Identification of Novel Human NK Cell Progenitor Subsets
Abstract
:1. Introduction
2. Results
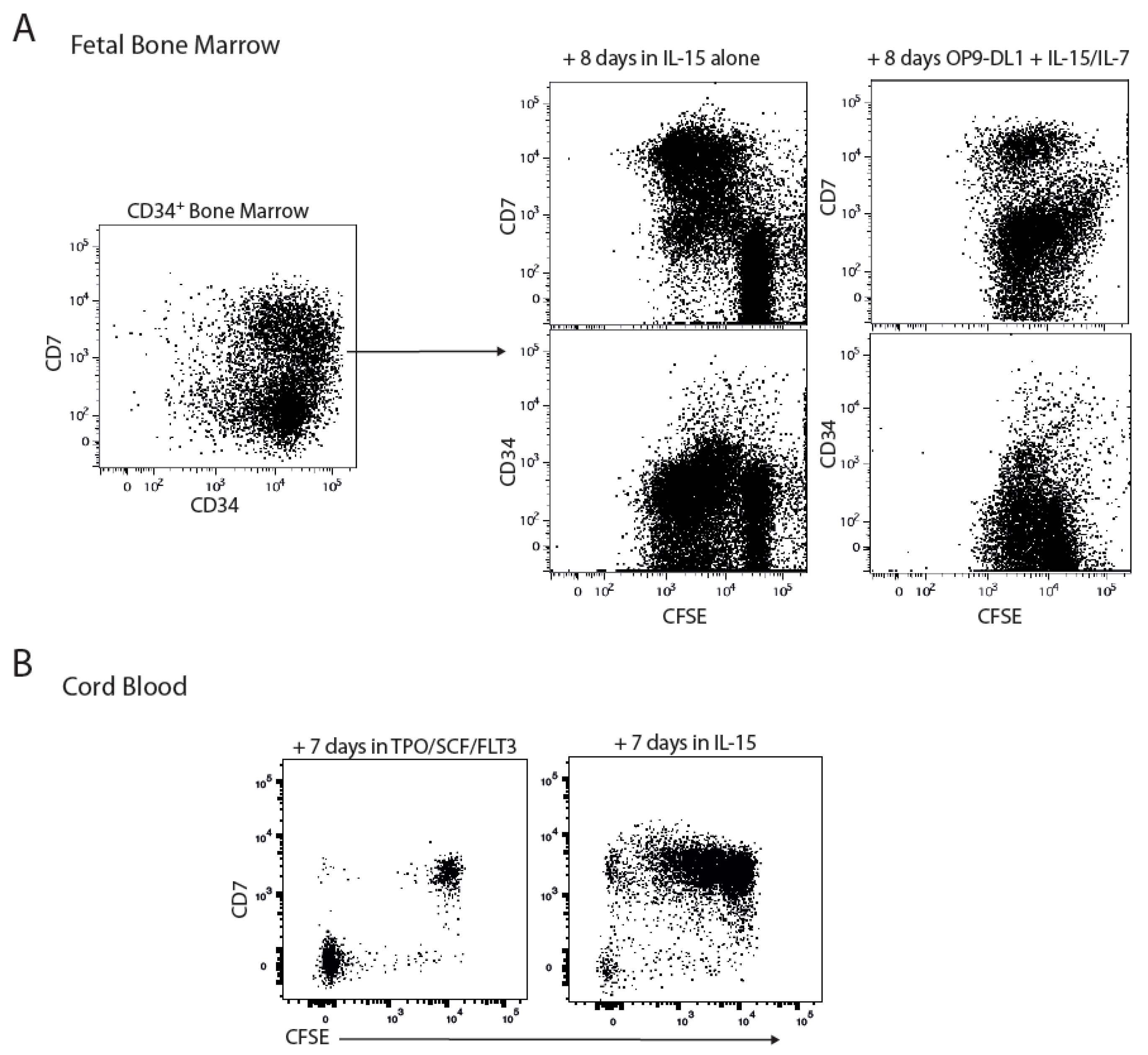
2.1. CD7 Identifies the IL-15 Responsive Population Amongst CD34+ Progenitors
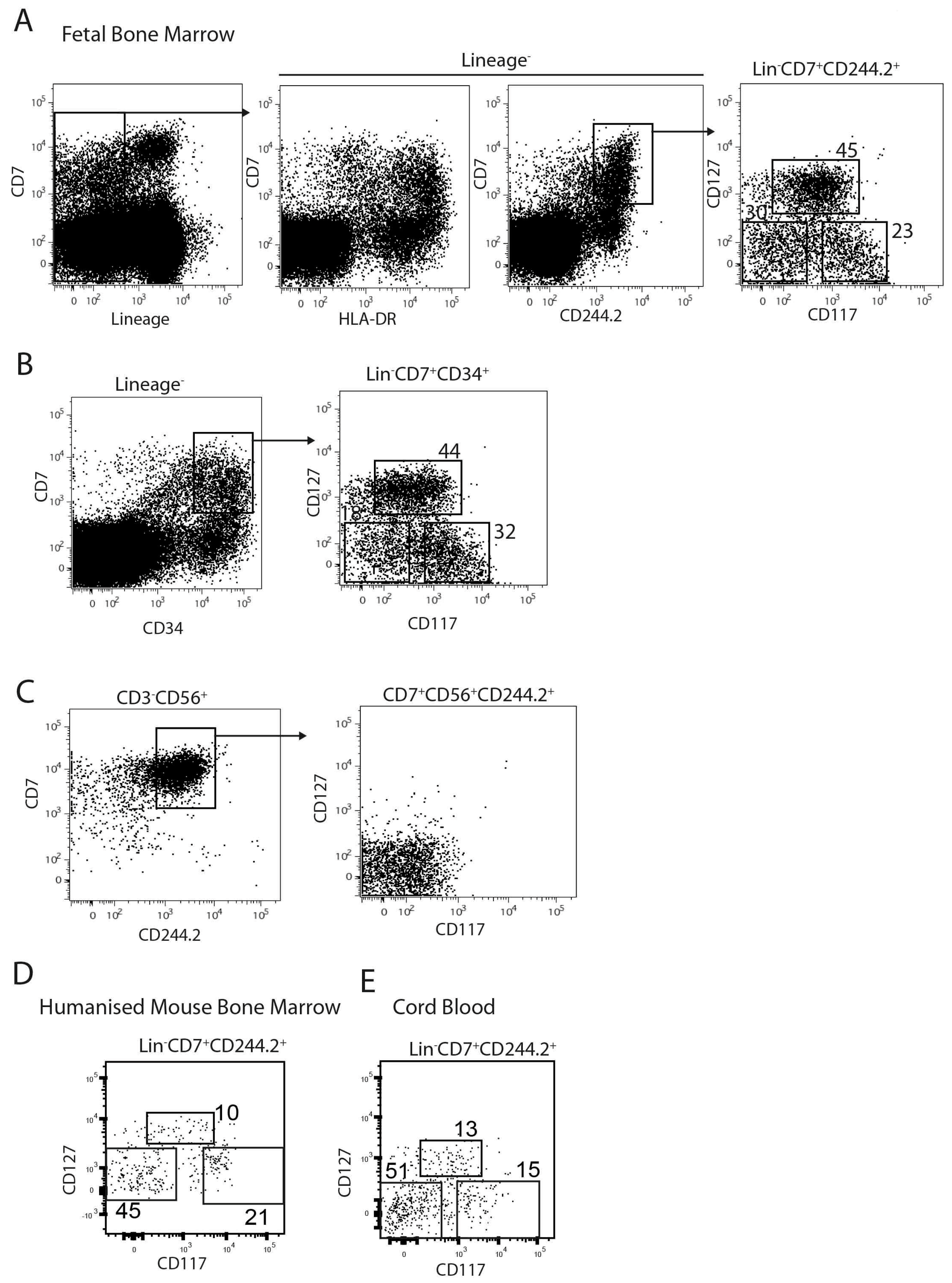
2.2. The Lin−CD7+ Fraction Consists of Multiple Subsets Defined by CD117 and CD127
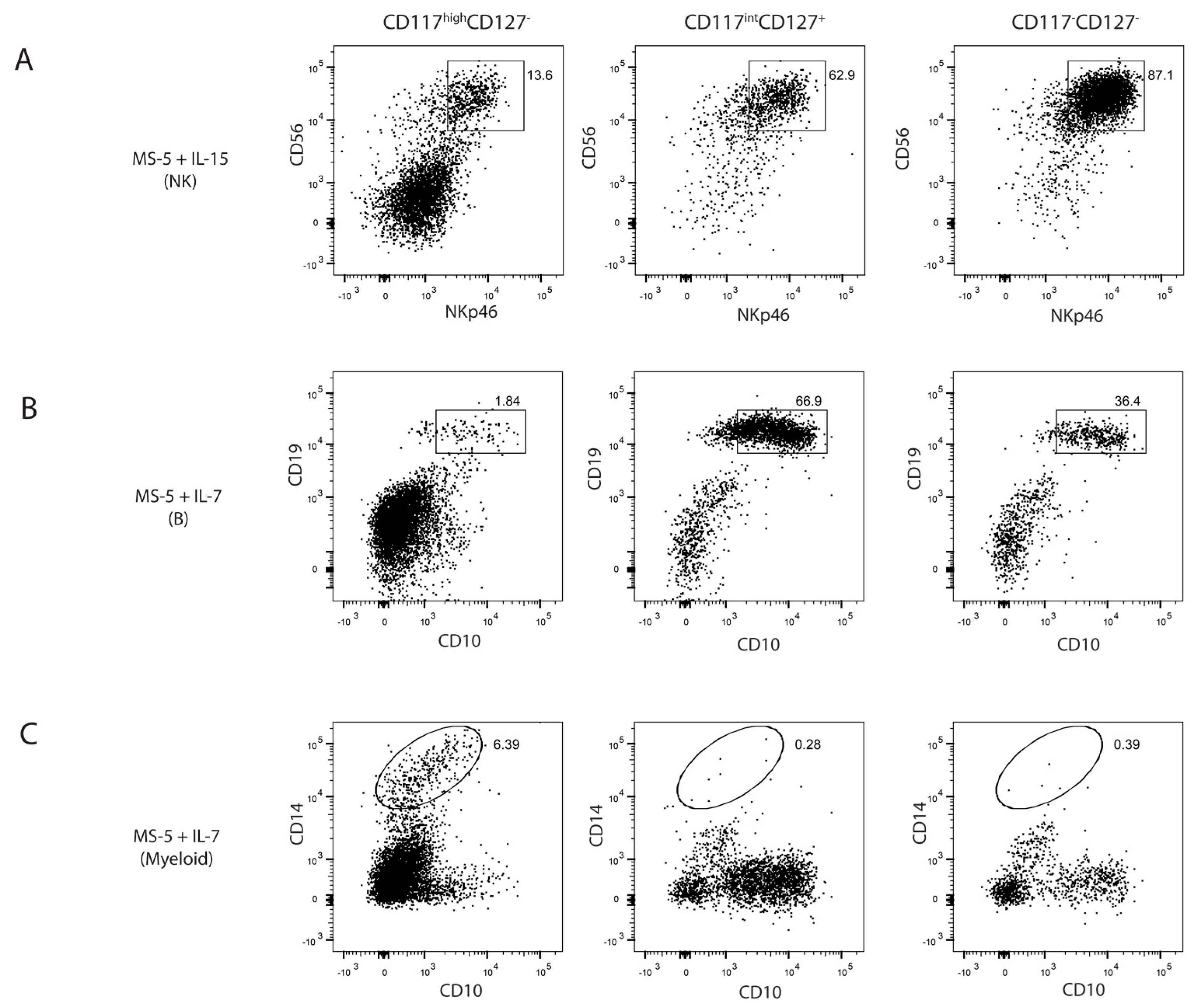
2.3. Differential Lineage Potential of Progenitors
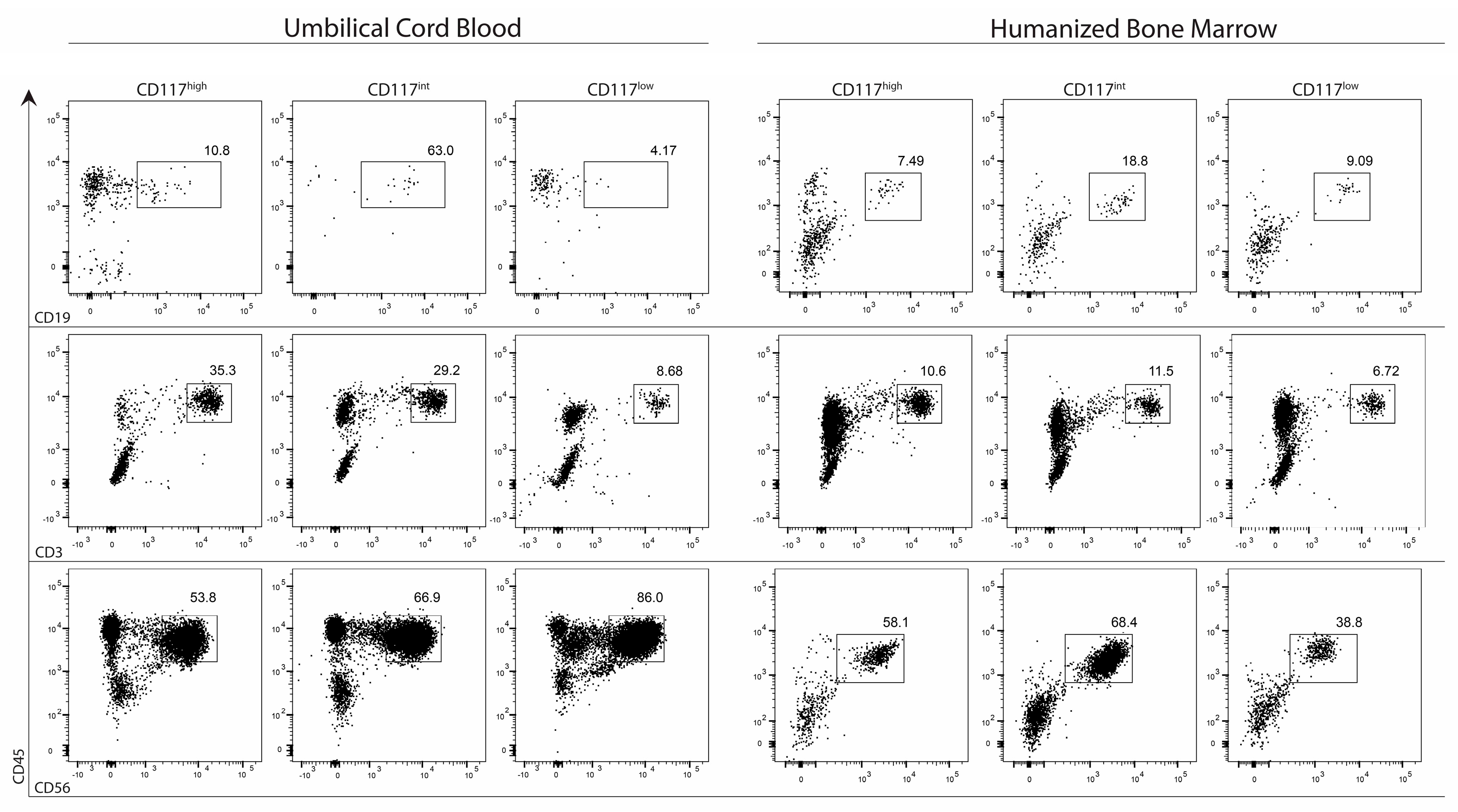
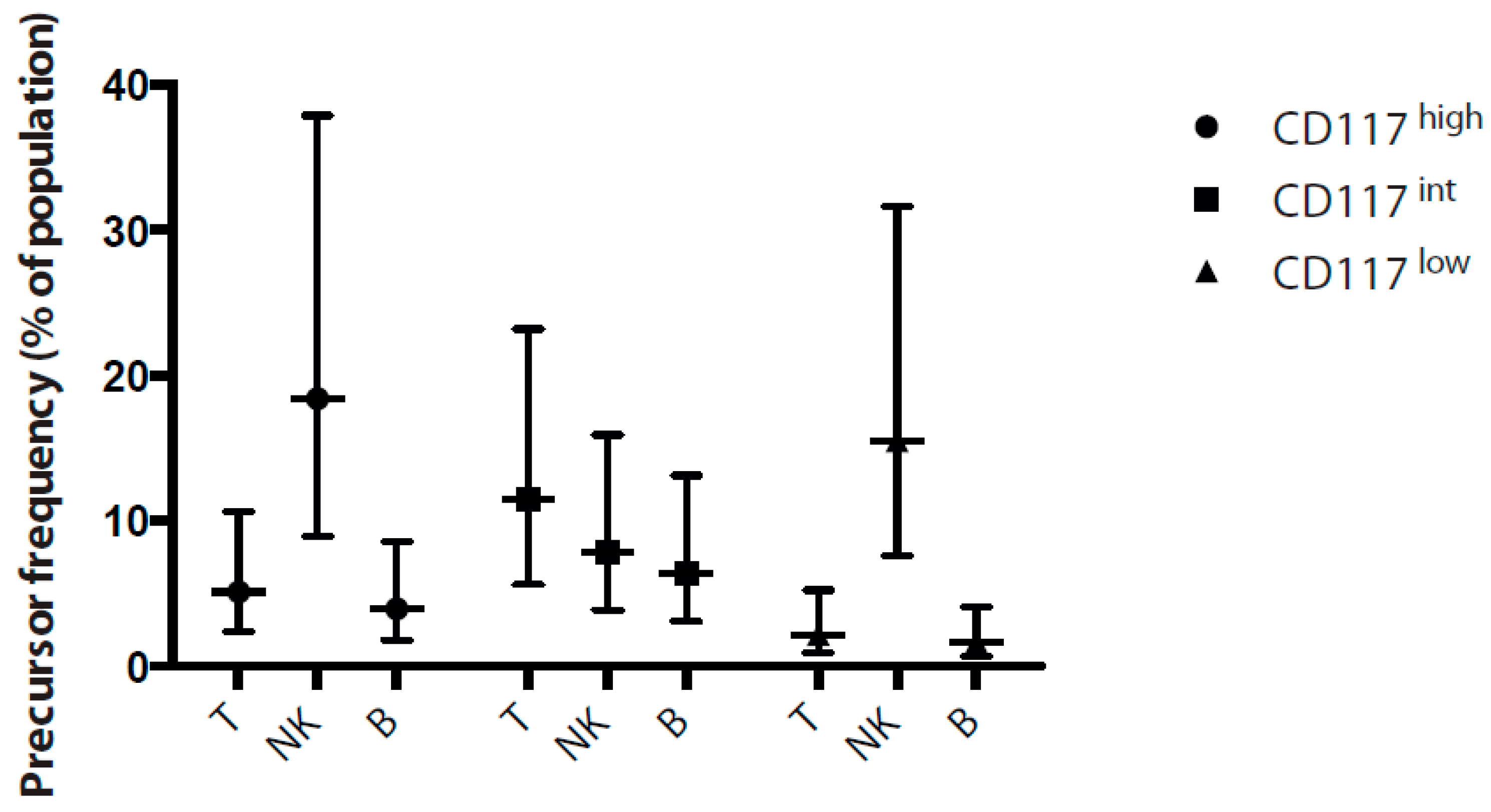
2.4. CD7+ Progenitor Subset Potential in HIS BM and Cord Blood
2.5. Frequency of Lymphoid Progenitors within CD7 Populations
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. In Vitro Assays of Progenitor Potential
4.3. Flow Cytometry Analysis for Cell-Surface and Intracellular Markers
4.4. Cell Preparation
4.5. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mori, Y.; Iwasaki, H.; Kohno, K.; Yoshimoto, G.; Kikushige, Y.; Okeda, A.; Miyamoto, T. Identification of the human eosinophil lineage-committed progenitor: Revision of phenotypic definition of the human common myeloid progenitor. J. Exp. Med. 2009, 206, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Manz, M.G.; Miyamoto, T.; Akashi, K.; Weissman, I.L. