Abstract
Pediatric cancers rarely exhibit recurrent mutational events when compared to most adult cancers. This poses a challenge in understanding how cancers initiate, progress, and metastasize in early childhood. Also, due to limited detected driver mutations, it is difficult to benchmark key genes for drug development. In this review, we use neuroblastoma, a pediatric solid tumor of neural crest origin, as a paradigm for exploring “big data” applications in pediatric oncology. Computational strategies derived from big data science–network- and machine learning-based modeling and drug repositioning—hold the promise of shedding new light on the molecular mechanisms driving neuroblastoma pathogenesis and identifying potential therapeutics to combat this devastating disease. These strategies integrate robust data input, from genomic and transcriptomic studies, clinical data, and in vivo and in vitro experimental models specific to neuroblastoma and other types of cancers that closely mimic its biological characteristics. We discuss contexts in which “big data” and computational approaches, especially network-based modeling, may advance neuroblastoma research, describe currently available data and resources, and propose future models of strategic data collection and analyses for neuroblastoma and other related diseases.
1. Introduction
Many research approaches to pediatric cancers have been adapted from adult oncology research. However, due to manifold differences in cellular basis and molecular mechanisms between the adult and pediatric cancers, these approaches may need to be critically evaluated for and tailored to the pediatric oncology research context. A common model of oncology research involves identifying genetic alterations in a type or subtype of cancer, investigating their roles in tumorigenesis, progression, or drug resistance, and exploiting them or their pathways as therapeutic targets. However, many pediatric cancers have a striking paucity of recurrent mutations when compared to most adult cancers [1,2], and require new research perspectives to better understand their specific etiologies and vulnerabilities. Furthermore, just as pediatric cancers differ from adult cancers biologically, they also present unique obstacles to research. Unlike adult cancers, most pediatric cancers are exceedingly rare, and therefore subject to limited funding support and fewer potential subjects for clinical research [1]. As pediatric cancers can be more biologically and clinically heterogeneous than adult cancers, generating the study power necessary to understand the complexity of these pediatric diseases is a daunting task [1,3]. Hence, research methods involving large-scale “big data” approaches may hold the key to addressing these challenges.
New technologies, including next-generation sequencing, have recently enabled the rapid generation, analyses, and dissemination of large biological datasets. In oncology, specifically, high-throughput and large-scale “big data” methods have led to exciting and clinically relevant discoveries. For example, these technologies recently led to the identification of frequent mutations in anaplastic lymphoma kinase (ALK) gene in both familial and sporadic neuroblastoma cases; as a result, ALK inhibitors are currently in clinical trials with promising preliminary results [4,5,6]. Using neuroblastoma as a paradigm of pediatric cancer, the aim of this review is to broadly engage with a few important facets of “big data” in a pediatric research setting. It is the hope of the authors that the ideas proposed in this review can be a helpful starting point for neuroblastoma researchers or pediatric oncologists who are interested in augmenting their existing research with techniques derived from “big data” science.
2. Current State of Big Data Use in Neuroblastoma Research
Neuroblastoma is the most common extracranial solid tumor in children [7,8,9,10]. It accounts for 7% of malignancies diagnosed in children between the ages of 0 and 14 [9,11], and nearly 15% of pediatric cancer-related mortality [9]. Neuroblastoma arises from malignant neuroblasts descended from neural crest cells. During development, sympathoadrenal (SA) lineage of neural crest progenitor cells migrate to form the peripheral sympathetic nervous system (PSNS) in the body [12,13]. Transformation of undifferentiated SA progenitor cells results in tumor formation in the adrenal medulla in the abdomen and sympathetic ganglia along the sympathetic chains (Table 1). In the advanced stages, neuroblastoma cells metastasize widely to the bone marrow, bone, lymph nodes, liver, intracranial and orbital sites, lung, central nervous system, and skin (Table 1) [1,9] resulting in long-term survival rates less than 40%, even with intensive treatment [5]. Interestingly, a subset of neuroblastoma patients (stage 4S) presenting with widespread metastases usually spontaneously regress with minimal or no treatment (Table 1) [11,14,15,16]. Such diverse clinical phenotypes of neuroblastomas implicate its heterogeneous and complex biological bases and provide compelling rationale for further study.

Table 1.
Treatment and prognosis of neuroblastoma patients by risk group and staging. Prognosis is 5-year event-free survival [3,9,11,15,17]. MYCN: V-Myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog.
2.1. Generation of Neuroblastoma Staging Systems and Their Importance in Guiding Big Data Generation and Downstream Analyses
The complexity of neuroblastoma development is staggering, thus necessitating new approaches not only to stratify patients, but also to guide big data generation and comparative analyses. Understanding disease etiology and progression requires collection of spatial data, as migration is implicated in disease onset and progression; temporal data, as development involves many time-stratified and -dependent phases; and other high-dimensional cell data, as the developing organism is a dynamic collection of simultaneously co-evolving cells and cell types. Research consortia devoted to staging the disease played an integral role in the field’s earliest attempts to collect such data and generate large-scale clinical neuroblastoma databases. The International Neuroblastoma Staging Series (INSS) (Table 1) stratifies neuroblastoma by risk level, tumor location and dissemination, and MYCN (V-Myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog) amplification, using data from consortia in the United States, Europe, and Japan [15]. In part, INSS staging relied on surgical observations and extent of resection, and, therefore was subject to certain biases, however, it also recommends what tests to perform and which types of data to collect [18]. A more recent classification from the International Neuroblastoma Risk Group (INRG) Task Force involved collecting clinical data and biosamples from more than 8800 cases in North America, Australia, Europe, and Japan [16]. The INRG staging incorporates medical imaging data, adding significant data volume and variety, in staging and risk stratification to avoid the aforementioned surgical biases. Importantly, the INRG moves towards a more molecular and genetic classification of disease, incorporating such additional measurements as general tumor ploidy, chromosome 11q aberration, and classification of MYCN amplification as focal or diffuse in the tumor population [16]. The INRG Task Force exemplifies how careful curation of large datasets can revolutionize care [19], e.g., comparative analyses between the “4S” under INSS staging scheme with other neuroblastoma stages can advance our understanding how tumor regress.
2.2. Recent Discoveries Made Possible by Large-Scale Data Analyses
The evolution of neuroblastoma staging reflects the general trend of incorporating next-generation technologies into neuroblastoma research. Prior to high-throughput genomic sequencing, amplification of the oncogenic transcription factor MYCN was identified in 20% of neuroblastoma cases and associated significantly with high-risk disease and poor outcome [9,20,21,22,23,24,25]. With recent integrative genomic analyses, including whole-exome, genome, and transcriptome sequencing, genome-wide association studies (GWAS) and array comparative genomic hybridization (array-CGH) analyses [7,20,21,26,27,28,29,30,31,32], an increasing number of genetic and chromosomal alterations have been discovered. In our current review, we would like to briefly highlight key findings—in terms of genetic mutations and epigenetic alterations, chromosomal alterations, predisposition alleles, and aberrations in micro and long non-coding RNAs—generated from large-scale data analyses.
