A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder
Abstract
1. Introduction
2. Results
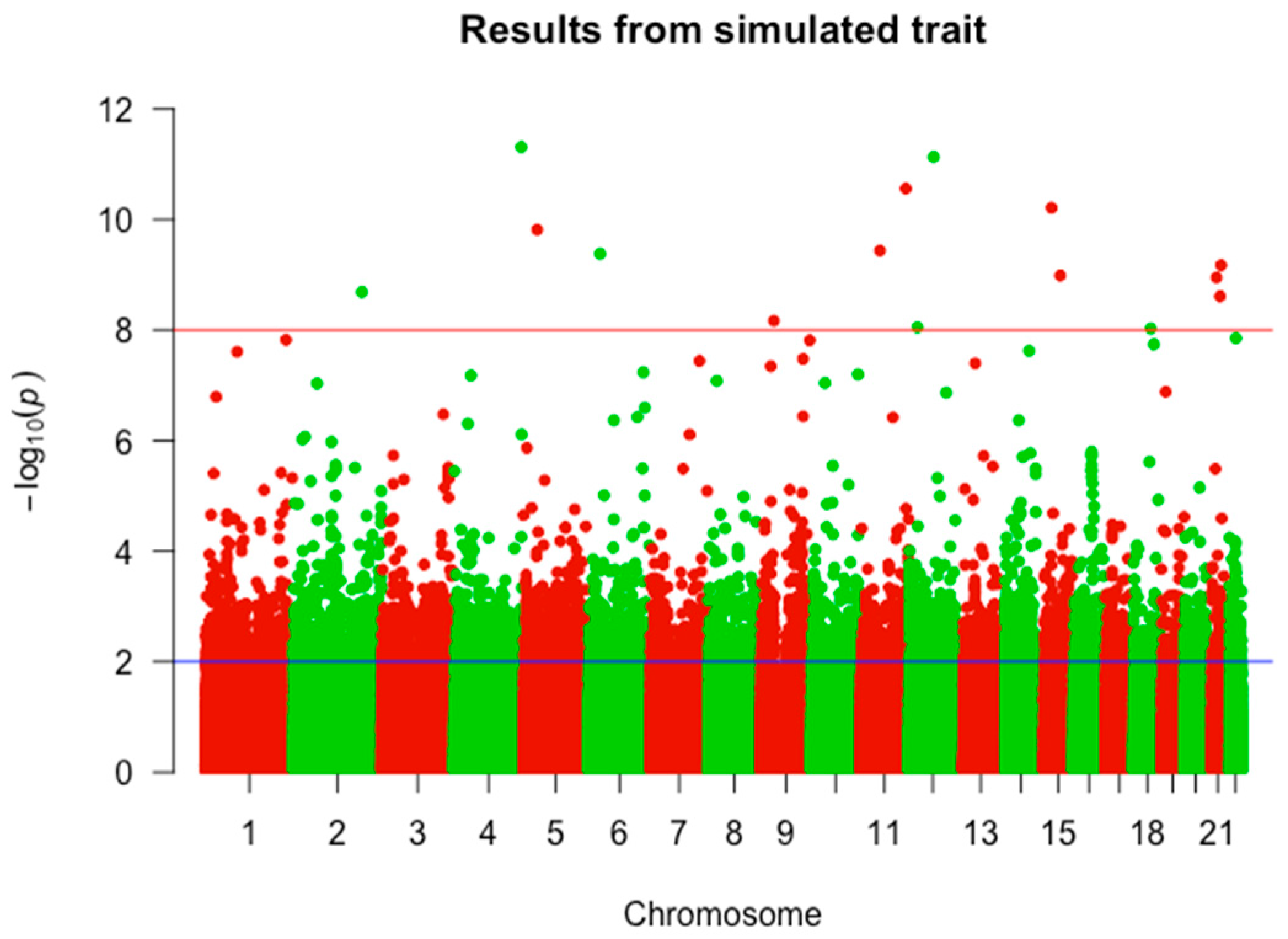
2.1. GWAS Results
2.2. Gene Functional Analysis
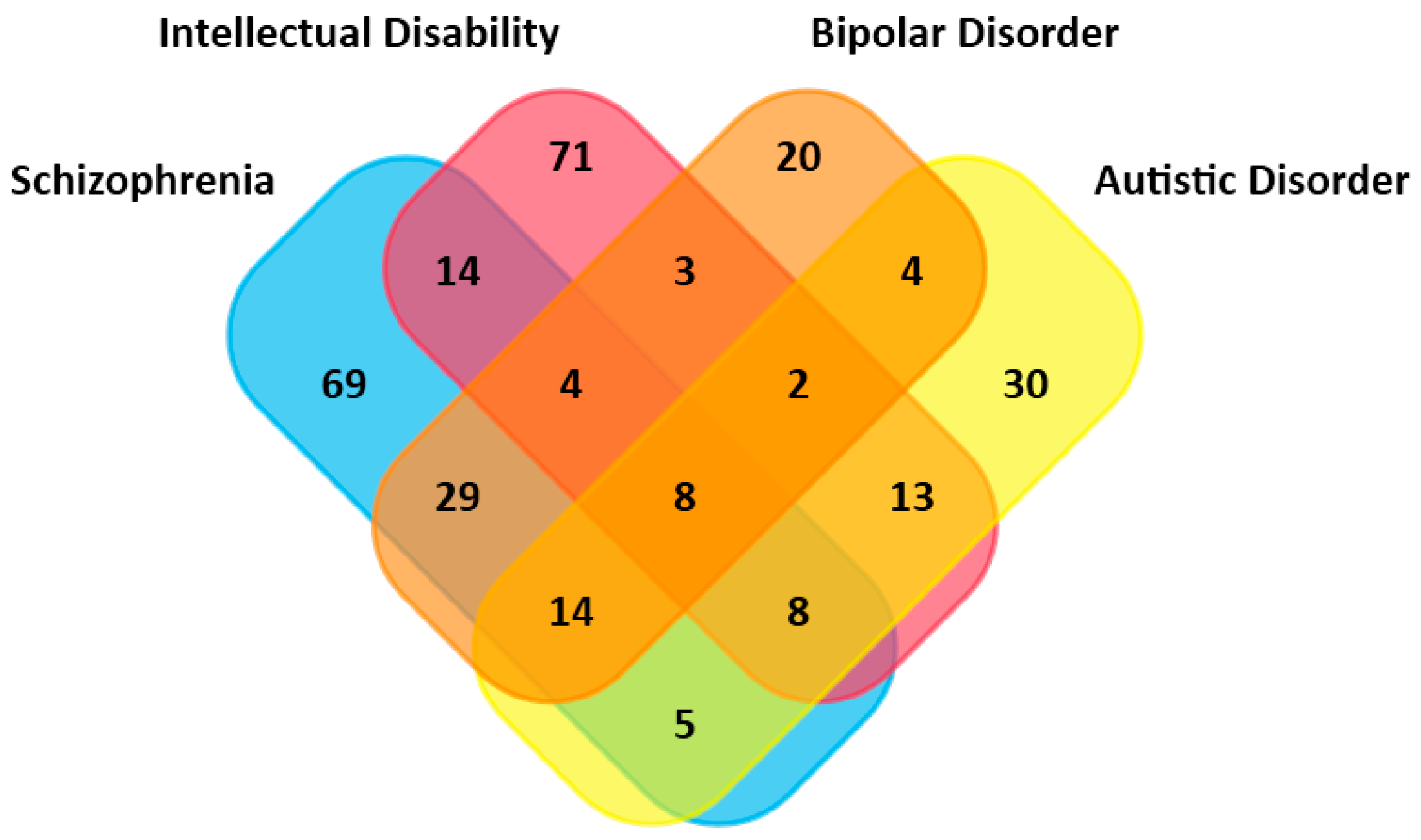
2.3. Overlapped Genes in Different Mental Illnesses
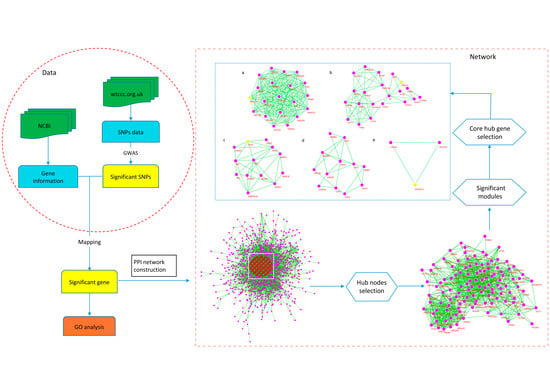
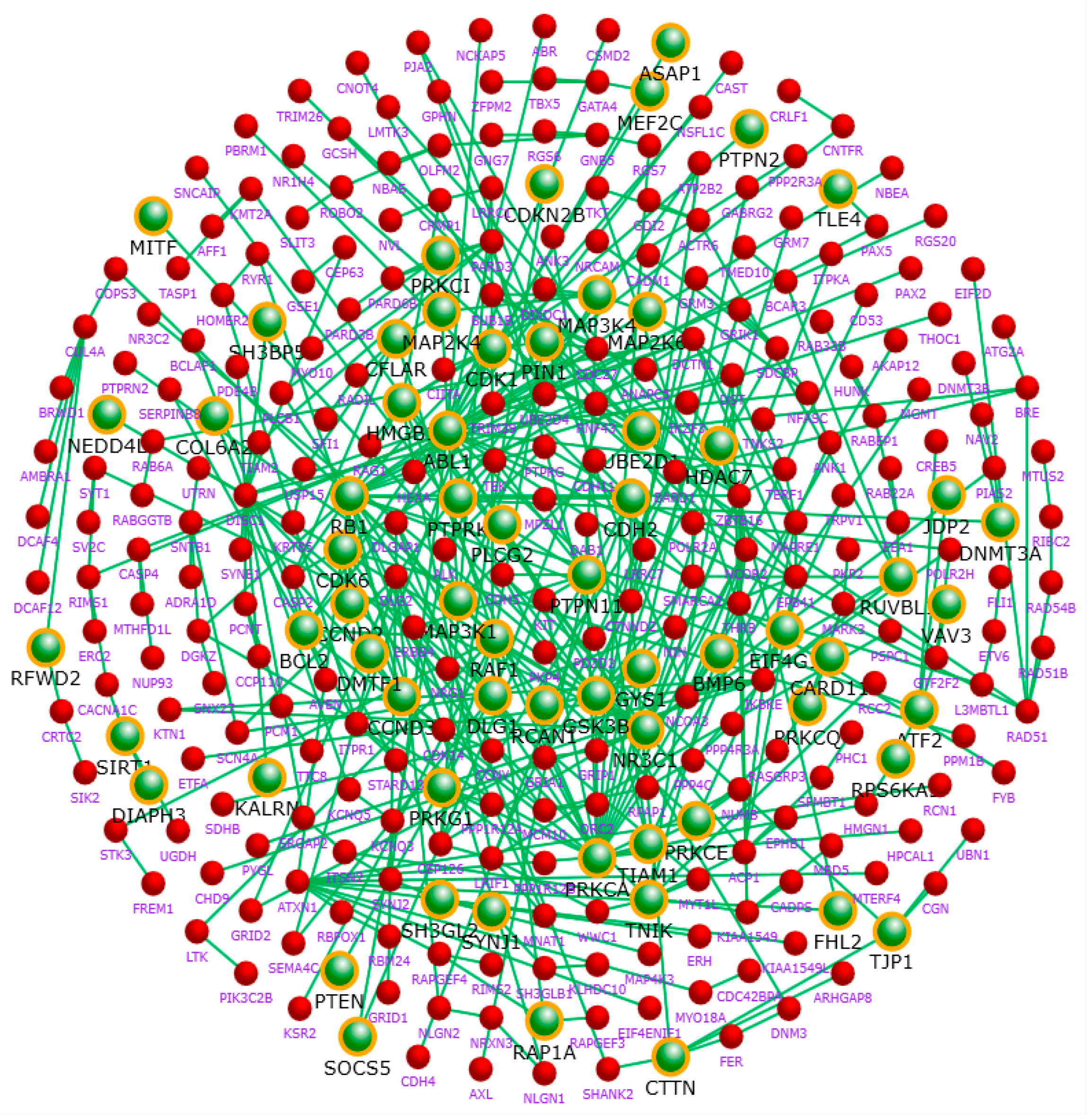
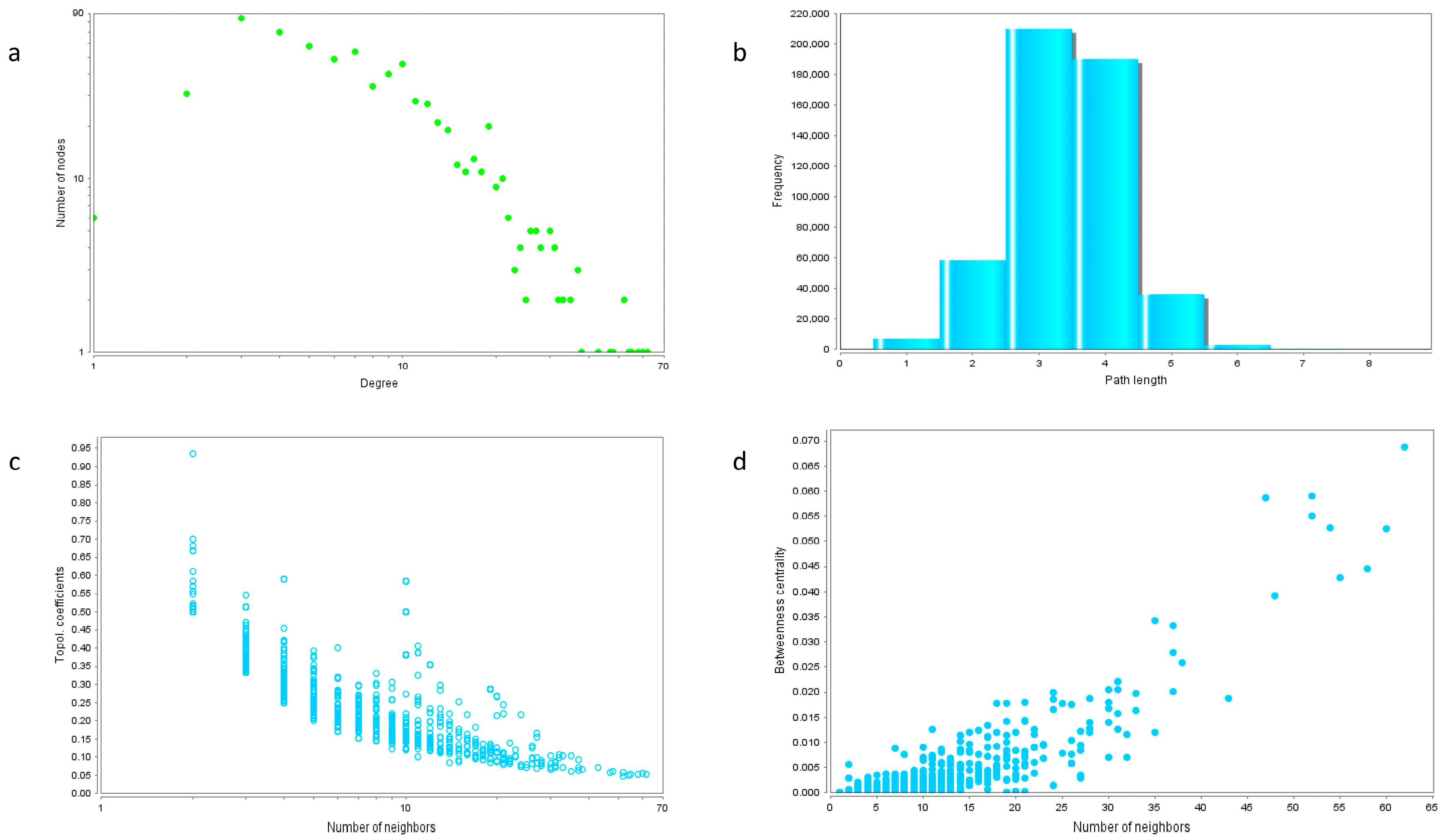
2.4. Protein Interaction Network
2.5. Hub Genes of the Network
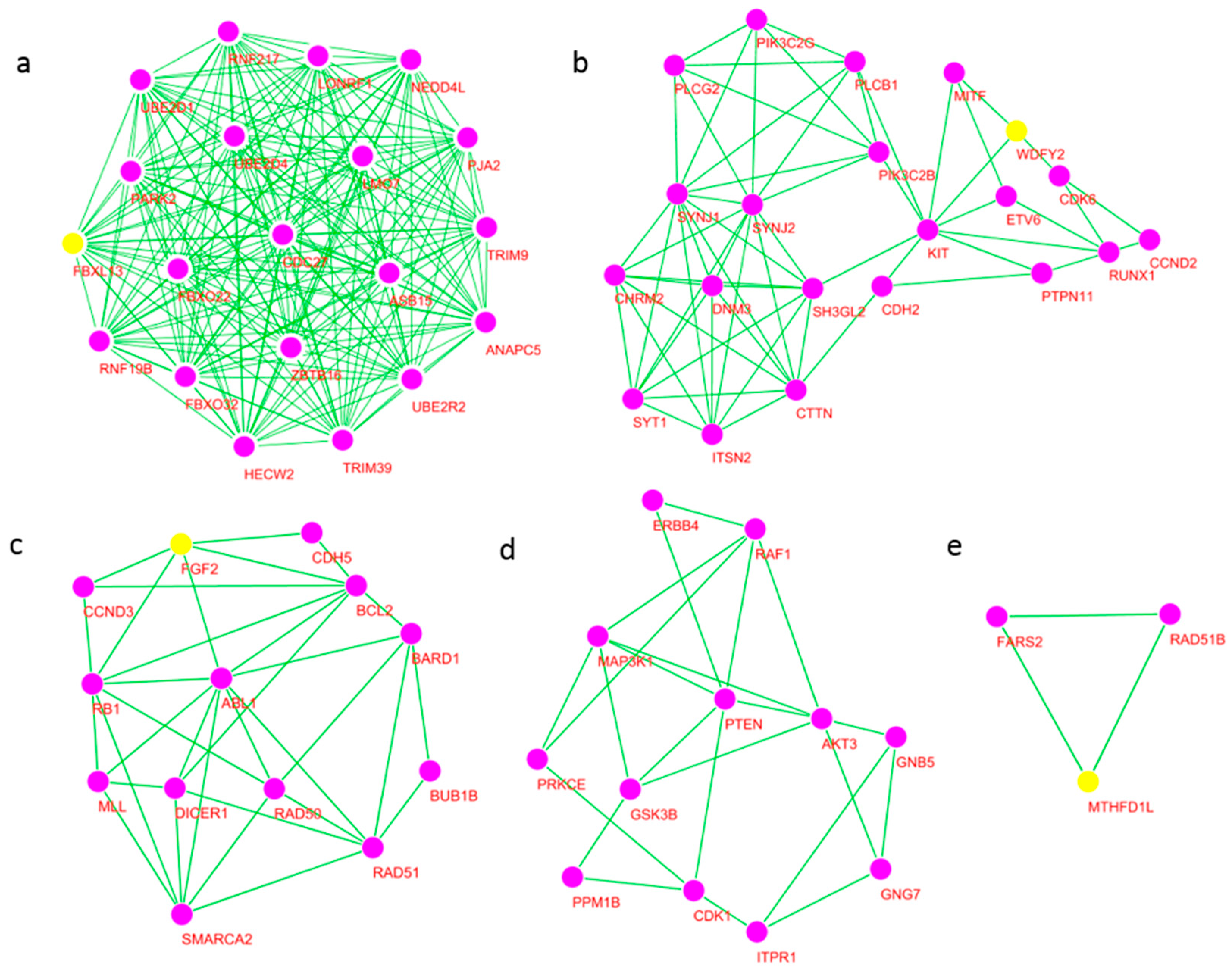
2.6. Significant Modules of the Network and Core Hub Genes
3. Discussion
3.1. Most BD Risk Gene Products Are Located in the Nervous System
3.2. Intense Overlappings of Genes Associated with BD and Other Mental Disorders
3.3. Core Hub Genes Give New Insights of BD
3.4. Effectiveness of GWAS Followed by Gene Network Analysis
4. Materials and Methods
4.1. Bipolar Disorder Datasets
4.2. Screening of Risk SNPs
4.3. Mapping Significant Risk SNPs to Genes
4.4. Gene Function and Disease Enrichment Analysis
4.5. Protein Network Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BD | bipolar disorder |
| GWAS | genome-wide association study |
| SNP | single nucleotide polymorphism |
| WTCCC | the World Healthcare Case Control Association |
Appendix A
| GENE | Degree | Ensembl | UniProtKB |
|---|---|---|---|
| CDK1 | 62 | ENSG00000170312 | P06493 |
| PTEN | 61 | ENSG00000171862 | P60484 |
| BCL2 | 60 | ENSG00000171791 | P10415 |
| POLR2A | 55 | ENSG00000181222 | P24928 |
| SMARCA2 | 55 | ENSG00000080503 | P5153 |
| GSK3B | 54 | ENSG00000082701 | P49841 |
| ABL1 | 53 | ENSG00000097007 | P00519 |
| PRKCA | 50 | ENSG00000154229 | P17252 |
| FGF2 | 48 | ENSG00000138685 | P09038 |
| RB1 | 45 | ENSG00000139687 | P06400 |
| KIT | 40 | ENSG00000157404 | P10721 |
| RAD51 | 38 | ENSG00000051180 | Q06609 |
| SIRT1 | 38 | ENSG00000096717 | Q96EB6 |
| UBE2D1 | 37 | ENSG00000072401 | P51668 |
| DLG1 | 36 | ENSG00000075711 | Q12959 |
| CDC27 | 35 | ENSG00000004897 | P30260 |
| NEDD4L | 35 | ENSG00000049759 | Q96PU5 |
| PRKG1 | 35 | ENSG00000185532 | Q13976 |
| RAP1A | 34 | ENSG00000116473 | P62834 |
| CDH2 | 33 | ENSG00000170558 | P19022 |
| GNB5 | 33 | ENSG00000069966 | O14775 |
| MAPK6 | 33 | ENSG00000069956 | Q16659 |
| GNG7 | 32 | ENSG00000176533 | O60262 |
| PTPN11 | 32 | ENSG00000179295 | Q06124 |
| ZBTB16 | 32 | ENSG00000109906 | Q05516 |
| ADCY8 | 31 | ENSG00000155897 | P40145 |
| DICER1 | 31 | ENSG00000100697 | Q9UPY3 |
| SYNJ1 | 31 | ENSG00000159082 | O43426 |
| CACNA1C | 30 | ENSG00000151067 | Q13936 |
| CTTN | 30 | ENSG00000085733 | Q14247 |
| DLG2 | 30 | ENSG00000150672 | Q15700 |
| MAP3K1 | 30 | ENSG00000095015 | Q13233 |
| RIT2 | 30 | ENSG00000152214 | Q99578 |
| ANAPC5 | 28 | ENSG00000089053 | Q9UJX4 |
| PLCB1 | 28 | ENSG00000182621 | Q9NQ66 |
| RAF1 | 28 | ENSG00000132155 | P04049 |
| PARK2 | 27 | ENSG00000185345 | O60260 |
| PLCG2 | 27 | ENSG00000197943 | P16885 |
| PNPLA6 | 27 | ENSG00000032444 | Q8IY17 |
| SYNJ2 | 27 | ENSG00000078269 | O15056 |
| UBE2R2 | 27 | ENSG00000107341 | Q712K3 |
| CACNA1D | 26 | ENSG00000157388 | Q01668 |
| CDK6 | 26 | ENSG00000105810 | Q00534 |
| CHRM2 | 26 | ENSG00000181072 | P08172 |
| MTHFD1L | 26 | ENSG00000120254 | Q6UB35 |
| GRIA1 | 25 | ENSG00000155511 | P42261 |
| POLR2H | 25 | ENSG00000163882 | P52434 |
| TJP1 | 25 | ENSG00000104067 | Q07157 |
| MAPRE1 | 24 | ENSG00000101367 | Q15691 |
| RUNX1 | 24 | ENSG00000159216 | Q01196 |
| UBE2D4 | 24 | ENSG00000078967 | Q9Y2X8 |
| EHHADH | 23 | ENSG00000113790 | Q08426 |
| IQCB1 | 23 | ENSG00000173226 | Q15051 |
| PPM1B | 23 | ENSG00000138032 | O75688 |
| PPP4C | 23 | ENSG00000149923 | P60510 |
| RAD50 | 23 | ENSG00000113522 | Q92878 |
| SH3GL2 | 23 | ENSG00000107295 | Q99962 |
| DCTN1 | 22 | ENSG00000204843 | Q14203 |
| ERBB4 | 22 | ENSG00000178568 | Q15303 |
| FBXO32 | 22 | ENSG00000156804 | Q969P5 |
| ITPR1 | 22 | ENSG00000150995 | Q14643 |
| MLL | 22 | ENSG00000118058 | Q03164 |
| NCOR2 | 22 | ENSG00000196498 | Q9Y618 |
| PRKCE | 22 | ENSG00000171132 | Q02156 |
| RAD51B | 22 | ENSG00000182185 | O15315 |
| ACTN4 | 21 | ENSG00000130402 | O43707 |
| CCND2 | 21 | ENSG00000118971 | P30279 |
| CDH5 | 21 | ENSG00000179776 | P33151 |
| CUL4A | 21 | ENSG00000139842 | Q13619 |
| EFCAB13 | 21 | ENSG00000178852 | Q8IY85 |
| LMO7 | 21 | ENSG00000136153 | Q8WWI1 |
| MITF | 21 | ENSG00000187098 | O75030 |
| TRIM9 | 21 | ENSG00000100505 | Q9C026 |
| CCND3 | 20 | ENSG00000112576 | P30281 |
| EPHB1 | 20 | ENSG00000154928 | P54762 |
| FARS2 | 20 | ENSG00000145982 | O95363 |
| FBXO22 | 20 | ENSG00000167196 | Q8NEZ5 |
| FLT3 | 20 | ENSG00000122025 | P36888 |
| GATA4 | 20 | ENSG00000136574 | P43694 |
| ITSN2 | 20 | ENSG00000198399 | Q9NZM3 |
| KIF18A | 20 | ENSG00000121621 | Q8NI77 |
| LONRF1 | 20 | ENSG00000154359 | Q17RB8 |
| NCOA3 | 20 | ENSG00000124151 | Q9Y6Q9 |
| PCNT | 20 | ENSG00000160299 | O95613 |
| PJA2 | 20 | ENSG00000198961 | O43164 |
| SYT1 | 20 | ENSG00000067715 | P21579 |