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. USA 2002, 99, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Galy, A.; Travis, M.; Cen, D.; Chen, B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 1995, 3, 459–473. [Google Scholar] [CrossRef]
- Galy, A.H.; Cen, D.; Travis, M.; Chen, S.; Chen, B.P. Delineation of T-progenitor cell activity within the CD34+ compartment of adult bone marrow. Blood 1995, 85, 2770–2778. [Google Scholar] [PubMed]
- Hao, Q.L.; Zhu, J.; Price, M.A.; Payne, K.J.; Barsky, L.W.; Crooks, G.M. Identification of a novel, human multilymphoid progenitor in cord blood. Blood 2001, 97, 3683–3690. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Niiro, H.; Iino, T.; Yoshida, S.; Saito, N.; Onohara, S.; Harada, M. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood 2007, 110, 3591–3660. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.J.; Muench, M.O.; Roncarolo, M.G.; Lanier, L.L.; Phillips, J.H. Identification of a common T/natural killer cell progenitor in human fetal thymus. J. Exp. Med. 1994, 180, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Renoux, V.M.; Zriwil, A.; Peitzsch, C.; Michaëlsson, J.; Friberg, D.; Soneji, S.; Sitnicka, E. Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity 2015, 43, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Kovanen, P.E.; Leonard, W.J. Cytokines and immunodeficiency diseases: Critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol. Rev. 2004, 202, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Puthalakath, H.; Gunn, P.; Naik, E.; Michalak, E.M.; Smyth, M.J.; Motoyama, N. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat. Immunol. 2007, 8, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Legrand, N.; Alves, N.L.; Jaron, B.; Weijer, K.; Plet, A.; Di Santo, J.P. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009, 206, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Mrozek, E.; Anderson, P.; Caligiuri, M.A. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood 1996, 87, 2632–2640. [Google Scholar] [PubMed]