2.2.1. Genetic Mutations and Epigenetic Alterations
Unlike adult cancers, which are characterized by many acquired somatic mutations, neuroblastoma exhibits surprisingly few recurrent mutations conserved across high-risk cases, as assessed in several large-scale sequencing studies [33]. Among those frequently mutated genes, activated mutations in the ALK kinase and loss-of-function mutations in the PHOX2B (paired-like homeobox 2b) transcription factor account for ~80% of hereditary neuroblastomas [9,28,33,34,35]. ALK acquired somatic mutations also have been reported in ~14% of all high-risk cases [33,35]. Recently, we and others showed the cooperative oncogenic effect of MYCN and activated ALK in prompting neuroblastomagenesis in both transgenic mouse and zebrafish models of neuroblastoma [36,37,38]; our results further confirm ALK is a key oncogenic driver, in collaboration with MYCN, in neuroblastoma pathogenesis. In addition, recurrent somatic mutations have also been found in the genes contributing to RAS-MAPK pathway activation, PTPN11 (protein tyrosine phosphatase, non-receptor type 11), NRAS (neuroblastoma RAS viral oncogene homolog), BRAF (B-Raf proto-oncogene, serine/threonine kinase) and NF1 (neurofibromin 1) [34,37]; chromatin remodeling, such as ATRX (α thalassemia/mental retardation syndrome X-Linked) and ARID1A/B (AT-Rich interaction domain 1A/B) [3,14,35,38]; Rac/Rho pathway regulation, such as TIAM1 (T-cell lymphoma invasion and metastasis 1), DLC1 (deleted in liver cancer 1), ARHGAP10 (Rho GTPase activating protein 10), ATRX; and others, such as MYCN, TP53 (tumor protein P53) [33,35] and so on.
2.2.2. Chromosomal Alterations
Besides MYCN amplification (the hallmark of high-risk neuroblastoma), chromosomal alterations—specifically copy number variants at 1p, 11p, 2p, 3p, 4p, 6p, 6q, 1q, 11q and 17q—have been identified using array-CGH and single nucleotide polymorphism (SNP) analyses [3,5,36,37,39,40,41,42,43]. Loss of heterozygosity of chromosomes 1p and 11q are associated with increased disease severity, with loss of 11q notably being inversely associated with MYCN amplification [3,5]. Chromosome rearrangements resulting in an increased activation of TERT (telomerase reverse transcriptase) have recently been identified in ~23% of high-stage neuroblastoma with very poor prognosis but no MYCN amplification [44]. Structural defects caused by chromothripsis have been found to recurrently affect ATRX, ODZ3 (odd oz/ten-M homolog 3 (Drosophila)), PTPRD (protein tyrosine phosphatase, receptor type D), and CSMD1 (CUB and sushi multiple domains 1) in high-risk neuroblastoma [35].
2.2.3. Predisposition Alleles
GWAS approach has been successfully applied to identify common polymorphic alleles that associate with a predisposition to neuroblastoma. The most common polymorphic alleles have been identified at the LIM-domain-only gene LMO1 locus which is significantly associated with advanced stage neuroblastoma [39,45]. Very recently, we have found a polymorphism in the first intron of LMO1 that can influence neuroblastoma susceptibility by affecting differential GATA transcription factor binding [46]. Other recently identified common SNPs which confer higher susceptibility to neuroblastoma include variants in BARD1 (BRCA1-associated RING domain 1) [47], a non-protein coding CASC15 (cancer susceptibility candidate 15) [48], DUSP12 (dual specificity phosphatase 12) [41], DDX4 (DEAD box polypeptide 4) [41], IL31RA (interleukin 31 receptor A) [41], HSD17B12 (hydroxysteroid 17b dehydrogenase 12) [41], LIN28B (lin-28 homolog B) [36,49], HACE1 (HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1) [36], FLJ44180 [46], NEFL (neurofilament, light polypeptide) [50], NBPF23 (neuroblastoma breakpoint family, member 23) [37], and TP53 [51].
2.2.4. Implications of Micro and Long Non-Coding RNAs and Other Factors in Neuroblastoma
Functional genetic regulators of neuroblastoma development and progression also include micro RNAs (miRNAs) [52,53,54], long non-coding RNAs (lncRNAs) [25], alternative splicing programs [24], fragile sites [55] and genome-wide methylation [56,57,58,59,60,61,62,63,64,65]. miRNAs have been implicated in neuroblastoma metastasis [52], MYCN regulation [66,67], and cell differentiation [68]; thus they may serve as novel therapeutic targets, especially in high-risk, MYCN-amplified patients [53,69,70]. For example, Das et al., created a novel method of analyzing DNA methylation and miRNA expression, and were able to identify a panel of epigenetically regulated miRNAs that contribute to disease pathogenesis [54]. Zhang et al., found that MYCN directly regulates the expression of key splicing factors PTBP1 (polypyrimidine tract binding protein 1) and HNRNPA1 (heterogeneous nuclear ribonucleoprotein A1) in high-risk neuroblastoma, leading to increased cell proliferation and poor overall survival [24]. Long noncoding RNA—NBAT-1 (Neuroblastoma Associated Transcript 1)—has been found to epigenetically down-regulate tumorigenic factors and promote differentiation of tumor cells [25]. Both miRNAs and lncRNAs have been implicated in cell differentiation and development [71,72,73], further emphasizing the importance of the developmental context of this disease. There are a number of databases where information on lncRNAs [74,75,76,77] and miRNAs [78,79] and disease associations can be found (see also Table 2).

Table 2.
A reference table of key resources for neuroblastoma big data.
2.3. Unique Features of Neuroblastoma Development, Metastasis, and Regression Warrant Further “Big Data” Analysis
2.3.1. Embryonal Tumor Derived from Aberrant Neural Crest Development
While the roles of genetic components are important for understanding and treating cancers, they offer insight into just one of the potential mechanisms of disease pathogenesis. Since neuroblastoma occurs so early in life, and usually only early in life, the developmental context of disease development must be considered as well. A model of pediatric cancers described by Marshall et al., suggests that pediatric cancers can be linked back to some defect in signaling crucial to embryonic development [10]. Although cell death is a necessary part of embryonic development as many more cells are being produced than are actually needed for growth and organogenesis [10], neuroblastomas may derive from neuroblasts that persist despite apoptotic and cell cycle arrest signals, and subsequently adopt tumorigenic phenotypes [10].