| TRIM39 | 20 | ENSG00000204599 | Q9HCM9 |
| WDFY2 | 20 | ENSG00000139668 | Q96P53 |
| AK4 | 19 | ENSG00000162433 | P27144 |
| ASB15 | 19 | ENSG00000146809 | Q8WXK1 |
| ATF2 | 19 | ENSG00000115966 | P15336 |
| BUB1B | 19 | ENSG00000156970 | O60566 |
| DHX15 | 19 | ENSG00000109606 | O43143 |
| DNM3 | 19 | ENSG00000197959 | Q9UQ16 |
| ETV6 | 19 | ENSG00000139083 | P41212 |
| FBXL13 | 19 | ENSG00000161040 | Q8NEE6 |
| HECW2 | 19 | ENSG00000138411 | Q9P2P5 |
| MEF2C | 19 | ENSG00000081189 | Q06413 |
| NR3C1 | 19 | ENSG00000113580 | P04150 |
| PDE4D | 19 | ENSG00000113448 | Q08499 |
| RNF19B | 19 | ENSG00000116514 | Q6ZMZ0 |
| RNF217 | 19 | ENSG00000146373 | Q8TC41 |
| RXFP2 | 19 | ENSG00000133105 | Q8WXD0 |
| RYR1 | 19 | ENSG00000196218 | P21817 |
| THBS2 | 19 | ENSG00000186340 | P35442 |
| AKT3 | 18 | ENSG00000117020 | Q9Y243 |
| BARD1 | 18 | ENSG00000138376 | Q99728 |
| CTNNA2 | 18 | ENSG00000066032 | P26232 |
| HDAC7 | 18 | ENSG00000061273 | Q8WUI4 |
| ITGAV | 18 | ENSG00000138448 | P06756 |
| PARD3 | 18 | ENSG00000148498 | Q8TEW0 |
| PCSK2 | 18 | ENSG00000125851 | P16519 |
| PIK3C2B | 18 | ENSG00000133056 | O00750 |
| PIK3C2G | 18 | ENSG00000139144 | O75747 |
| UBQLN1 | 18 | ENSG00000135018 | Q9UMX0 |
References
- Craddock, N.; Sklar, P. Genetics of bipolar disorder. Lancet 2013, 381, 1654–1662. [Google Scholar] [CrossRef]
- Akiskal, H.S.; Bourgeois, M.L.; Angst, J.; Post, R.; Moller, H.; Hirschfeld, R. Re-evaluating the prevalence of and diagnostic composition within the broad clinical spectrum of bipolar disorders. J. Affect. Disord. 2000, 59, S5–S30. [Google Scholar] [CrossRef]
- Kessler, R.C.; Akiskal, H.S.; Ames, M.; Birnbaum, H.; Greenberg, P.; Hirschfeld, R.M.; Jin, R.; Merikangas, K.R.; Simon, G.E.; Wang, P.S. Prevalence and effects of mood disorders on work performance in a nationally representative sample of U.S. workers. Am. J. Psychiatry 2006, 163, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Grande, I.; Berk, M.; Birmaher, B.; Vieta, E. Bipolar disorder. Lancet 2016, 387, 1561–1572. [Google Scholar] [CrossRef]
- Smoller, J.W.; Finn, C.T. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet. Part C Semin. Med. Genet. 2003, 123C, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.H.; Smoller, J.W. The genetics of bipolar disorder. Neuroscience 2009, 164, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Daly, M.J.; O’Donovan, M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012, 13, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Lescai, F.; Als, T.D.; Li, Q.; Nyegaard, M.; Andorsdottir, G.; Biskopsto, M.; Hedemand, A.; Fiorentino, A.; O’Brien, N.; Jarram, A.; et al. Whole-exome sequencing of individuals from an isolated population implicates rare risk variants in bipolar disorder. Transl. Psychiatry 2017, 7, e1034. [Google Scholar] [CrossRef] [PubMed]
- Sklar, P.; Smoller, J.W.; Fan, J.; Ferreira, M.A.; Perlis, R.H.; Chambert, K.; Nimgaonkar, V.L.; McQueen, M.B.; Faraone, S.V.; Kirby, A.; et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry 2008, 13, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.; O’Donovan, M.C.; Meng, Y.A.; Jones, I.R.; Ruderfer, D.M.; Jones, L.; Fan, J.; Kirov, G.; Perlis, R.H.; Green, E.K.; et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008, 40, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Muglia, P.; Kong, X.Q.; Guan, W.; Flickinger, M.; Upmanyu, R.; Tozzi, F.; Li, J.Z.; Burmeister, M.; Absher, D.; et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl. Acad. Sci. USA 2009, 106, 7501–7506. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Cohen-Woods, S.; Chen, Q.; Noor, A.; Knight, J.; Hosang, G.; Parikh, S.V.; De Luca, V.; Tozzi, F.; Muglia, P.; et al. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med. Genet. 2014, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.E.; Akula, N.; Cabanero, M.; Cardona, I.; Corona, W.; Klemens, B.; Schulze, T.G.; Cichon, S.; Rietschel, M.; Nothen, M.M.; et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry 2008, 13, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.W.; Medland, S.E.; Verweij, K.J.; Lee, S.H.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Montgomery, G.W.; Wright, M.J.; Martin, N.G. Educational attainment: A genome wide association study in 9538 Australians. PLoS ONE 2011, 6, e20128. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, P.; Rijsdijk, F.; Andrew, M.; Sham, P.; Katz, R.; Cardno, A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry 2003, 60, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, M.; Mao, F.; Shao, M.; Zhao, B.; Song, Z.; Shao, C.; Gong, Y. Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis. Models Mech. 2013, 6, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Le-Niculescu, H.; Levey, D.F.; Ayalew, M.; Palmer, L.; Gavrin, L.M.; Jain, N.; Winiger, E.; Bhosrekar, S.; Shankar, G.; Radel, M.; et al. Discovery and validation of blood biomarkers for suicidality. Mol. Psychiatry 2013, 18, 1249–1264. