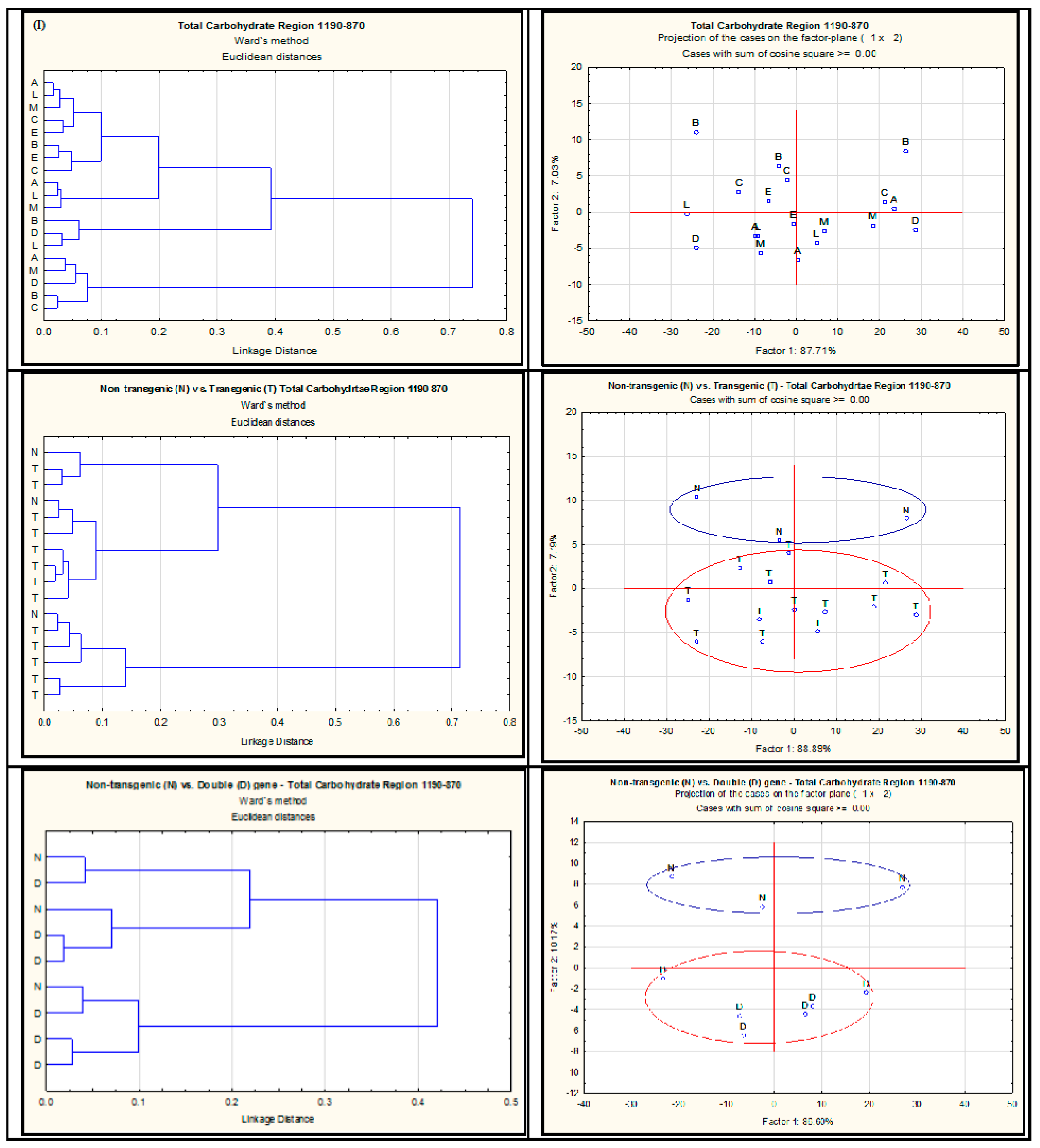
| Alfalfa Population | Protein Amide Height | Protein Amide Area | Protein Fine Structure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (Baseline ~1710–1487) | (Baseline ~1710–1487) | (Baseline ~1710–1487) | ||||||||
| Amide I Height (~1650 cm−1) | Amide II Height (~1550 cm−1) | Ratio of Amide I/II Heights | Amide I Area | Amide II Area | Amide I and II Total Area | Ratio of Amide I/II Area | α-Helix Height (~1650 cm−1) | β-Sheet Height (~1596 cm−1) | Ratio of α-Helix/β-Sheet Height | |
| C1 | 0.029 | 0.019 | 1.536 | 1.598 a,b | 2.197 a,b | 3.795 a,b | 0.725 b,c | 0.027 a,b | 0.027 | 0.983 b |
| Lc1 | 0.024 | 0.015 | 1.533 | 1.261 b | 1.729 b | 2.990 b | 0.729 b,c | 0.023 b | 0.02 | 1.109 b |
| Lc3 | 0.028 | 0.018 | 1.519 | 1.518 a,b | 2.041 a,b | 3.558 a,b | 0.744 a,b,c | 0.025 a,b | 0.023 | 1.088 b |
| Lc1C1 | 0.029 | 0.019 | 1.582 | 1.658 a | 2.160 a,b | 3.817 a,b | 0.768 a | 0.029 a,b | 0.041 | 1.048 b |
| Lc3C1 | 0.027 | 0.016 | 1.657 | 1.452 a,b | 2.017 a,b | 3.469 a,b | 0.718 c,d | 0.025 a,b | 0.024 | 1.045 b |
| NT | 0.029 | 0.025 | 1.497 | 1.579 a,b | 2.279 a | 3.858 a | 0.694 d | 0.027 a,b | 0.027 | 1.012 b |
| ACGL | 0.030 | 0.019 | 1.572 | 1.589 a,b | 2.122 a,b | 3.711 a,b | 0.750 a,b | 0.029 a | 0.022 | 1.319 a |
| SEM | 0.0014 | 0.0032 | 0.0386 | 0.0821 | 0.1133 | 0.1944 | 0.0074 | 0.0014 | 0.006 | 0.0302 |
| p value | 0.08 | 0.38 | 0.07 | 0.04 | 0.04 | 0.049 | <0.01 | 0.04 | 0.29 | <0.01 |
| Contrast p value | ||||||||||
| Single vs. Double | 0.23 | 0.99 | 0.01 | 0.21 | 0.35 | 0.29 | 0.15 | 0.09 | 0.11 | 0.64 |
| NT vs. Trans | 0.34 | 0.02 | 0.08 | 0.32 | 0.03 | 0.09 | <0.01 | 0.36 | 0.93 | 0.16 |
| NT vs. Single | 0.20 | 0.03 | 0.44 | 0.18 | 0.02 | 0.05 | <0.01 | 0.15 | 0.63 | 0.14 |
| NT vs. Double | 0.81 | 0.04 | 0.01 | 0.79 | 0.13 | 0.33 | <0.01 | 0.96 | 0.38 | 0.31 |
| C1 vs. Lc | 0.08 | 0.63 | 0.83 | 0.05 | 0.03 | 0.04 | 0.21 | 0.13 | 0.52 | <0.01 |
| ACGL vs. Single | 0.04 | 0.66 | 0.35 | 0.18 | 0.32 | 0.25 | 0.06 | 0.01 | 0.86 | <0.01 |