Neural crest cells form between the surface ectoderm and developing neural tube during neurulation [13,90]. They then migrate and differentiate to form many diverse cell types and tissues, such as the neurons and glia of the enteric, sympathetic, and parasympathetic nervous systems, skeletal and connective tissues in the face, pigment cells, and the adrenal medulla [10,13]. Throughout this migration neural crest cells ubiquitously express MYCN, which induces them to proliferate, expand, and maintains stem cell characteristics [90,91,92,93]. Therefore, the normal function of MYCN is remarkably similar phenotypically to the abnormal function of tumors. Usually, expression of MYCN is stopped by signals that induce differentiation of neural crest cells into neuroblasts and other derivatives and is rarely expressed in fully differentiated cells [21,23,64,93,94,95,96,97]. If cells do not differentiate, because of excessive expression of MYCN, they generally undergo apoptosis [10]. Neuroblastoma may develop when neuroblasts evade this cell death signaling (e.g., through loss of CASPASE-8 or activation of ALK) [90,98,99,100,101,102], and subsequently adopt tumorigenic phenotypes.
While MYCN amplification is the most common genetic event associated with high-risk neuroblastoma, over half of high-risk patients don’t harbor this alteration [9]. As such, alternative mechanisms underlying neuroblastoma development could involve pathways that are independent of MYCN signaling; this gap in knowledge may potentially be addressed through the use of computational strategies. In addition, signaling and interactions between neuroblasts and other cell types may be of additional interest, since paracrine interaction within the somites, neural tube, notochord, and dorsal aorta are vital for normal neural crest cell specification, migration and differentiation [13,90]. Alterations in the microenvironment, especially signaling from other types of cells surrounding neural crest could lead to its abnormal development, which might not be clearly identified by examining neuroblastoma samples alone. This potential etiology can be investigated through recently developed computational techniques that model paracrine signaling, elucidate contributions of the tumor microenvironment to disease, and propose treatments targeting the tumor-stroma interface [103,104].
2.3.2. Wide-Spread Metastasis in High-Risk Neuroblastomas
Since 90% of cancer mortality results from metastatic lesions [105] and over 50% of neuroblastoma patients especially those are older than 1 year of age with MYCN amplification present wide-spread metastasis at diagnosis [106], it is important to understand the pathogenesis of neuroblastoma metastasis. Neuroblastoma metastasis occurs overwhelmingly in sites of normal neuroblast migration that leads to the development of multifocal hyperplasia [14]. Very few studies have compared single metastases to primary tumors [107], and none have looked at multiple metastases from single individuals. Despite the well-documented barriers to autopsy collection in pediatric patients [108,109], an important analysis would involve constructing phylogenies of multiple tumors such as genomic or transcriptomic data from single post-mortem individuals [110]. Additionally, in vivo animal models [111,112] with neuroblastoma cells labeled with fluorescent reporter genes can be useful to visually track the dissemination and metastases of tumor cell subpopulations to delineate the molecular mechanisms underlying neuroblastoma metastasis.
2.3.3. Spontaneous Regression of Stage 4S Neuroblastomas
Spontaneous regression or remission has been reported in many pediatric and adult cancer types; one estimate has its pan-cancer incidence as 1/80,000 [113]. Conversely, neuroblastoma has a significantly higher rate: one study suggests 25% of neuroblastoma cases experience spontaneous remission or regression [114]. Of note, stage 4S cases, which comprise about 10% of neuroblastoma cases, generally result in complete remission with minimal or no intervention [9,14]. Therefore, the 4S subtype is a very valuable model in understanding—and potentially inducing—regression in other neuroblastoma classifications and other cancers. Brodeur et al., propose neurotrophin growth factor (NGF) signaling, immune intervention and epigenetic regulation as potential mechanisms for regression [14]. While differential gene expression has been detected between 4S and non-4S neuroblastoma subtypes, no significant different molecular signatures have been found between regressing and non-regressing individuals within the 4S subtype. Both of these interfaces warrant further study, possibly through experiments with in vivo animal models and large scale data. The development of novel computational tools to decipher context-specific molecular events that signify whether tumors remit because of certain factors, including tumor cell death or differentiation and intra- or extracellular signaling, can be a promising avenue for novel drug development [14].
3. The Promise of Available Big Data Resources in Neuroblastoma Research
The concept of “big data” is relatively new in many fields, including the biological sciences. Big data can be described as a catch-all term for situations in data science and analytics that have four Vs: volume, velocity, variety, and veracity [115]. In rare diseases like neuroblastoma, high data volume (petabyte scale) is not often seen, if ever, since there are few affected individuals from which to sample. There are, however, other aspects of the big data paradigm that are directly applicable. The use of animal models and cell lines that closely mimic the biology of human neuroblastoma reduces uncertainty and provides veracity. Data velocity—conventionally referring to streaming data requiring immediate analysis and decision-making—informs solutions to the technical challenges involved with high-throughput computing pipelines that rapidly integrate new information with shared data repositories and produce candidate hypotheses for immediate validation. Integrating disparate data types (e.g., genomic, imaging) from different models into a single analysis produce data variety that can be integral to unraveling the complexity of disease like neuroblastoma. Strategic evaluation of these characteristics will benefit any data-driven research setting.
Understanding the implementation—in addition to the qualities—of big data derived methods in biological research is critical. Figure 1 outlines a high-throughput workflow highlighting relevant data types, databases, and computational techniques that can be used by neuroblastoma researchers. Importantly, big data analytics is an iterative process, and interfaces between biological context and analytical technology must be continually considered. The processes of decontextualizing data from its native context for use in generalized databases, and subsequently re-contextualizing data for use in novel contexts are non-trivial. To execute these processes effectively, data must be strategically collected, contexts must be appropriate for data use, and computational techniques must be available to bridge the two.
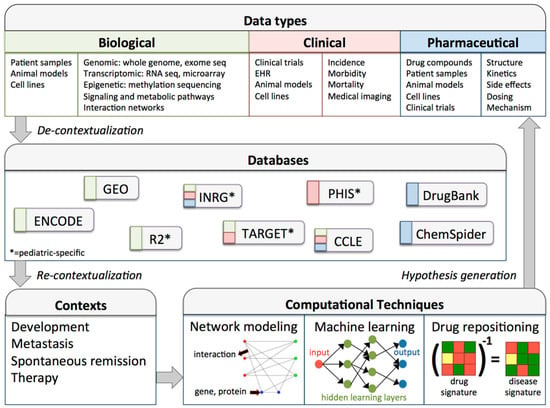
Figure 1.
Data-driven research workflow for neuroblastoma or other pediatric cancers. The data types panel enumerates data sources (e.g., animal models) and types (e.g., RNA sequencing) for three categories of biological data. Following collection, data may be de-contextualized for inclusion in databases. De-contextualization involves properly formatting and annotating datasets, so that they are standardized, accessible, and useful for re-contextualization into new research contexts. Note that this workflow is iterative, and can therefore benefit from continued improvement of data infrastructure and data collections, development of more accurate and comprehensive data analysis tools, and advances in basic and translational research and therapeutic applications [35,118,119,120,121,122,123,124]. CCLE, cancer cell line encyclopedia; ENCODE, encyclopedia of DNA elements; GEO, gene expression omnibus; EHR: electronic health record; INRG, international neuroblastoma risk group; PHIS, pediatric health information system; R2, genomics analysis and visualization platform; TARGET, therapeutically applicable research to generate effective treatments.