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Rivera, F.; Sharifi-Hannauer, P.; Martinez-Agosto, J.A. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: Case report and review. Am. J. Med. Genet. Part A 2010, 152A, 2459–2467. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Green, M.; Corson, T.W.; Perova, T.; Li, P.P.; Warsh, J.J. Bcl-2 SNP rs956572 associates with disrupted intracellular calcium homeostasis in bipolar I disorder. Bipolar Disord. 2011, 13, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, D.V.; de Klerk, E.; Westra, H.J.; Mastrokolias, A.; Amini, S.; Ariyurek, Y.; Jansen, R.; Penninx, B.W.; Hottenga, J.J.; Willemsen, G.; et al. DeepSAGE reveals genetic variants associated with alternative polyadenylation and expression of coding and non-coding transcripts. PLoS Genet. 2013, 9, e1003594. [Google Scholar] [CrossRef]
- Silberberg, G.; Baruch, K.; Navon, R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal. Biochem. 2009, 391, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Gedvilaite, E.; Badner, J.A.; Erdman, C.; Baird, L.; Matsunami, N.; Leppert, M.; Xing, J.; Byerley, W. A Rare Variant in CACNA1D Segregates with 7 Bipolar I Disorder Cases in a Large Pedigree. Mol. Neuropsychiatry 2016, 2, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Kabir, Z.D.; Martinez-Rivera, A.; Rajadhyaksha, A.M. From Gene to Behavior: L-Type Calcium Channel Mechanisms Underlying Neuropsychiatric Symptoms. Neurotherapeutics 2017, 14, 588–613. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rivera, A.; Hao, J.; Tropea, T.F.; Giordano, T.P.; Kosovsky, M.; Rice, R.C.; Lee, A.; Huganir, R.L.; Striessnig, J.; Addy, N.A.; et al. Enhancing VTA Cav1.3 L-type Ca2+ channel activity promotes cocaine and mood-related behaviors via overlapping AMPA receptor mechanisms in the nucleus accumbens. Mol. Psychiatry 2017, 22, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Teschler, S.; Bartkuhn, M.; Kunzel, N.; Schmidt, C.; Kiehl, S.; Dammann, G.; Dammann, R. Aberrant methylation of gene associated CpG sites occurs in borderline personality disorder. PLoS ONE 2013, 8, e84180. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Xiong, L.; Fathalli, F.; Benkelfat, C.; Tabbane, K.; Danics, Z.; Labelle, A.; Lal, S.; Krebs, M.O.; Rouleau, G.; et al. Association study of the trinucleotide repeat polymorphism within SMARCA2 and schizophrenia. BMC Genet. 2006, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Ishiguro, H.; Yazaki, S.; Horiuchi, Y.; Arai, M.; Niizato, K.; Iritani, S.; Itokawa, M.; Inada, T.; Iwata, N.; et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet. 2009, 18, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Benes, F.M.; Lim, B.; Subburaju, S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc. Natl. Acad. Sci. USA 2009, 106, 11731–11736. [Google Scholar] [CrossRef] [PubMed]
- Vine, A.E.; McQuillin, A.; Bass, N.J.; Pereira, A.; Kandaswamy, R.; Robinson, M.; Lawrence, J.; Anjorin, A.; Sklar, P.; Gurling, H.M.; et al. No evidence for excess runs of homozygosity in bipolar disorder. Psychiatr. Genet. 2009, 19, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Serretti, A.; Colombo, C.; Lorenzi, C.; Tubazio, V.; Smeraldi, E. A glycogen synthase kinase 3-beta promoter gene single nucleotide polymorphism is associated with age at onset and response to total sleep deprivation in bipolar depression. Neurosci. Lett. 2004, 368, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Begum, A.N.; Jones, M.R.; Oh, M.S.; Beech, W.K.; Beech, B.H.; Yang, F.; Chen, P.; Ubeda, O.J.; Kim, P.C.; et al. GSK3 inhibitors show benefits in an Alzheimer’s disease (AD) model of neurodegeneration but adverse effects in control animals. Neurobiol. Dis. 2009, 33, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jope, R.S. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology 2010, 35, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Bollettini, I.; Barberi, I.; Radaelli, D.; Poletti, S.; Locatelli, C.; Pirovano, A.; Lorenzi, C.; Falini, A.; Colombo, C.; et al. Lithium and GSK3-beta promoter gene variants influence white matter microstructure in bipolar disorder. Neuropsychopharmacology 2013, 38, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Cannon, D.M.; Klaver, J.K.; Gandhi, S.K.; Solorio, G.; Peck, S.A.; Erickson, K.; Akula, N.; Savitz, J.; Eckelman, W.C.; Furey, M.L.; et al. Genetic variation in cholinergic muscarinic-2 receptor gene modulates M2 receptor binding in vivo and accounts for reduced binding in bipolar disorder. Mol. Psychiatry 2011, 16, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Munkholm, K.; Peijs, L.; Vinberg, M.; Kessing, L.V. A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl. Psychiatry 2015, 5, e614. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Hayashi-Takagi, A.; Toyota, T.; Yoshikawa, T.; Iwamoto, K. Gene expression analysis in lymphoblastoid cells as a potential biomarker of bipolar disorder. J. Hum. Genet. 2011, 56, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Munkholm, K.; Peijs, L.; Kessing, L.V.; Vinberg, M. Reduced mRNA expression of PTGDS in peripheral blood mononuclear cells of rapid-cycling bipolar disorder patients compared with healthy control subjects. Int. J. Neuropsychopharmacol. 2014, 18. [Google Scholar] [CrossRef] [PubMed]