- Carotta, S.; Pang, S.H.; Nutt, S.L.; Belz, G.T. Identification of the earliest NK-cell precursor in the mouse BM. Blood 2011, 117, 5449–5452. [Google Scholar] [CrossRef] [PubMed]
- Delconte, R.B.; Shi, W.; Sathe, P.; Ushiki, T.; Seillet, C.; Minnich, M.; Busslinger, M. The Helix-Loop-Helix Protein ID2 Governs NK Cell Fate by Tuning Their Sensitivity to Interleukin-15. Immunity 2016, 44, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Rosmaraki, E.E.; Douagi, I.; Roth, C.; Colucci, F.; Cumano, A.; Di Santo, J.P. Identification of committed NK cell progenitors in adult murine bone marrow. Eur. J. Immunol. 2001, 31, 1900–1909. [Google Scholar] [CrossRef]
- Carayol, G.; Robin, C.; Bourhis, J.H.; Bennaceur-Griscelli, A.; Chouaib, S.; Coulombel, L.; Caignard, A. NK cells differentiated from bone marrow, cord blood and peripheral blood stem cells exhibit similar phenotype and functions. Eur. J. Immunol. 1998, 28, 1991–2002. [Google Scholar] [CrossRef]
- Kyoizumi, S.; Kubo, Y.; Kajimura, J.; Yoshida, K.; Hayashi, T.; Nakachi, K.; Kusunoki, Y. Fate Decision Between Group 3 Innate Lymphoid and Conventional NK Cell Lineages by Notch Signaling in Human Circulating Hematopoietic Progenitors. J. Immunol. 2017, 199, 2777–2793. [Google Scholar] [CrossRef] [PubMed]