3.1. Pitfalls in Clinical Data Collection
While optimal for research, high-throughput and high-fidelity pediatric cancer data can be difficult to collect. For one, pediatric cancers have lower incidence than adult cancers, and therefore fewer individuals to enroll in studies; in the US per 1 million individuals, neuroblastoma has an incidence of 21.1 and breast cancer, 123,700 [116]. While a large percentage of children with cancer are enrolled in clinical trials, certain pediatric groups—especially adolescents and low-income and non-white children—face barriers to participation [117]. Additionally, rates of autopsies performed on all patients in the US is low, occurring in about 5% of hospital deaths, but may be lower in pediatric patients due to the intrusive, insensitive perception of autopsies, unawareness of research studies, and the reluctance of physicians to ask bereaved family members [1,108]. Nevertheless, pathological samples gathered through autopsy can be important sources of high-quality data [109], and may even provide a small, positive experience for family members during their time of grief if the scientific value of the autopsy is emphasized [108]. We have assembled a table of databases where patient-derived data has been compiled (Table 2); though not comprehensive, it serves as a starting reference for researchers. The following sub-sections—animal models and cancers with related qualities to neuroblastoma—will describe alternate sources of potentially high-volume data that may serve to validate hypotheses generated by patient data, or explore the novel contexts supplied above.
3.2. Animal Models for Data Generation and Validation
Animal models provide several advantages to researchers; due to ease of use compared to human subjects and strong data-sharing communities, model organisms can be the basis for high volume and velocity data. For neuroblastoma, however, the distinctly important advantage of animal models is veracity in simulating the developmental context of neuroblastoma onset and progression. Unlike cell lines, which lack vascularity, immune involvement, paracrine signaling from other cell types, and other important factors contributing to tumor biology, animal models can better recapitulate human disease [112,125].
Neuroblastoma has several autochthonous or companion animal models, the most notable being MYCN- and ALK-expressing mice and zebrafish (Table 3) [98,102]. In the transgenic mouse or zebrafish model overexpressing MYCN in sympathoadrenal cells, animals developed tumors in the peripheral sympathetic nervous system, similar to neuroblastoma development in humans, and histological comparisons confirmed the similarity of these tumors to human tumors [102,126,127]. Combinations of gene alterations can also be investigated by crossing these models, such as between MYCN and ALK, nf1 loss or Caspase 8 loss [101,102,128]. Additionally, tumors from neuroblastoma patients can be xenografted into both zebrafish and mice, providing a way to capture the heterogeneity of human disease [94,129,130,131,132].

Table 3.
Animal models of neuroblastoma. Table adapted from Zhu, 2016 [127], and expanded. ALK: anaplastic lymphoma kinase; CGH: comparative genomic hybridization.
Zebrafish have an advantage over mouse models due to their small size, high fecundity, and ease of maintenance; additionally, their transparent embryos allow for easy observation of cancer progression and metastasis [127,133]. Conversely, mice more closely mimic human biology, and can be better analogs for investigating epigenetics [9], and drug therapies. Importantly, the Pediatric Preclinical Testing Program (PPTP) aims to expedite pediatric drug development by testing potential agents in mouse patient-derived xenografts (PDXs) [129]. While not close analogues of disease, Drosophila melanogaster models have been used to investigate the possible role of stemness in neuroblastoma [134,135]. Although in vitro models can be poorer proxies for disease than in vivo, cell lines generate high-quality and high-volume sequencing data and, in future, may better represent human disease through some developing technologies [136]. The Cellosaurus database [137] has a robust catalog of neuroblastoma-derived lines, as does Thiele’s 1998 review [138].
3.3. Similar Features between Neuroblastoma and Other Pediatric Solid Tumors
Grouping diverse cancers based on similar pathology, histology, or molecular biology can enrich oncological research efforts. A notable example involves the coordinated research efforts between ovarian and breast cancer researchers to identify and investigate the BRCA1/2 (breast and ovarian cancer susceptibility protein 1/2) [139]. We believe similar approaches will be fruitful for neuroblastoma by increasing the volume of data available for analysis and provide insights for other pediatric cancers. Therefore, important axes for comparison between neuroblastoma and other types of pediatric solid tumors are summarized in Table 4.

Table 4.
Similarities among neuroblastoma and other pediatric solid tumors. A cancer is marked pediatric if it has high incidence among all children with cancer, or has high pediatric incidence relative to adult. Small round blue cell tumors are cancers with a similar histologic appearance (highly nucleated, mesenchymal) that are often difficult to distinguish, especially in pediatric patients. If a subset of patients present with mutations or amplifications, a cancer is marked as being MYCN or ALK modified. While spontaneous regression is a rare event in all cancers, some experience slightly higher rates of treatment-independent full or partial regression of tumors and are included in the table as experiencing “spontaneous regression.” (*) Spontaneous regression cases of retinoblastomas may actually be benign retinomas, and therefore misclassified [96,97,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154].
4. Modeling Neuroblastoma-Derived Big Data
The rationale for modeling neuroblastoma with big data is to obtain new knowledge via hypothesis-free models without relying on prior biological knowledge. As neuroblastoma can be considered a complex trait (i.e., the disease results from interactions among multiple molecular factors), one of the key purposes of integrating multi-omics big data is to uncover genotype-phenotype interactions [155] that give rise to underlying neuroblastoma pathogenesis. The immense complexity and variability of big data is mirrored in the computational models used to extract meaning from it. Often the first line of analysis on biological big data involves statistical modeling and/or clustering to find significantly different or similar items. For genomic data, genome-wide association studies (GWAS) are an example of statistical interrogation of large datasets [156]. Clustering is more often used on RNA microarrays or sequencing to find groups of genes that co-express [157]. Finally, mathematical and rule-based modeling can simulate complex and unwieldy biological systems through a combination of experimental data and theoretical equations [158,159]. The remainder of the review will present strategies used to integrate big data and focus on three additional computational techniques—network modeling, machine learning, and drug repositioning—that are especially applicable to addressing the current research contexts in neuroblastoma described above.
4.1. Data Integration
Integrating different types or layers of big data offers many advantages and facilitates efforts to pinpoint genetic and molecular factors that play critical roles in regulating disease outcomes. Such integration compensates for missing or incomplete data coverage from technology that generates one single data type. In addition, when multiple sources of evidence from different data types point to a similar observation, confidence in findings is increased and false positives reduced. Furthermore, integrating different omics data allows for a more thorough and comprehensive understanding of how biological processes are regulated at different layers (i.e., from genetic mutations to epigenetic regulation, to transcription, to RNA processing, to protein synthesis, to protein modifications) and how malfunction at any of these layers causes neuroblastomas.