- Eszlari, N.; Kovacs, D.; Petschner, P.; Pap, D.; Gonda, X.; Elliott, R.; Anderson, I.M.; Deakin, J.F.; Bagdy, G.; Juhasz, G. Distinct effects of folate pathway genes MTHFR and MTHFD1L on ruminative response style: A potential risk mechanism for depression. Transl. Psychiatry 2016, 6, e745. [Google Scholar] [CrossRef] [PubMed]
- Kittel-Schneider, S.; Lorenz, C.; Auer, J.; Weissflog, L.; Reif, A. DGKH genetic risk variant influences gene expression in bipolar affective disorder. J. Affect. Disord. 2016, 198, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.S.; Williams, N.M.; Moskvina, V.; Russell, E.; Norton, N.; Williams, H.J.; Peirce, T.; Georgieva, L.; Dwyer, S.; Grozeva, D.; et al. Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol. Psychiatry 2010, 15, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Kerner, B.; Jasinska, A.J.; DeYoung, J.; Almonte, M.; Choi, O.W.; Freimer, N.B. Polymorphisms in the GRIA1 gene region in psychotic bipolar disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009, 150B, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhang, C.; Shao, X.; Liu, Q.; Qian, Y.; Feng, L.; Chen, J.; Zha, Y.; Zhang, Q.; Jiang, X. Enhancement of nose-to-brain delivery of basic fibroblast growth factor for improving rat memory impairments induced by co-injection of beta-amyloid and ibotenic acid into the bilateral hippocampus. Int. J. Pharm. 2012, 423, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Numata, S.; Iga, J.; Nakataki, M.; Tayoshi, S.; Tanahashi, T.; Itakura, M.; Ueno, S.; Ohmori, T. Positive association of the pericentrin (PCNT) gene with major depressive disorder in the Japanese population. J. Psychiatry Neurosci. 2009, 34, 195–198. [Google Scholar] [PubMed]
- De Ligt, J.; Willemsen, M.H.; van Bon, B.W.; Kleefstra, T.; Yntema, H.G.; Kroes, T.; Vulto-van Silfhout, A.T.; Koolen, D.A.; de Vries, P.; Gilissen, C.; et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012, 367, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; Zheng, T.; Baron, M.; Gilliam, T.C.; Rzhetsky, A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008, 18, 1150–1162. [Google Scholar] [CrossRef] [PubMed]
- Drago, A.; Crisafulli, C.; Sidoti, A.; Calabro, M.; Serretti, A. The microtubule-associated molecular pathways may be genetically disrupted in patients with Bipolar Disorder. Insights from the molecular cascades. J. Affect. Disord. 2016, 190, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Cupertino, R.B.; Kappel, D.B.; Bandeira, C.E.; Schuch, J.B.; da Silva, B.S.; Muller, D.; Bau, C.H.; Mota, N.R. SNARE complex in developmental psychiatry: Neurotransmitter exocytosis and beyond. J. Neural Transm. 2016, 123, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Serretti, A.; Mandelli, L. The genetics of bipolar disorder: Genome ‘hot regions’, genes, new potential candidates and future directions. Mol. Psychiatry 2008, 13, 742–771. [Google Scholar] [CrossRef] [PubMed]
- Kishi, T.; Fukuo, Y.; Kitajima, T.; Okochi, T.; Yamanouchi, Y.; Kinoshita, Y.; Kawashima, K.; Inada, T.; Kunugi, H.; Kato, T.; et al. SIRT1 gene, schizophrenia and bipolar disorder in the Japanese population: An association study. Genes Brain Behav. 2011, 10, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Herskovits, A.Z.; Guarente, L. SIRT1 in neurodevelopment and brain senescence. Neuron 2014, 81, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Nivoli, A.; Porcelli, S.; Albani, D.; Forloni, G.; Fusco, F.; Colom, F.; Vieta, E.; Serretti, A. Association between Sirtuin 1 Gene rs10997870 Polymorphism and Suicide Behaviors in Bipolar Disorder. Neuropsychobiology 2016, 74, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moskvina, V.; Craddock, N.; Holmans, P.; Nikolov, I.; Pahwa, J.S.; Green, E.; Wellcome Trust Case Control Consortium; Owen, M.J.; O’Donovan, M.C. Gene-wide analyses of genome-wide association data sets: Evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol. Psychiatry 2009, 14, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Smalheiser, N.R.; Lugli, G.; Rizavi, H.S.; Torvik, V.I.; Turecki, G.; Dwivedi, Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE 2012, 7, e33201. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, Y. Emerging role of microRNAs in major depressive disorder: Diagnosis and therapeutic implications. Dialogues Clin. Neurosci. 2014, 16, 43–61. [Google Scholar] [PubMed]