- Sathe, P.; Huntington, N.D.; (The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC 3052, Australia). Personal communication, 2017.
- Huntington, N.D.; Vosshenrich, C.A.; Di Santo, J.P. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 2007, 7, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D. The unconventional expression of IL-15 and its role in NK cell homeostasis. Immunol. Cell Biol. 2014, 92, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Rabinowich, H.; Pricop, L.; Herberman, R.B.; Whiteside, T.L. Expression and function of CD7 molecule on human natural killer cells. J. Immunol. 1994, 152, 517–526. [Google Scholar] [PubMed]
- Huntington, N.D.; Alves, N.L.; Legrand, N.; Lim, A.; Strick-Marchand, H.; Mention, J.J.; Guzman, C. IL-15 transpresentation promotes both human T-cell reconstitution and T-cell-dependent antibody responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6217–6222. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Xu, Y.; Nutt, S.L.; Tarlinton, D.M. A requirement for CD45 distinguishes Ly49D-mediated cytokine and chemokine production from killing in primary natural killer cells. J. Exp. Med. 2005, 201, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sathe, P.; Pang, S.H.M.; Delconte, R.; Elwood, N.; Huntington, N.D. Identification of Novel Human NK Cell Progenitor Subsets. Int. J. Mol. Sci. 2017, 18, 2716. https://doi.org/10.3390/ijms18122716
Sathe P, Pang SHM, Delconte R, Elwood N, Huntington ND. Identification of Novel Human NK Cell Progenitor Subsets. International Journal of Molecular Sciences. 2017; 18(12):2716. https://doi.org/10.3390/ijms18122716
Chicago/Turabian StyleSathe, Priyanka, Swee Heng Milon Pang, Rebecca Delconte, Ngaire Elwood, and Nicholas D. Huntington. 2017. "Identification of Novel Human NK Cell Progenitor Subsets" International Journal of Molecular Sciences 18, no. 12: 2716. https://doi.org/10.3390/ijms18122716
APA StyleSathe, P., Pang, S. H. M., Delconte, R., Elwood, N., & Huntington, N. D. (2017). Identification of Novel Human NK Cell Progenitor Subsets. International Journal of Molecular Sciences, 18(12), 2716. https://doi.org/10.3390/ijms18122716