Although it is still a challenge to identify the best approach to integrate big data derived from different types and scales in a meaningful way, in general, there are two main strategies: meta-dimensional analysis and multi-stage analysis [160]. Meta-dimensional analysis integrates multiple datasets in a single study via three approaches: concatenating multiple data matrices from different omics data into one large input matrix before model construction (concatenation-based) [161]; transforming each data type into an intermediate form such as a graph matrix that represents a network before multiple data are combined (transformation-based) [162]; or generating different models by using different types of data as training sets before integrating into a final model, thus allowing independent analysis for each data type (model-based) [163]. Unlike meta-dimensional analysis, multi-stage analysis divides data analysis into multiple steps, capturing associations between enriched signals between different data types at each step. One example is genomic variation analysis where SNPs that associate with a phenotypic trait identified from GWAS are tested for their correlations with another omics datasets, such as transcriptomic data [163]. The associated SNPs are called expression quantitative trait loci (eQTLs) and genes associated with these eQTLs are in turn tested for correlations with the phenotype of interest.
4.2. Network-Based Modeling
Although network approaches have been applied to big data—most notably in social networks research [164]—their utility in biological research includes pathway analysis and integration of diverse data to model complex systems. We believe that network modeling algorithms—which have rarely been applied to neuroblastoma—are uniquely suited to provide insights into neuroblastoma development and treatment.
Network computation generally requires two types of data: (1) context- or phenotype-specific biological information (usually genomic, transcriptomic, metabolomic, or epigenomic); and (2) interaction information (usually curated protein-protein/gene-gene interactions, gene sets/signatures, or pathways) [165,166]. Recent expansion and more accurate annotation of protein-protein interaction (PPI) networks at both the genome and proteome scale [167] has facilitated discovery of novel functional crosstalk between genes. Thus far, PPI networks represent the most characterized biological network, although other type of networks, such as miRNA-gene networks [168], are being constructed. Computational modeling of cancer had been performed using PPI networks to decipher the effects of mutated genes on affected biological pathways [165,169]. Researchers can overlay biological data onto the network scaffold and investigate the structure or function of a given network (see Network panel of Figure 1 for an example of a gene interaction network). The databases shown in Figure 1 can be helpful sources for biological data to overlay onto curated networks; updated gene and protein interactions can be found in KEGG [170], BioGRID [171], and STRING [172], among others [173]. The commercial ingenuity pathway analysis and open-source network analysis tool (NeAT) online interfaces provide diverse network analyses in one integrated platform [174,175]. Additional platforms can analyze and visualize transcriptomic [176], metabolomic [177], or epigenetic data [178].
In addition to modeling approaches that use pre-constructed networks, other approaches use data to reverse-engineer networks. These approaches infer interactions between data through Pearson correlation coefficients, Bayes probabilistic models, and information-theoretic approaches such as mutual information [179]. The major advantages of reverse engineering methods are that no prior knowledge is required and novel cause-effect relationships can be inferred. Many of these methods have proven to possess the power to dissect gene regulatory modules in bacteria [180], yeasts [181], and human diseases [182], as well as specific biological process such as hematopoietic stem cell differentiation [183]. Reverse engineering approaches may be particularly suited to dissect novel key regulators driving neuroblastoma etiology and progression using large-scale multi-omics data. For example, we recently developed NetDecoder, a context-based network modeling tool that integrates both PPI network-based and reverse engineering-based approaches via a novel process-guided flow algorithm [176]. Our results showed that NetDecoder is capable of identifying genes that play critical roles under different disease contexts as evidenced by our breast cancer, dyslipidemia and Alzheimer’s disease case studies. For instance, we found the tumor suppressor gene TP53 plays different regulatory roles in breast cancers versus dyslipidemia, indicating how the very same gene can cause different disease phenotypes under different pathological contexts. Context-dependent network modeling may be particularly important for understanding neuroblastoma etiology, given poor disease outcome may be shaped by MYCN amplification and MYCN-independent molecular mechanisms may drive higher risk among patients without MYCN amplification.
In addition, dynamical modeling using mathematical approaches, which facilitate modeling of cancer progression [184] and altered activities of signaling pathways [185], may prove useful in neuroblastoma modeling in the future. For example, dynamical modeling helped uncover the importance of phosphorylation events and effects due to gain or loss of protein-protein interactions in establishing novel pathway crosstalk. Using mass action law, our previous studies showed that dynamical modeling is a powerful approach to identify the “Achilles heel” of a cancer network, or genes sensitive to perturbations that may alter disease outcome [186]. Dynamical modeling may be applied to neuroblastomas, once key regulatory modules have been discerned via network-based modeling. For example, focusing on a particular pathway cascade such as how differential activities of Ras-MAPK cascade cause relapse [87] to decipher regulatory properties of neuroblastoma-specific molecular crosstalk may prove to be fruitful.
4.3. Machine Learning-Based Modeling
The term “machine learning” refers to a group of computational algorithms that can perform pattern recognition, classification, and prediction on novel data by “learning” from existing data (i.e., training set). In other words, machine-learning algorithms are mathematical mapping methods that aim to learn or uncover any underlying patterns (or features) embedded in the data. Machine learning has been applied in many areas of biological and medical sciences [187,188]. Currently, the most commonly used machine learning approaches in biology are support vector machines (SVMs) and artificial neural networks (ANNs). Recent advances in deep learning [189], specifically a more sophisticated ANN model with “deeper” neuron layers, spurred a number of applications in biology. For example, ANN can be used to predict sequence specificities of DNA- and RNA-binding proteins [190], effects of noncoding variants [191], alternative splicing [192], and quantitative structure-activity relationships of drugs [193] (Figure 1). These deep learning models have proven capable of outperforming other machine learning methods and identifying more complex features from data. Also, incorporation of deep reinforcement learning methods can facilitate improved performance in finding an optimal solution for a given task [194]. However, to achieve such levels of complexity, deep learning techniques generally require a higher volume of data and more computational time than other machine learning algorithms.
When using machine learning on biological big data, many different data types (genomic, transcriptomic, epigenomic, etc.) may be integrated into a single model. However, these data often have large dimension (e.g., whole exome sequencing has ~20,000 data points), and require many times more training data to build an appropriate model. Therefore, to make machine learning algorithms feasible for researchers, data collection can be minimized by reducing the dimension of input data. This dimensionality reduction can be done before or after data integration with Principle Component Analysis (PCA), or after data integration with feature selection algorithms [195]. Both of these strategies provide information about data sets by scoring variables on how much or little they contribute to a model.
Network modeling—specifically reverse-engineered networks—can be combined with machine learning. One method, Mode-of-action by network identification (MNI) reverse-engineers a network model of regulatory interactions using a training set of multidimensional biological data such as transcriptomics, proteomics, and metabolomics in order to identify genetic components and network modules that correspond to a cellular state. The method generates a directed graph relating the amounts of biomolecules to each other via a set of ordinary differential equations (ODEs). When transcriptomic data is used as training data, regulatory influences between genes can be inferred. MNI was recently applied to decipher causal genes responsible for driving tumor progression in ductal carcinoma in situ (DCIS), a non-invasive lesion of breast cancer. MNI identified HoxA1 (homeobox A1) as the top candidate, and subsequent experimental work showed that in vitro silencing of HoxA1 can revert the cancer phenotype [196].