- Uezato, A.; Yamamoto, N.; Iwayama, Y.; Hiraoka, S.; Hiraaki, E.; Umino, A.; Haramo, E.; Umino, M.; Yoshikawa, T.; Nishikawa, T. Reduced cortical expression of a newly identified splicing variant of the DLG1 gene in patients with early-onset schizophrenia. Transl. Psychiatry 2015, 5, e654. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Kimura, H.; Wang, C.; Ishizuka, K.; Kushima, I.; Arioka, Y.; Yoshimi, A.; Nakamura, Y.; Shiino, T.; Oya-Ito, T.; et al. Resequencing and Association Analysis of Six PSD-95-Related Genes as Possible Susceptibility Genes for Schizophrenia and Autism Spectrum Disorders. Sci. Rep. 2016, 6, 27491. [Google Scholar] [CrossRef] [PubMed]
- Hunsberger, J.G.; Chibane, F.L.; Elkahloun, A.G.; Henderson, R.; Singh, R.; Lawson, J.; Cruceanu, C.; Nagarajan, V.; Turecki, G.; Squassina, A.; et al. Novel integrative genomic tool for interrogating lithium response in bipolar disorder. Transl. Psychiatry 2015, 5, e504. [Google Scholar] [CrossRef] [PubMed]
- Le-Niculescu, H.; Patel, S.D.; Bhat, M.; Kuczenski, R.; Faraone, S.V.; Tsuang, M.T.; McMahon, F.J.; Schork, N.J.; Nurnberger, J.I., Jr.; Niculescu, A.B., 3rd. Convergent functional genomics of genome-wide association data for bipolar disorder: Comprehensive identification of candidate genes, pathways and mechanisms. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009, 150B, 155–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ross, C.A.; Wang, N.; Huo, Y.; MacKinnon, D.F.; Potash, J.B.; Simpson, S.G.; McMahon, F.J.; DePaulo, J.R., Jr.; McInnis, M.G. NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur. J. Hum. Genet. 2001, 9, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, R.; Jaffe, A.E.; Hyde, T.M.; Kleinman, J.E.; Weinberger, D.R. Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am. J. Psychiatry 2014, 171, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Webb, B.T.; Jia, P.; Bigdeli, T.B.; Maher, B.S.; van den Oord, E.; Bergen, S.E.; Amdur, R.L.; O’Neill, F.A.; Walsh, D.; et al. Association study of 167 candidate genes for schizophrenia selected by a multi-domain evidence-based prioritization algorithm and neurodevelopmental hypothesis. PLoS ONE 2013, 8, e67776. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Watanabe, Y.; Someya, T.; Araki, K.; Shibuya, M.; Niizato, K.; Oshima, K.; Kunii, Y.; Yabe, H.; Matsumoto, J.; et al. Assessment of copy number variations in the brain genome of schizophrenia patients. Mol. Cytogenet. 2015, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Moya, P.R.; Dodman, N.H.; Timpano, K.R.; Rubenstein, L.M.; Rana, Z.; Fried, R.L.; Reichardt, L.F.; Heiman, G.A.; Tischfield, J.A.; King, R.A.; et al. Rare missense neuronal cadherin gene (CDH2) variants in specific obsessive-compulsive disorder and Tourette disorder phenotypes. Eur. J. Hum. Genet. (EJHG) 2013, 21, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Lodder, E.M.; De Nittis, P.; Koopman, C.D.; Wiszniewski, W.; Moura de Souza, C.F.; Lahrouchi, N.; Guex, N.; Napolioni, V.; Tessadori, F.; Beekman, L.; et al. GNB5 Mutations Cause an Autosomal-Recessive Multisystem Syndrome with Sinus Bradycardia and Cognitive Disability. Am. J. Hum. Genet. 2016, 99, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, J.; Huang, K.; Ji, W.; Wang, T.; Li, T.; Wang, Y.; Wang, H.; He, L.; Feng, G.; et al. Analysis of association between common SNPs in ErbB4 and bipolar affective disorder, major depressive disorder and schizophrenia in the Han Chinese population. Prog. Neuro-psychopharmacol. Biol. Psychiatry 2012, 36, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Goes, F.S.; Rongione, M.; Chen, Y.C.; Karchin, R.; Elhaik, E.; Bipolar Genome, S.; Potash, J.B. Exonic DNA sequencing of ERBB4 in bipolar disorder. PLoS ONE 2011, 6, e20242. [Google Scholar] [CrossRef] [PubMed]
- Knijff, E.M.; Ruwhof, C.; de Wit, H.J.; Kupka, R.W.; Vonk, R.; Akkerhuis, G.W.; Nolen, W.A.; Drexhage, H.A. Monocyte-derived dendritic cells in bipolar disorder. Biol. Psychiatry 2006, 59, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Ebstein, F.; Lange, N.; Urban, S.; Seifert, U.; Kruger, E.; Kloetzel, P.M. Maturation of human dendritic cells is accompanied by functional remodelling of the ubiquitin-proteasome system. Int. J. Biochem. Cell Biol. 2009, 41, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Padmos, R.C.; Hillegers, M.H.; Knijff, E.M.; Vonk, R.; Bouvy, A.; Staal, F.J.; de Ridder, D.; Kupka, R.W.; Nolen, W.A.; Drexhage, H.A. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch. Gen. Psychiatry 2008, 65, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Badner, J.A.; Koller, D.; Foroud, T.; Edenberg, H.; Nurnberger, J.I., Jr.; Zandi, P.P.; Willour, V.L.; McMahon, F.J.; Potash, J.B.; Hamshere, M.; et al. Genome-wide linkage analysis of 972 bipolar pedigrees using single-nucleotide polymorphisms. Mol. Psychiatry 2012, 17, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, D.; Kuroda, K.; Tanaka, M.; Namba, T.; Iizuka, Y.; Taya, S.; Shinoda, T.; Hikita, T.; Muraoka, S.; Iizuka, M.; et al. Disrupted-in-schizophrenia 1 regulates transport of ITPR1 mRNA for synaptic plasticity. Nat. Neurosci. 2015, 18, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.C.; Javidfar, B.; Pothula, V.; Ibi, D.; Shen, E.Y.; Peter, C.J.; Bicks, L.K.; Fehr, T.; Jiang, Y.; Brennand, K.J.; et al. MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol. Psychiatry 2017. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, W.J.; Pulido, R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta 2013, 1832, 1673–1696. [Google Scholar] [CrossRef] [PubMed]
