The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint
Abstract
1. Introduction
2. Current Knowledge on MicroRNAs Involved in PD-1/PD-L1 Regulation
2.1. MicroRNAs That Regulate PD-1/PD-L1 Directly
2.1.1. MicroRNAs Regulating PD-1 Expression
2.1.2. MicroRNAs Regulating PD-L1 Expression
2.2. Potential PD-1/PD-L1 Regulatory microRNAs
2.2.1. MicroRNAs Regulating IFN-γ Expression
2.2.2. MicroRNAs Regulating IFNGR Expression
2.2.3. MicroRNAs Regulating STAT1 Expression
2.2.4. MicroRNAs Regulating IRF1 Expression
2.2.5. MicroRNAs Regulating PTEN Expression
2.2.6. MicroRNAs Regulatingm TOR Expression
2.2.7. MicroRNAs Regulating Eukaryotic Translation Initiation Factor 4B (EIF4B) Expression
2.2.8. MicroRNAs Regulating SHP2 Expression
2.2.9. MicroRNAs Regulating c-Fos Protein (c-Fos) Expression
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Hirano, F.; Kaneko, K.; Tamura, H.; Dong, H.; Wang, S.; Ichikawa, M.; Rietz, C.; Flies, D.B.; Lau, J.S.; Zhu, G.; et al. Blockade of B7-H1 and PD-1 by Monoclonal Antibodies Potentiates Cancer Therapeutic Immunity. Cancer Res. 2005, 65, 1089–1096. [Google Scholar] [PubMed]
- Eppihimer, M.J.; Gunn, J.; Freeman, G.J.; Greenfield, E.A.; Chernova, T.; Erickson, J.; Leonard, J.P. Expression and Regulation of the PD-L1 Immunoinhibitory Molecule on Microvascular Endothelial Cells. Microcirculation 2002, 9, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Mühlbauer, M.; Fleck, M.; Schütz, C.; Weiss, T.; Froh, M.; Blank, C.; Schölmerich, J.; Hellerbrand, C. PD-L1 is induced in hepatocytes by viral infection and by interferon-α and -γ and mediates T cell apoptosis. J. Hepatol. 2006, 45, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hamrouni, A.; Wolowiec, D.; Coiteux, V.; Kuliczkowski, K.; Hetuin, D.; Saudemont, A.; Quesnel, B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-γ and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood 2007, 110, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, Y.; Xiang, J.; Chen, Z.; Wang, L.; Lu, L.; Qian, S. Interferon-γ triggers hepatic stellate cell-mediated immune regulation through MEK/ERK signaling pathway. Clin. Dev. Immunol. 2013, 2013, 389807. [Google Scholar] [CrossRef] [PubMed]
- Parsa, A.T.; Waldron, J.S.; Panner, A.; Crane, C.A.; Parney, I.F.; Barry, J.J.; Cachola, K.E.; Murray, J.C.; Tihan, T.; Jensen, M.C.; et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007, 13, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Ogasawara, K.; Takaoka, A.; Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001, 19, 623–655. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Lander, E.S.; Rogers, J.; Waterson, R.H. Finishing the euchromatic sequence of the human genome International Human Genome Sequencing Consortium Nature 2004 431 931 45. Nature 2003, 431, 931–945. [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.J.; Matzke, M.A. The small RNA world. J. Cell Sci. 2003, 23(116Pt), 4689–4693. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Calin, G.A. MicroRNAs in cancer: From developmental genes in worms to their clinical application inpatients. Br. J. Cancer 2015, 113, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Johnston, N.; Zheng, X.; Wang, H.; Zhang, X.; Gao, D.; Min, W. MiR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 2016, 7, 53735–53750. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Nduom, E.; Kong, L.Y.; Wang, F.; Xu, S.; Gabrusiewicz, K.; Alum, A.; Fuller, G.; Calin, G.; Heimberger, A.B. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. J. Immunother. Cancer 2013, 1, 177. [Google Scholar] [CrossRef]