| ACGL vs. Double | 0.30 | 0.67 | 0.31 | 0.74 | 0.80 | 0.77 | 0.47 | 0.21 | 0.16 | <0.01 |
| Alfalfa Population | Structural Carbohydrates (StCHO) Profile | Cellulosic Compound Profile | Total Carbohydrate (TCHO) Profile | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak 1 Height (~1410) | Peak 2 Height (~1370) | Peak 3 Height (~1240) | Total Area | Height (~1240) | Area | Peak 1 Height (~1153) | Peak 2 Height (~1080) | Peak 3 Height (~1020) | Peak 1 Area | Peak 2 Area | Peak 3 Area | Total Area | |
| Baseline | Baseline | Baseline | Baselines | ||||||||||
| ~1487–1190 | ~1290–1190 | ~1190–880 | ~1190–1128 | ~1128–1055 | ~1055–880 | ~1190–880 | |||||||
| C1 | 0.014 | 0.012 | 0.010 | 2.532 | 0.007 | 0.322 | 0.016 a,b | 0.052 | 0.064 a,b | 0.705 | 2.962 | 4.759 a | 8.426 |
| Lc1 | 0.013 | 0.011 | 0.010 | 2.318 | 0.008 | 0.363 | 0.015 a,b | 0.053 | 0.064 a,b,c | 0.695 | 3.120 | 4.629 a,b | 8.444 |
| Lc3 | 0.014 | 0.012 | 0.010 | 2.539 | 0.007 | 0.359 | 0.016 a,b | 0.054 | 0.066 a,b | 0.746 | 3.192 | 4.727 a | 8.665 |
| Lc1C1 | 0.015 | 0.012 | 0.010 | 2.527 | 0.008 | 0.351 | 0.016 a,b | 0.054 | 0.064 a,b | 0.720 | 3.296 | 4.508 a,b | 8.524 |
| Lc3C1 | 0.013 | 0.011 | 0.010 | 2.343 | 0.007 | 0.335 | 0.014 b | 0.047 | 0.055 b,c | 0.654 | 2.809 | 4.152 a,b | 7.614 |
| NT | 0.014 | 0.014 | 0.011 | 2.713 | 0.008 | 0.351 | 0.017 a | 0.053 | 0.068 a | 0.750 | 3.070 | 4.972 a | 8.810 |
| ACGL | 0.012 | 0.011 | 0.010 | 2.233 | 0.008 | 0.375 | 0.015 a,b | 0.051 | 0.051 c | 0.690 | 3.288 | 3.736 b | 7.713 |
| SEM | 0.0007 | 0.0009 | 0.0005 | 0.122 | 0.0004 | 0.0185 | 0.0007 | 0.0024 | 0.0028 | 0.0319 | 0.1369 | 0.2153 | 0.3808 |
| p value | 0.07 | 0.09 | 0.49 | 0.07 | 0.61 | 0.48 | 0.04 | 0.37 | <0.01 | 0.34 | 0.13 | <0.01 | 0.16 |
| Contrast p value | |||||||||||||
| Single vs. Double | 0.60 | 0.85 | 0.68 | 0.81 | 0.94 | 0.78 | 0.16 | 0.32 | 0.08 | 0.34 | 0.76 | 0.07 | 0.22 |
| NT vs. Trans | 0.54 | 0.02 | 0.05 | 0.03 | 0.42 | 0.78 | 0.07 | 0.51 | 0.09 | 0.15 | 0.97 | 0.05 | 0.21 |