- Arey, R.N.; Enwright, J.F., 3rd; Spencer, S.M.; Falcon, E.; Ozburn, A.R.; Ghose, S.; Tamminga, C.; McClung, C.A. An important role for cholecystokinin, a CLOCK target gene, in the development and treatment of manic-like behaviors. Mol. Psychiatry 2014, 19, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Spijker, A.T.; van Rossum, E.F.; Hoencamp, E.; DeRijk, R.H.; Haffmans, J.; Blom, M.; Manenschijn, L.; Koper, J.W.; Lamberts, S.W.; Zitman, F.G. Functional polymorphism of the glucocorticoid receptor gene associates with mania and hypomania in bipolar disorder. Bipolar Disord. 2009, 11, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Spijker, S.; Van Zanten, J.S.; De Jong, S.; Penninx, B.W.; van Dyck, R.; Zitman, F.G.; Smit, J.H.; Ylstra, B.; Smit, A.B.; Hoogendijk, W.J. Stimulated gene expression profiles as a blood marker of major depressive disorder. Biol. Psychiatry 2010, 68, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, J.A.; Carter, B.S.; Meng, F.; Turner, D.L.; Dai, M.; Schatzberg, A.F.; Barchas, J.D.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J. Psychiatr. Res. 2016, 82, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Muhleisen, T.W.; Leber, M.; Schulze, T.G.; Strohmaier, J.; Degenhardt, F.; Treutlein, J.; Mattheisen, M.; Forstner, A.J.; Schumacher, J.; Breuer, R.; et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 2014, 5, 3339. [Google Scholar] [CrossRef] [PubMed]
- Perlis, R.H.; Huang, J.; Purcell, S.; Fava, M.; Rush, A.J.; Sullivan, P.F.; Hamilton, S.P.; McMahon, F.J.; Schulze, T.G.; Potash, J.B.; et al. Genome-wide association study of suicide attempts in mood disorder patients. Am. J. Psychiatry 2010, 167, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Gouvea, E.S.; Ota, V.K.; Noto, C.; Santoro, M.L.; Spindola, L.M.; Moretti, P.N.; Carvalho, C.M.; Xavier, G.; Rios, A.C.; Sato, J.R.; et al. Gene expression alterations related to mania and psychosis in peripheral blood of patients with a first episode of psychosis. Transl. Psychiatry 2016, 6, e908. [Google Scholar] [CrossRef] [PubMed]
- Pulay, A.J.; Rethelyi, J.M. Multimarker analysis suggests the involvement of BDNF signaling and microRNA biosynthesis in suicidal behavior. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2016, 171, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Shelley, W.B.; Shelley, E.D. A dermatologic diary. Portrait of a practice. Cutis 1992, 50, 179–186. [Google Scholar] [PubMed]
- Detera-Wadleigh, S.D.; Badner, J.A.; Berrettini, W.H.; Yoshikawa, T.; Goldin, L.R.; Turner, G.; Rollins, D.Y.; Moses, T.; Sanders, A.R.; Karkera, J.D.; et al. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc. Natl. Acad. Sci. USA 1999, 96, 5604–5609. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Guan, F.; Papolos, D.F.; Lau, S.; Klein, M.; Fann, C.S.; Lachman, H.M. Mutation analysis of SYNJ1: A possible candidate gene for chromosome 21q22-linked bipolar disorder. Mol. Psychiatry 2001, 6, 387–395. [Google Scholar] [CrossRef] [PubMed]
- McQuillin, A.; Bass, N.J.; Kalsi, G.; Lawrence, J.; Puri, V.; Choudhury, K.; Detera-Wadleigh, S.D.; Curtis, D.; Gurling, H.M. Fine mapping of a susceptibility locus for bipolar and genetically related unipolar affective disorders, to a region containing the C21ORF29 and TRPM2 genes on chromosome 21q22.3. Mol. Psychiatry 2006, 11, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Ruderfer, D.M.; Fanous, A.H.; Ripke, S.; McQuillin, A.; Amdur, R.L.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium; Cross-Disorder Working Group of the Psychiatric Genomics Consortium; Gejman, P.V.; O’Donovan, M.C.; et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry 2014, 19, 1017–1024. [Google Scholar] [PubMed]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011, 43, 977–983. [Google Scholar]
- Ruzicka, W.B.; Subburaju, S.; Benes, F.M. Circuit- and Diagnosis-Specific DNA Methylation Changes at gamma-Aminobutyric Acid-Related Genes in Postmortem Human Hippocampus in Schizophrenia and Bipolar Disorder. JAMA Psychiatry 2015, 72, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Benes, F.M.; Lim, B.; Matzilevich, D.; Walsh, J.P.; Subburaju, S.; Minns, M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc. Natl. Acad. Sci. USA 2007, 104, 10164–10169. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.; Lionel, A.C.; Cohen-Woods, S.; Moghimi, N.; Rucker, J.; Fennell, A.; Thiruvahindrapuram, B.; Kaufman, L.; Degagne, B.; Wei, J.; et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2014, 165B, 303–313. [Google Scholar] [CrossRef] [PubMed]