- Zhang, G.; Na, L.; Zhu, L.; Zhu, Q.; Fang, L.; Yang, C.; Han, Q.; Yi, L.; Zhou, Z.; Liu, Z. MicroRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget 2015, 6, 18933–18944. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.C.; Cheng, Y.Y.; Williams, M.; Kirschner, M.B.; Madore, J.; Lum, T.; Sarun, K.H.; Linton, A.; Mccaughan, B.; Klebe, S.; et al. Tumour suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J. Thorac. Oncol. 2017, 12, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; Trabulo, S.M.; Vallespinos, M.; Raj, D.; Kheir, T.B.; Lin, M.L.; Begum, J.; Baker, A.M.; Amgheib, A.; Saif, J.; et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget 2017, 8, 21609–21625. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yu, H.; Yi, S.; Peng, X.; Peng, S.; Xiao, Z.; Liu, R.; Tang, A.; Li, X.; Liu, F.; et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016, 7, 45370–45384. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. MiR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Di, W.; Xie, G.; Yin, Y.; Zhao, E.; Tao, K.; Li, R. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 2017, 8, 28125–28134. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef] [PubMed]
- Holla, S.; Stephenvictor, E.; Prakhar, P.; Sharma, M.; Saha, C.; Udupa, V.; Kaveri, S.V.; Bayry, J. Mycobacteria-responsive sonic hedgehog signaling mediates programmed death-ligand 1- and prostaglandin E2-induced regulatory T cell expansion. Sci. Rep. 2016, 6, 24193. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; Ouyang, J.; Chen, J.; et al. MiR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.Y.; Zhou, R.; Hu, G.; Liu, J.; Sosnowska, D.; Drescher, K.M.; Dong, H.; Chen, X.M. Cryptosporidium parvum Induces B7-H1 Expression in Cholangiocytes by Downregulating MicroRNA-513. J. Infect. Dis. 2010, 201, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Sang, W.; Zhang, C.; Zhang, D.; Wang, Y.; Sun, C.; Niu, M.; Sun, X.; Zhou, C.; Zeng, L.; Pan, B.; et al. MicroRNA-181a, a Potential Diagnosis Marker, Alleviates Acute Graft Versus Host Disease by Regulating IFN-γ Production. Am. J. Hematol. 2015, 90, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, Y.; Teng, M.; Zhang, D.; Li, L.; Liu, Y. Signal Transducers and Activators of Transcription-1 (STAT1) Regulates microRNA Transcription in Interferon γ-Stimulated HeLa Cells. PLoS ONE 2010, 5, e11794. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Jiang, H.; Gao, Y.; Zhao, Y.; Dai, L.; Xiong, Q.; Xu, Y.; Zhao, Z.; Zhang, J. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon γ receptor 1 gene. J. Appl. Genet. 2011, 52, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Jacobsen, A.B.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-145 Targets YES and STAT1 in Colon Cancer Cells. PLoS ONE 2010, 5, e8836. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef] [PubMed]