CellNet is another network-based classification system with proven ability to classify cellular states based on their gene regulatory network (GRN) status [197,198]. CellNet was originally used to assess cellular states using their respective gene regulatory networks that act as major molecular determinants of cell type identity. GRNs were first reconstructed using a reverse engineering algorithm and then used as training data using Naïve Bayesian method. The study showed CellNet reverse engineered GRN-suggested neurons derived from directed differentiation of embryonic stem cells (ESCs) achieved a higher classification score than neurons derived from direct conversion from fibroblasts. CellNet also has the capability to infer causal genetic factors in driving cellular states such as Pou2af1 (POU class 2 associating factor 1) and Ebf1 (early B-cell factor 1) in B cells and Foxa1 (forkhead box a1), Hnf4α (hepatocyte nuclear factor 4 alpha), Cdx2 (caudal type homeobox 2) in hepatocytes. Thus, CellNet has the power to elucidate cell states via GRNs and identify potential regulators within GRNs that are responsible for cell state maintenance or transition. We anticipate that machine learning-integrated reverse engineering methods, such as MNI and CellNet, will hold the power to uncover causal genetic components that drive different stages of neuroblastoma.
4.4. Network-Guided Drug Discovery and Repositioning
Biological networks can also be efficient tools for identifying and evaluating drug targets. For example, Qin et al., used network flow algorithms to predict the sensitivity of cell lines to MAPK pathway inhibitors given as mono or combination therapy, based on copy number and mutational data for adult cancer cell lines in The Cancer Genome Atlas (TCGA) [199]. While the usual metric for selecting drug targets is gene and protein alteration [200], these network models can also identify genes and proteins that are necessary for disease progression or cell survival, but are not altered in any way. Given many of the altered genes in neuroblastoma, like ALK, have important roles in development of non-tumor cells [201], targeting other tumor-specific vulnerabilities could potentially reduce late effects from treatment.
The treatment of pediatric cancers is challenging due to the young age of the patients. The median age of children diagnosed with neuroblastoma is 17 months [9], therefore an ideal treatment would be non-invasive and minimize potential late effects. Treatment of neuroblastoma varies according to the stage of the disease (Table 1). However, children who receive treatment for aggressive disease can suffer from many treatment-related complications, such as hearing loss, cardiac toxicity, infertility, and increased risk of developing second cancers due to chemotherapy [11,202]. Because of these risks, more targeted and less harmful treatments are indicated. However, drug discovery in pediatric patients can involve many hurdles. For example, pediatric oncology patients are often excluded from clinical trials until drugs are shown to be effective at treating adult cancers [1]. This current method of drug discovery does not take into account the manifold differences between adult and pediatric cancers, such as the developmental context and low rate of recurrent mutations of pediatric cancers. Drug repositioning can leverage big biological, chemical, and pharmaceutical data to identify promising new neuroblastoma drug targets and combinations, and estimate their efficacy and safety in silico.
Prescribing drugs for treatment of other diseases beyond their previously approved uses is called drug repositioning or repurposing. This approach is especially useful in pediatric oncology research, since using previously pediatrically approved drugs in clinical trials can allow researchers to bypass Phase I safety trials. Several treatments for neuroblastoma have been repositioned, such as isotretinoin, originally an acne treatment, which is now a treatment for several other cancers, and an important component of multimodal therapy for advanced neuroblastoma patients [203]. Unlike many of the computational techniques above, the main object of performing drug repositioning is not to model or gain deep biological insights into neuroblastoma; rather, the approach seeks to rapidly bring treatments into clinic, by reducing the logistical burden of drug development and testing.
Drug repositioning algorithms follow a few broad computational paradigms: similarity measures, machine learning/mathematical modeling, and text mining. Similarity measures look for correlations between drug structure, functional category, mechanism of action, side effects, or transcriptomic or proteomic signatures. For example, if a drug shares a similar transcriptional profile with the inverse signature of aggressive neuroblastoma tumors (illustrated in the drug repositioning panel of Figure 1), it can be hypothesized that the drug could be effective. In addition, machine learning and mathematical models can pool many types of data, to find multi-dimensional similarity between drugs and diseases. One study examined the chemical structure, molecular activity, and side effects of drugs simultaneously to predict which drugs might behave similarly. Lastly, text mining methods look for associations between drugs and symptoms, genotypes, mechanism of action, or vice versa, within large publication databases to suggest novel drug-disease pairings [204].
A promising direction of drug repositioning is investigating the safety and efficacy of novel drug combinations, rather than single agents [205,206]. While additional safety trials are usually needed to test drugs in combination, combination therapies are, on the whole, more deleterious to cancer tissues than normal cells [207,208]. Of note, network-based drug repositioning algorithms have shown promise in repurposing drugs, both as monotherapies and in combination [209].
These drug repositioning strategies rely on robust clinical, genotypic, and drug structural data. A review by Ryall et al. [209], supplies an impressive list of such resources for drug repositioning, in addition to those listed in Figure 1. Nevertheless, many of these resources have scarce representation of pediatric drugs and diseases; for pediatric drug dosing, safety, and indications, researchers may access The Children’s Pharmacy Collective™, a novel online database, and the Harriet Lane Handbook, in print [210].
4.5. Future Modeling Challenges and Therapeutic Discovery
The ultimate goal of performing computational modeling on neuroblastoma using big data is to illuminate disease etiology and develop effective therapy. During the past few years, advances in network medicine [211] and systems pharmacology [212] have helped make strides toward this goal. However, as there are very rare recurrent somatic mutations detected in neuroblastoma, genetic approaches that aim to find druggable targets based on mutations alone are unlikely to yield fruitful drug discovery. For example, a mini-driver model of polygenic cancer etiology [213] proposes that many mutations found in cancers do not exhibit major driver effects but instead exert weak tumor-promoting effects. In addition, recent mathematical modeling work by Gatenby and colleagues suggests “never mutated” genes play a crucial role in cancer [200], indicating that most key genes may go undetected if only “mutation criterion” is applied to define cancer genes. Furthermore, epigenetic factors [214], RNA mis-splicing [215] or alternative splicing [216] and the presence of differentially expressed non-coding RNA species [217] have been reported to contribute to cancer etiology, and these processes are not necessarily affected by major mutations detected in most cancers. Such evidence challenges us to rethink how we could better model neuroblastomas at the network and systems level by integrating different types of big data. More sophisticated modeling approaches such as deep learning, which can integrate big data generated from patients and animal models, may be best suited to model the complexity of neuroblastoma etiology.