- MacLaren, E.J.; Charlesworth, P.; Coba, M.P.; Grant, S.G. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell. Neurosci. 2011, 47, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Hannah, J.; Zhou, P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 2015, 573, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.R.; Floyd, K.; Law, A.J. PKBgamma/AKT3 loss-of-function causes learning and memory deficits and deregulation of AKT/mTORC2 signaling: Relevance for schizophrenia. PLoS ONE 2017, 12, e0175993. [Google Scholar] [CrossRef] [PubMed]
- Hicks, C.; Asfour, R.; Pannuti, A.; Miele, L. An integrative genomics approach to biomarker discovery in breast cancer. Cancer Inform. 2011, 10, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Emamalizadeh, B.; Jamshidi, J.; Movafagh, A.; Ohadi, M.; Khaniani, M.S.; Kazeminasab, S.; Biglarian, A.; Taghavi, S.; Motallebi, M.; Fazeli, A.; et al. RIT2 Polymorphisms: Is There a Differential Association? Mol. Neurobiol. 2017, 54, 2234–2240. [Google Scholar] [CrossRef] [PubMed]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 2013, 381, 1371–1379. [Google Scholar]
- Vakalopoulos, C. The effect of deficient muscarinic signaling on commonly reported biochemical effects in schizophrenia and convergence with genetic susceptibility loci in explaining symptom dimensions of psychosis. Front. Pharmacol. 2014, 5, 277. [Google Scholar] [CrossRef] [PubMed]
- Lo Vasco, V.R.; Longo, L.; Polonia, P. Phosphoinositide-specific Phospholipase C beta1 gene deletion in bipolar disorder affected patient. J. Cell Commun. Signal. 2013, 7, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ban, H.J.; Kim, S.C.; Seo, J.; Kang, H.B.; Choi, J.K. Genetic and metabolic characterization of insomnia. PLoS ONE 2011, 6, e18455. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, T.; Ikeda, M.; Glatt, S.J.; Tsutsumi, A.; Kikuyama, H.; Kawamura, Y.; Nishida, N.; Miyagawa, T.; Hashimoto, R.; Takeda, M.; et al. Genome-wide association study of atypical psychosis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2013, 162B, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Zhou, R.; Wang, Y.; Li, X.; Li, J.; Chen, G.; Guitart, X.; Manji, H.K. Altered levels of extracellular signal-regulated kinase signaling proteins in postmortem frontal cortex of individuals with mood disorders and schizophrenia. J. Affect. Disord. 2010, 124, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, J.Y.; Park, H.J.; Kim, J.W.; Chung, J.H. Association study between polymorphisms of the PARD3 gene and schizophrenia. Exp. Ther. Med. 2012, 3, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Ling, W.; Jiang, J.; Hu, J.; Fan, J.; Guo, X.; Huang, G.; Xie, X.; Long, J. Association of EPHB1 rs11918092 and EFNB2 rs9520087 with psychopathological symptoms of schizophrenia in Chinese Zhuang and Han populations. Asia-Pac. Psychiatry 2016, 8, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, N.; Zhao, X.; Ross, C.A.; O’Shea, K.S.; McInnis, M.G. Gene expression alterations in bipolar disorder postmortem brains. Bipolar Disord. 2013, 15, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Geddes, J.R.; Miklowitz, D.J. Treatment of bipolar disorder. Lancet 2013, 381, 1672–1682. [Google Scholar] [CrossRef]
- Doherty, J.L.; Owen, M.J. Genomic insights into the overlap between psychiatric disorders: Implications for research and clinical practice. Genome Med. 2014, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Tsao, T.T.; Yang, K.C.; Lin, H.; Kuo, Y.L.; Hsu, C.H.; Lee, W.K.; Huang, K.C.; Kao, C.Y. Construction and analysis of the protein–protein interaction networks for schizophrenia, bipolar disorder, and major depression. BMC Bioinform. 2011, 12, S20. [Google Scholar] [CrossRef] [PubMed]
- Fritzius, T.; Burkard, G.; Haas, E.; Heinrich, J.; Schweneker, M.; Bosse, M.; Zimmermann, S.; Frey, A.D.; Caelers, A.; Bachmann, A.S.; et al. A WD-FYVE protein binds to the kinases Akt and PKCzeta/lambda. Biochem. J. 2006, 399, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Karege, F.; Perroud, N.; Schurhoff, F.; Meary, A.; Marillier, G.; Burkhardt, S.; Ballmann, E.; Fernandez, R.; Jamain, S.; Leboyer, M.; et al. Association of AKT1 gene variants and protein expression in both schizophrenia and bipolar disorder. Genes Brain Behav. 2010, 9, 503–511. [Google Scholar] [PubMed]
- Clarke, G.M.; Anderson, C.A.; Pettersson, F.H.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Basic statistical analysis in genetic case–control studies. Nat. Protoc. 2011, 6, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar]
- Wellcome Trust Case Control Consortium. Available online: https://www.wtccc.org.uk/info/access_to_data_samples.html (accessed on 15 December 2017).