- Moles, R.; Bellon, M.; Nicot, C. STAT1: A Novel Target of miR-150 and miR-223 Is Involved in the Proliferation of HTLV-I-Transformed and ATL Cells. Neoplasia 2015, 17, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, B.; Feng, M.; Ouyang, H.; Zheng, M.; Ye, Q.; Nie, Q.; Zhang, X. MicroRNA-23b Promotes Avian Leukosis Virus Subgroup J (ALV-J) Replication by Targeting IRF1. Sci. Rep. 2015, 5, 10294. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Tian, H.; Liu, L.; Zhang, X.S.; Li, W.Q.; Deng, Y.M.; Yao, G.D.; Yin, M.M.; Sun, F. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010, 1, e94. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Liu, L.; Li, J.; Yan, M.; Lin, H.; Liu, Y.; Chu, D.; Tu, H.; Gu, A.; Yao, M. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget 2015, 6, 30239–30250. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, T.; Zhang, B.; Li, H.; Wu, Q.; Yang, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem. Biophys. Res. Commun. 2013, 434, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, L.; Zou, L.; Yang, P.; Wu, R.; Mao, Y.; Zhou, H.; Li, R.; Wang, K.; Wang, W.; et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol. 2014, 75, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wu, X.; Liu, B.; Wang, C.; Liu, Y.; Zhou, Q.; Xu, K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim. Biophys. Acta 2012, 1822, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhou, H.; Xiao, H.; Liu, Z.; Tian, H.; Zhou, T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig. Dis. Sci. 2014, 59, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Liang, Z.; Huang, J.; Zhao, R.; Su, C.; Wang, S.; Wang, X.; Zhang, R.; Lee, M.H.; Yang, H. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 2012, 11, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V. MicroRNA expression profiling in human ovarian cancer: MiR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Han, L.; Zhang, A.; Yue, X.; Wang, G.; Jia, Z.; Pu, P.; Zhang, Q.; Kang, C. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10, 367. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, J.; Zhong, L.; Wang, L.; Liu, Y.; Wang, Y.; Peng, L.; Guo, B. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene 2014, 535, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lai, L.; Chen, Q.; Song, Y.; Xu, S.; Ma, F.; Wang, X.; Wang, J.; Yu, H.; Cao, X.; et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J. Immunol. 2012, 188, 5500–5510. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zeng, Q.; Xu, W.; Jiao, L.; Chen, Y.; Zhang, Z.; Wu, C.; Jin, T.; Pan, A.; Wei, R.; et al. MiRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol. Cancer Ther. 2013, 12, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Shao, N.N.; Fan, L.; Ma, X.C.; Pu, F.F.; Shao, Z.W. Effect of microRNA-101 on proliferation and apoptosis of human osteosarcoma cells by targeting mTOR. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014, 34, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Huang, H.J.; He, C.N.; Wang, K.Y. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR). Int. J. Gynecol. Cancer 2013, 23, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Fu, G.B.; Tao, Z.; Ouyang, J.; Kong, F.; Jiang, B.H.; Wan, X.; Chen, K. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget 2015, 6, 26457–26471. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.H.; Wang, S.L.; Zhao, J.T.; Lin, Z.J.; Chen, L.Y.; Su, R.; Xie, S.T.; Bing, Z.C.; Xu, B. MiR-150 exerts antileukemia activityin vitroandin vivothrough regulating genes in multiple pathways. Cell Death Dis. 2016, 7, e2371. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.T.; Xu, M.; Xu, C.X.; Song, Z.G.; Jin, H. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin. Cancer Res. 2014, 20, 4705–4716. [Google Scholar] [CrossRef] [PubMed]
- Courboulin, A.; Paulin, R.; Giguère, N.J.; Saksouk, N.; Perreault, T.; Meloche, J.; Paquet, E.R.; Biardel, S.; Provencher, S.; Côté, J.; et al. Role for miR-204 in human pulmonary arterial hypertension. J. Exp. Med. 2011, 208, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Rongzhen, H.E.; Xia, H.; Wei, Y.U.; Song, W.U. MicroRNA-101 has a suppressive role in osteosarcoma cells through the targeting of c-FOS. Exp. Ther. Med. 2016, 11, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Katsumi, T.; Ninomiya, M.; Nishina, T.; Mizuno, K.; Tomita, K.; Haga, H.; Okumoto, K.; Saito, T.; Shimosegawa, T.; Ueno, Y. MiR-139-5p is associated with inflammatory regulation through c-FOS suppression, and contributes to the progression of primary biliary cholangitis. Lab. Investig. 2016, 96, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Dunand-Sauthier, I.; Santiago-Raber, M.L.; Capponi, L.; Vejnar, C.E.; Schaad, O.; Irla, M.; Seguín-Estévez, Q.; Descombes, P.; Zdobnov, E.M.; Acha-Orbea, H.; et al. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood 2011, 117, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Gong, Y.; Yuan, J.; Zhang, W.; Zhao, G.; LI, H.; Sun, A.; Hu, K.; Zou, Y.; Ge, J. MicroRNA-181a represses ox-LDL-stimulated inflammatory response in dendritic cell by targeting c-Fos. J. Lipid Res. 2012, 53, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Wang, Y.; Luo, H.; Yao, L.; Wang, L.; Wang, J.; Zhang, J.; Wang, H.; Shi, Y.; Yin, Y.; et al. Involvement of FOS-mediated miR-181b/miR-21 signalling in the progression of malignant gliomas. Eur. J. Cancer 2013, 49, 3055–3063. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, X.; Xu, X.; Hu, Z.; Wu, J.; Zhu, Y.; Chen, H.; Mao, Y.; Lin, Y.; Luo, J.; et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem. Biophys. Res. Commun. 2013, 441, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yang, J.; Li, J.; Wang, X.; Chen, Y.; Huang, S.; Chen, J.L. EIF4B is a convergent target and critical effector of oncogenic Pim and PI3K/Akt/mTOR signaling pathways in Abl transformants. Oncotarget 2016, 7, 10073–10089. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Liu, B.; Zang, L.E.; Jiang, H. MiR-631/ZAP70: A novel axis in the migration and invasion of prostate cancer cells. Biochem. Biophys. Res. Commun. 2015, 469, 345–351. [Google Scholar] [CrossRef] [PubMed]
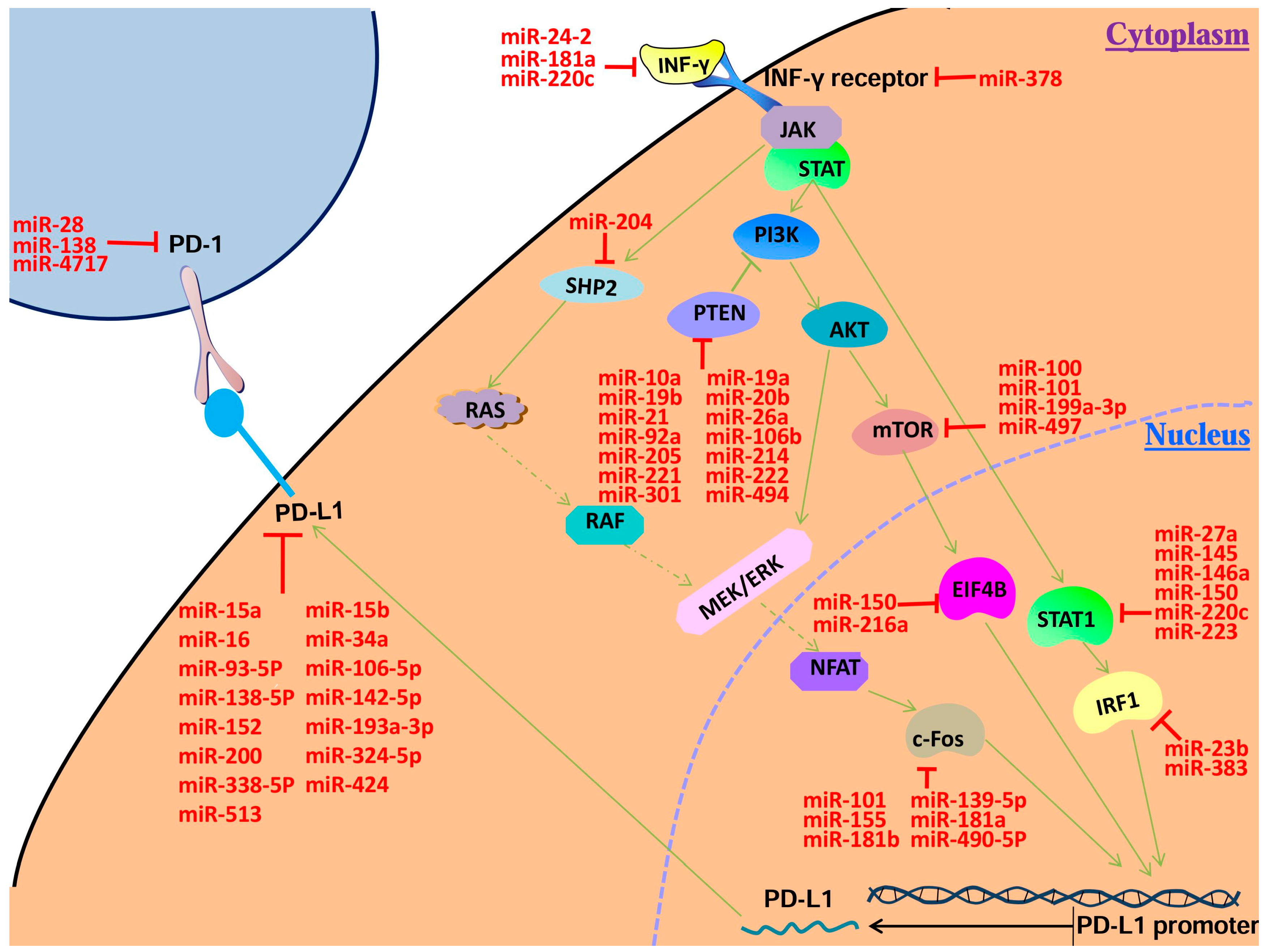
| Target mRNA | miRNA | Host | Reference |
|---|---|---|---|
| PD-1 | miR-28 | melanoma | Li et al. [17] |
| miR-138 | glioblastoma | Wei et al. [18] | |