Another challenge that must be addressed: tumors frequently develop resistance to chemotherapeutics, rendering currently available drugs ineffective. Re-sensitizing tumors by modulating the network that regulates drug response may hold promise when tumors acquire resistance to existing drugs. Toward that end, our group recently developed Phenotyping Mapping (P-Map), a network-based tool that helps researchers to chart drug response networks [205]. Using anthracyclines and taxanes as case studies, we showed that it is possible to manipulate drug response phenotypes by perturbing key genes in a drug response network. Our study together with others (such as Chen et al. [218]) provide examples of efforts to rescue drug resistance via co-administration of compounds that modulate the activities of drug response networks.
5. Conclusions
Neuroblastoma is a common and complex pediatric cancer with extreme heterogeneity both at the clinical and molecular level [1]. Given many pediatric cancers share similar biology as well as research challenges, we seek to highlight the challenges and potential utility of using big data to further our understanding of neuroblastoma and related pediatric cancers and to develop effective drug treatments. The developmental context in which neuroblastoma arises appears to be particularly important, with many genes associated with aggressive disease (MYCN, ALK) driving cell proliferation and neuritogenesis [7,8,43,219]. Therefore, using animal models in which context is conserved is very important for generating more next-generation sequencing generated multi-omics data. Network models and drug repositioning can be leveraged to understand neuroblastoma and develop better treatment options.
The current review highlights the importance of collecting, annotating, and sharing data within the neuroblastoma research community, as well as the pediatric oncology communities. As new computational tools and perspectives arise, these data may be continually mined to generate new hypotheses and validate previously stated results. The biological complexity of disease must be matched by complexity of data and analysis in order to improve treatment at all stages of neuroblastoma.
Acknowledgments
This work was supported by grants K99/R00CA178189 (Shizhen Zhu) from the National Cancer Institute; young investigator awards from Alex’s Lemonade Stand Foundation (Shizhen Zhu) and the CureSearch for Children’s Cancer Foundation (Shizhen Zhu); a V Scholar award from the V Foundation for Cancer Research (Shizhen Zhu) and pilot project awards from the Fraternal Order of Eagles (Shizhen Zhu) and the Mayo Center for Biomedical Discovery (Shizhen Zhu); support from the Mayo Clinic Cancer Centerand Center for Individualized Medicine (Shizhen Zhu).
Author Contributions
Brittany M. Salazar and Emily A. Balczewski contributed equally to this work. Brittany M. Salazar, Emily A. Balczewski, Choong Yong Ung and Shizhen Zhu contributed to the conception and design of this work. Brittany M. Salazar and Emily A. Balczewski contributed to the acquisition of information. Brittany M. Salazar, Emily A. Balczewski, and Choong Yong Ung drafted the manuscript. Shizhen Zhu edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adamson, P.C.; Houghton, P.J.; Perilongo, G.; Pritchard-Jones, K. Drug discovery in paediatric oncology: Roadblocks to progress. Nat. Rev. Clin. Oncol. 2014, 11, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Bosse, K.R.; Maris, J.M. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer 2016, 122, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Barone, G.; Anderson, J.; Pearson, A.D.; Petrie, K.; Chesler, L. New strategies in neuroblastoma: Therapeutic targeting of MYCN and ALK. Clin. Cancer Res. 2013, 19, 5814–5821. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; Committee, C.O.G.N. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.H.; Schulte, S.; Heukamp, L.C.; Astrahantseff, K.; Stephan, H.; Fischer, M.; Schramm, A.; Eggert, A. Targeted therapy for neuroblastoma: ALK inhibitors. Klin. Padiatr. 2013, 225, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Dyer, M.A. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef] [PubMed]
- De Preter, K.; Vandesompele, J.; Heimann, P.; Yigit, N.; Beckman, S.; Schramm, A.; Eggert, A.; Stallings, R.L.; Benoit, Y.; Renard, M.; et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol. 2006, 7, R84. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2210. [Google Scholar] [CrossRef]
- Marshall, G.M.; Carter, D.R.; Cheung, B.B.; Liu, T.; Mateos, M.K.; Meyerowitz, J.G.; Weiss, W.A. The prenatal origins of cancer. Nat. Rev. Cancer 2014, 14, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Cancer in Children & Adolescents. Cancer Facts Figures 2014, 1, 25–42. [Google Scholar]
- Blackburn, J.S.; Langenau, D.M. Zebrafish as a model to assess cancer heterogeneity, progression and relapse. Dis. Model. Mech. 2014, 7, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F. Developmental Biology; Sinauer Associates Inc.: Sunderland, MA, USA, 2013. [Google Scholar]
- Brodeur, G.M.; Bagatell, R. Mechanisms of neuroblastoma regression. Nat. Rev. Clin. Oncol. 2014, 11, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Seeger, R.C.; Barrett, A.; Berthold, F.; Castleberry, R.P.; D’Angio, G.; de Bernardi, B.; Evans, A.E.; Favrot, M.; Freeman, A.I.; et al. International criteria for diagnosis, staging, and response to treatment in patients with neuroblastoma. J. Clin. Oncol. 1988, 6, 1874–1881. [Google Scholar] [PubMed]
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; de Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [PubMed]
- Owens, C.; Irwin, M. Neuroblastoma: the impact of biology and cooperation leading to personalized treatments. Crit. Rev. Clin. Lab. Sci. 2012, 49, 85–115. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.J.; Krstic, A.; Halasz, M.; Schwarzl, T.; Fey, D.; Iljin, K.; Mehta, J.P.; Killick, K.; Whilde, J.; Turriziani, B.; et al. Integrative omics reveals MYCN as a global suppressor of cellular signalling and enables network-based therapeutic target discovery in neuroblastoma. Oncotarget 2015, 6, 43182–43201. [Google Scholar] [PubMed]