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Renteria, M.E.; Cortes, A.; Medland, S.E. Using PLINK for Genome-Wide Association Studies (GWAS) and data analysis. Methods Mol. Biol. 2013, 1019, 193–213. [Google Scholar] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [PubMed]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, R.; Linial, M.; Linial, N. Generative probabilistic models for protein–protein interaction networks—The biclique perspective. Bioinformatics 2011, 27, i142–i148. [Google Scholar] [CrossRef] [PubMed]
| Name | FDR | Gene Count |
|---|---|---|
| transferase activity, transferring phosphorus-containing groups | 5.67 × 10−5 | 118 |
| kinase activity | 5.67 × 10−5 | 103 |
| phosphotransferase activity, alcohol group as acceptor | 5.67 × 10−5 | 96 |
| protein serine/threonine kinase activity | 1.03 × 10−4 | 63 |
| protein kinase activity | 1.90 × 10−4 | 81 |
| GTPase regulator activity | 2.61 × 10−4 | 47 |
| signal transducer activity, downstream of receptor | 2.61 × 10−4 | 33 |
| GTPase activator activity | 4.57 × 10−4 | 43 |
| adenyl ribonucleotide binding | 7.16 × 10−4 | 154 |
| Name | FDR | Gene Count |
|---|---|---|
| synapse | 5.23 × 10−14 | 131 |
| postsynapse | 1.68 × 10−13 | 83 |
| synapse part | 7.30 × 10−13 | 110 |
| synaptic membrane | 1.89 × 10−12 | 62 |
| cell junction | 9.04 × 10−10 | 149 |
| postsynaptic embrane | 1.91 × 10−9 | 47 |
| neuron part | 2.94 × 10−9 | 178 |
| excitatory synapse | 7.86 × 10−9 | 49 |
| plasma membrane region | 8.87 × 10−9 | 130 |
| neuron projection | 8.89 × 10−9 | 144 |
| Name | FDR | Gene Count |
|---|---|---|
| neurogenesis | 1.28 × 10−7 | 186 |
| cell morphogenesis | 1.28 × 10−7 | 159 |
| generation of neurons | 1.28 × 10−7 | 176 |
| regulation of nervous system development | 1.28 × 10−7 | 116 |
| neuron differentiation | 3.55 × 10−7 | 162 |
| neuron development | 3.55 × 10−7 | 136 |
| cell projection morphogenesis | 3.61 × 10−7 | 116 |
| cellular component morphogenesis | 4.18 × 10−7 | 164 |
| cell projection organization | 4.63 × 10−7 | 163 |
| neuron projection morphogenesis | 6.25 × 10−7 | 88 |
| Hub Gene | Degree | Hub Gene | Degree | Hub Gene | Degree |
|---|---|---|---|---|---|
| CDK1 | 62 | PNPLA6 [16] | 27 | FARS2 | 20 |
| PTEN [17,18] | 61 | SYNJ2 | 27 | FBXO22 | 20 |
| BCL2 [19] | 60 | UBE2R2 [20] | 27 | FLT3 | 20 |
| POLR2A [21] | 55 | CACNA1D [22,23,24] | 26 | GATA4 [25] * | 20 |
| SMARCA2 [26,27] | 55 | CDK6 [28] | 26 | ITSN2 [29] | 20 |
| GSK3B [30,31,32,33] | 54 | CHRM2 [34] | 26 | KIF18A | 20 |
| ABL1 [35,36,37] | 53 | MTHFD1L [38] ** | 26 | LONRF1 | 20 |
| PRKCA [39,40] | 50 | GRIA1 [41] | 25 | NCOA3 | 20 |
| bFGF [42] ** | 48 | POLR2H | 25 | PCNT [43] | 20 |
| RB1 [44] * | 45 | TJP1 [45] * | 25 | PJA2 | 20 |
| KIT [11] | 40 | MAPRE1 [46] * | 24 | SYT1 [47] | 20 |
| RAD51 * | 38 | RUNX1 [48] | 24 | TRIM39 | 20 |
| SIRT1 [49,50,51] | 38 | UBE2D4 | 24 | WDFY2 [52] ** | 20 |
| UBE2D1 [53,54] | 37 | EHHADH | 23 | AK4 | 19 |
| DLG1 [55,56] | 36 | IQCB1 | 23 | ASB15 | 19 |
| CDC27 [57] | 35 | PPM1B [58] | 23 | ATF2 [29] | 19 |
| NEDD4L [59] | 35 | PPP4C [60] * | 23 | BUB1B | 19 |
| PRKG1 [61] * | 35 | RAD50 | 23 | DHX15 | 19 |
| RAP1A | 34 | SH3GL2 | 23 | DNM3 [62] * | 19 |
| CDH2 [63] * | 33 | DCTN1 | 22 | ETV6 | 19 |
| GNB5 [64] * | 33 | ERBB4 [65,66] | 22 | FBXL13 [67,68] ** | 19 |
| MAPK6 [69] | 33 | FBXO32 | 22 | HECW2 | 19 |
| GNG7 [70] | 32 | ITPR1 [71] * | 22 | MEF2C [72] * | 19 |
| PTPN11 [73] * | 32 | MLL [74] | 22 | NR3C1 [75] | 19 |
| ZBTB16 [76] | 32 | NCOR2 [77] | 22 | BDE4D | 19 |
| ADCY2 [78] | 31 | PRKCE [79] | 22 | RNF19B | 19 |
| DICER1 [80,81] | 31 | RAD51B | 22 | RNF217 | 19 |
| SYNJ1 [82,83,84,85] | 31 | ACTN4 | 21 | RXFP2 | 19 |
| CACNA1C [9,52,86,87] | 30 | CCND2 [88,89] | 21 | RYR1 | 19 |
| CTTN | 30 | CDH5 | 21 | THBS2 | 19 |
| DLG2 [90,91] | 30 | CUL4A [92] * | 21 | AKT3 [93] * | 18 |
| MAP3K1 [94] * | 30 | EFCAB13 | 21 | BARD1 | 18 |
| RIT2 [95] | 30 | LMO7 | 21 | CTNNA2 [11,96] | 18 |
| ANAPC5 [28] | 28 | MITF | 21 | HDAC7 | 18 |
| PLCB1 [97,98,99] | 28 | TRIM9 [100] | 21 | ITGAV | 18 |
| RAF1 [101] * | 28 | CCND3 | 20 | PARD3 [102] * | 18 |
| PARK2 [61] * | 27 | EPHB1 [103] * | 20 | PCSK2 | 18 |
| PLCG2 [13,39] | 27 |
| Cluster | Score | Nodes | Edges | Node IDs |
|---|---|---|---|---|
| 1 | 20 | 20 | 190 | ASB15, HECW2, UBE2D1, NEDD4L, ANAPC5, PJA2 TRIM39, UBE2R2, UBE2D4, CDC27, TRIM9, ZBTB16 LONRF1, PARK2, FBXL13 *, FBXO22, RNF19B, LMO7 RNF217, FBXO32 |
| 2 | 6.1 | 21 | 61 | SYNJ1, KIT, PIK3C2G, PTPN11, PIK3C2B, SYNJ2 RUNX1, ITSN2, PLCB1, CDH2, DNM3, SYT1, CTTN WDFY2 *, CHRM2, CCND2, MITF, PLCG2, CDK6, ETV6, SH3GL2 |
| 3 | 5.5 | 13 | 33 | MLL, bFGF(FGF2) *, BUB1B, BARD1, RB1, DICER1, RAD50, RAD51, BCL2, CDH5, SMARCA2, ABL1, CCND3 |
| 4 | 4.182 | 12 | 23 | AKT3, PTEN, ITPR1, PRKCE, GNB5, CDK1 ERBB4, GNG7, RAF1, GSK3B, PPM1B, MAP3K1 |
| 5 | 3 | 3 | 3 | FARS2, RAD51B, MTHFD1L * |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Yang, X.; Deng, X.; Ma, M.; Shu, K. A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder. Int. J. Mol. Sci. 2017, 18, 2763. https://doi.org/10.3390/ijms18122763
Xie Z, Yang X, Deng X, Ma M, Shu K. A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder. International Journal of Molecular Sciences. 2017; 18(12):2763. https://doi.org/10.3390/ijms18122763
Chicago/Turabian StyleXie, Zengyan, Xianyan Yang, Xiaoya Deng, Mingyue Ma, and Kunxian Shu. 2017. "A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder" International Journal of Molecular Sciences 18, no. 12: 2763. https://doi.org/10.3390/ijms18122763
APA StyleXie, Z., Yang, X., Deng, X., Ma, M., & Shu, K. (2017). A Genome-Wide Association Study and Complex Network Identify Four Core Hub Genes in Bipolar Disorder. International Journal of Molecular Sciences, 18(12), 2763. https://doi.org/10.3390/ijms18122763