| miR-4717 | HCC | Zhang et al. [19] | |
| PD-L1 | miR-15a | MPM | Kao et al. [20] |
| miR-15b | MPM | Kao et al. [20] | |
| miR-16 | MPM | Kao et al. [20] | |
| miR-34a | AML | Wang et al. [21] | |
| miR-93 | MSC | Cioffi et al. [22] | |
| miR-106b | MSC | Cioffi et al. [22] | |
| miR-138-5p | CRC | Zhao et al. [23] | |
| miR-142-5p | PC | Jia et al. [24] | |
| miR-152 | GC | Wang et al. [25] | |
| miR-193a-3p | MPM | Kao et al. [20] | |
| miR-200 | GC | Chen et al. [26] | |
| miR-324-5p | Treg | Holla et al. [27] | |
| miR-338-5p | Treg | Holla et al. [27] | |
| miR-424 | OC | Xu et al. [28] | |
| miR-513 | Cholangiocytes | Gong et al. [29] | |
| IFN-γ | miR-181a | aGVHD | Sang et al. [30] |
| miR-24-2 | HeLa | Wang et al. [31] | |
| miR-200c | HeLa | Wang et al. [31] | |
| IFNGR | miR-378 | CL | Ma et al. [32] |
| STAT1 | miR-27a | HeLa | Wang et al. [31] |
| miR-145 | CC | Gregersen et al. [33] | |
| miR-146a | Treg | Lu et al. [34] | |
| miR-150 | ATL | Moles et al. [35] | |
| miR-223 | ATL | Moles et al. [35] | |
| miR-200c | HeLa | Wang et al. [31] | |
| IRF1 | miR-23b | DF-1 | Li et al. [36] |
| miR-383 | TEC | Lian et al. [37] | |
| PTEN | miR-10a | NSCLC | Yu et al. [38] |
| miR-19a | GC | Wang et al. [39] | |
| miR-19b | GC | Wang et al. [39] | |
| miR-20b | CRC | Zhu et al. [40] | |
| miR-21 | CRC | Zhu et al. [40] | |
| miR-26a | LC | Liu et al. [41] | |
| miR-92a | CRC | Zhang et al. [42] | |
| miR-106b | GC | Yang et al. [43] | |
| miR-205 | NPC | Qu et al. [44] | |
| miR-214 | OC | Yang et al. [45] | |
| miR-221 | GC | Zhang et al. [46] | |
| miR-222 | GC | Zhang et al. [46] | |
| miR-301a | BC | Ma et al. [47] | |
| miR-494 | MDSCs | Liu et al. [48] | |
| MTOR | miR-100 | BLC | Xu et al. [49] |
| miR-101 | OTC | Lin et al. [50] | |
| miR-199a-3p | EEC | Wu et al. [51] | |
| miR-497 | OC | Xu et al. [52] | |
| EIF4B | miR-150 | HSC | Fang et al. [53] |
| miR-216a | NSCLC | Wang et al. [54] | |
| SHP2 | miR-204 | PASMCs | Courboulin et al. [55] |
| FOS | miR-101 | OS | Wang et al. [56] |
| miR-139-5p | PBC | Katsumi1 et al. [57] | |
| miR-155 | DCs | Dunand-Sauthier et al. [58] | |
| miR-181a | DCs | Wu et al. [59] | |
| miR-181b | MG | Tao et al. [60] | |
| miR-490-5p | BLC | Li et al. [61] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Lin, W.; Tang, X.; Li, S.; Guo, L.; Lin, Y.; Kwok, H.F. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int. J. Mol. Sci. 2017, 18, 2540. https://doi.org/10.3390/ijms18122540
Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y, Kwok HF. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. International Journal of Molecular Sciences. 2017; 18(12):2540. https://doi.org/10.3390/ijms18122540
Chicago/Turabian StyleWang, Qingshui, Wei Lin, Xiaoqiong Tang, Suhuan Li, Libin Guo, Yao Lin, and Hang Fai Kwok. 2017. "The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint" International Journal of Molecular Sciences 18, no. 12: 2540. https://doi.org/10.3390/ijms18122540
APA StyleWang, Q., Lin, W., Tang, X., Li, S., Guo, L., Lin, Y., & Kwok, H. F. (2017). The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. International Journal of Molecular Sciences, 18(12), 2540. https://doi.org/10.3390/ijms18122540




_Kwok.png)