| NT vs. Single | 0.45 | 0.03 | 0.10 | 0.06 | 0.48 | 0.87 | 0.24 | 0.79 | 0.33 | 0.32 | 0.89 | 0.25 | 0.47 |
| NT vs. Double | 0.78 | 0.03 | 0.05 | 0.04 | 0.45 | 0.69 | 0.02 | 0.28 | 0.02 | 0.08 | 0.91 | 0.01 | 0.08 |
| C1 vs. Lc | 0.26 | 0.41 | 0.74 | 0.51 | 0.15 | 0.10 | 0.66 | 0.55 | 0.83 | 0.71 | 0.27 | 0.77 | 0.79 |
| ACGL vs. Single | 0.04 | 0.24 | 0.81 | 0.11 | 0.37 | 0.21 | 0.18 | 0.42 | <0.01 | 0.50 | 0.22 | <0.01 | 0.08 |
| ACGL vs. Double | 0.02 | 0.32 | 0.93 | 0.17 | 0.35 | 0.15 | 0.83 | 0.99 | 0.01 | 0.94 | 0.16 | 0.02 | 0.44 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heendeniya, R.G.; Yu, P. Gene-Transformation-Induced Changes in Chemical Functional Group Features and Molecular Structure Conformation in Alfalfa Plants Co-Expressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes: Effects of Single-Gene and Two-Gene Insertion. Int. J. Mol. Sci. 2017, 18, 664. https://doi.org/10.3390/ijms18030664
Heendeniya RG, Yu P. Gene-Transformation-Induced Changes in Chemical Functional Group Features and Molecular Structure Conformation in Alfalfa Plants Co-Expressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes: Effects of Single-Gene and Two-Gene Insertion. International Journal of Molecular Sciences. 2017; 18(3):664. https://doi.org/10.3390/ijms18030664
Chicago/Turabian StyleHeendeniya, Ravindra G., and Peiqiang Yu. 2017. "Gene-Transformation-Induced Changes in Chemical Functional Group Features and Molecular Structure Conformation in Alfalfa Plants Co-Expressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes: Effects of Single-Gene and Two-Gene Insertion" International Journal of Molecular Sciences 18, no. 3: 664. https://doi.org/10.3390/ijms18030664
APA StyleHeendeniya, R. G., & Yu, P. (2017). Gene-Transformation-Induced Changes in Chemical Functional Group Features and Molecular Structure Conformation in Alfalfa Plants Co-Expressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes: Effects of Single-Gene and Two-Gene Insertion. International Journal of Molecular Sciences, 18(3), 664. https://doi.org/10.3390/ijms18030664