- Hsu, C.L.; Chang, H.Y.; Chang, J.Y.; Hsu, W.M.; Huang, H.C.; Juan, H.F. Unveiling MYCN regulatory networks in neuroblastoma via integrative analysis of heterogeneous genomics data. Oncotarget 2016, 7, 36293–36310. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.M.; Buckley, P.G.; Bryan, K.; Watters, K.M.; Koster, J.; van Sluis, P.; Molenaar, J.; Versteeg, R.; Stallings, R.L. Dissection of the oncogenic MYCN transcriptional network reveals a large set of clinically relevant cell cycle genes as drivers of neuroblastoma tumorigenesis. Mol. Carcinog. 2011, 50, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Westermark, U.K.; Wilhelm, M.; Frenzel, A.; Henriksson, M.A. The MYCN oncogene and differentiation in neuroblastoma. Semin. Cancer Biol. 2011, 21, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wei, J.S.; Li, S.Q.; Badgett, T.C.; Song, Y.K.; Agarwal, S.; Coarfa, C.; Tolman, C.; Hurd, L.; Liao, H.; et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett. 2016, 371, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Mitra, S.; Subhash, S.; Hertwig, F.; Kanduri, M.; Mishra, K.; Fransson, S.; Ganeshram, A.; Mondal, T.; Bandaru, S.; et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 2014, 26, 722–737. [Google Scholar] [CrossRef] [PubMed]
- De Preter, K.; de Brouwer, S.; van Maerken, T.; Pattyn, F.; Schramm, A.; Eggert, A.; Vandesompele, J.; Speleman, F. Meta-mining of neuroblastoma and neuroblast gene expression profiles reveals candidate therapeutic compounds. Clin. Cancer Res. 2009, 15, 3690–3696. [Google Scholar] [CrossRef] [PubMed]
- Decock, A.; Ongenaert, M.; Vandesompele, J.; Speleman, F. Neuroblastoma epigenetics: From candidate gene approaches to genome-wide screenings. Epigenetics 2011, 6, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Lequin, D.; Brugieres, L.; Ribeiro, A.; de Pontual, L.; Combaret, V.; Raynal, V.; Puisieux, A.; Schleiermacher, G.; Pierron, G.; et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008, 455, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Michels, E.; Vandesompele, J.; de Preter, K.; Hoebeeck, J.; Vermeulen, J.; Schramm, A.; Molenaar, J.J.; Menten, B.; Marques, B.; Stallings, R.L.; et al. ArrayCGH-based classification of neuroblastoma into genomic subgroups. Genes Chromosomes Cancer 2007, 46, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Laudenslager, M.; Khazi, D.; Carlisle, A.J.; Winter, C.L.; Rappaport, E.; Maris, J.M. Germline PHOX2B mutation in hereditary neuroblastoma. Am. J. Hum. Genet. 2004, 75, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Javanmardi, N.; Bernard, V.; Leroy, Q.; Cappo, J.; Rio Frio, T.; Pierron, G.; Lapouble, E.; Combaret, V.; Speleman, F.; et al. Emergence of new ALK mutations at relapse of neuroblastoma. J. Clin. Oncol. 2014, 32, 2727–2734. [Google Scholar] [CrossRef] [PubMed]
- Speleman, F.; de Preter, K.; Vandesompele, J. Neuroblastoma genetics and phenotype: A tale of heterogeneity. Semin. Cancer Biol. 2011, 21, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, E.F.; London, W.B.; Mosse, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Look, A.T.; Shimada, H.; et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005, 353, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Morozova, O.; Attiyeh, E.F.; Asgharzadeh, S.; Wei, J.S.; Auclair, D.; Carter, S.L.; Cibulskis, K.; Hanna, M.; Kiezun, A.; et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013, 45, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Diskin, S.J.; Capasso, M.; Schnepp, R.W.; Cole, K.A.; Attiyeh, E.F.; Hou, C.; Diamond, M.; Carpenter, E.L.; Winter, C.; Lee, H.; et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012, 44, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Diskin, S.J.; Hou, C.; Glessner, J.T.; Attiyeh, E.F.; Laudenslager, M.; Bosse, K.; Cole, K.; Mosse, Y.P.; Wood, A.; Lynch, J.E.; et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature 2009, 459, 987–991. [Google Scholar] [PubMed]
- Sausen, M.; Leary, R.J.; Jones, S.; Wu, J.; Reynolds, C.P.; Liu, X.; Blackford, A.; Parmigiani, G.; Diaz, L.A., Jr.; Papadopoulos, N.; et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 2013, 45, 12–17. [Google Scholar] [PubMed]
- Wang, K.; Diskin, S.J.; Zhang, H.; Attiyeh, E.F.; Winter, C.; Hou, C.; Schnepp, R.W.; Diamond, M.; Bosse, K.; Mayes, P.A.; et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature 2011, 469, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Mosse, Y.P.; Bradfield, J.P.; Hou, C.; Monni, S.; Scott, R.H.; Asgharzadeh, S.; Attiyeh, E.F.; Diskin, S.J.; Laudenslager, M.; et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Eng. J. Med. 2008, 358, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Nguyen le, B.; Diskin, S.J.; Capasso, M.; Wang, K.; Diamond, M.A.; Glessner, J.; Kim, C.; Attiyeh, E.F.; Mosse, Y.P.; Cole, K.; et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011, 7, e1002026. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Diskin, S.J.; Wasserman, N.; Rinaldi, K.; Attiyeh, E.F.; Cole, K.; Jagannathan, J.; Bhambhani, K.; Winter, C.; Maris, J.M. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer 2007, 46, 936–949. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; van Sluis, P.; Valentijn, L.J.; van der Ploeg, I.; Hamdi, M.; van Nes, J.; Westerman, B.A.; van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Valentijn, L.J.; Koster, J.; Zwijnenburg, D.A.; Hasselt, N.E.; van Sluis, P.; Volckmann, R.; van Noesel, M.M.; George, R.E.; Tytgat, G.A.; Molenaar, J.J.; et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 2015, 47, 1411–1414. [Google Scholar] [CrossRef] [PubMed]
- Oldridge, D.A.; Wood, A.C.; Weichert-Leahey, N.; Crimmins, I.; Sussman, R.; Winter, C.; McDaniel, L.D.; Diamond, M.; Hart, L.S.; Zhu, S.; et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 2015, 528, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Mosse, Y.P.; Bradfield, J.P.; Hou, C.; Monni, S.; Scott, R.H.; Asgharzadeh, S.; Attiyeh, E.F.; Diskin, S.J.; Laudenslager, M.; et al. A genome-wide association study identifies a susceptibility locus to clinically aggressive neuroblastoma at 6p22. N. Eng. J. Med. 2008, 24, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Capasso, M.; Devoto, M.; Hou, C.; Asgharzadeh, S.; Glessner, J.T.; Attiyeh, E.F.; Mosse, Y.P.; Kim, C.; Diskin, S.J.; Cole, K.A.; et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009, 41, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.R.; Penikis, A.; Oldridge, D.A.; Alvarez-Dominguez, J.R.; McDaniel, L.; Diamond, M.; Padovan, O.; Raman, P.; Li, Y.; Wei, J.S.; et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015, 75, 3155–3166. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Domingo-Fernandez, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Capasso, M.; Diskin, S.; Cimmino, F.; Acierno, G.; Totaro, F.; Petrosino, G.; Pezone, L.; Diamond, M.; McDaniel, L.; Hakonarson, H.; et al. Common genetic variants in NEFL influence gene expression and neuroblastoma risk. Cancer Res. 2014, 74, 6913–6924. [Google Scholar] [CrossRef] [PubMed]
- Diskin, S.J.; Capasso, M.; Diamond, M.; Oldridge, D.A.; Conkrite, K.; Bosse, K.R.; Russell, M.R.; Iolascon, A.; Hakonarson, H.; Devoto, M.; et al. Rare variants in TP53 and susceptibility to neuroblastoma. J. Natl. Cancer Inst. 2014, 106, dju047. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Yang, T.; Kuang, Y.; Kong, B.; Yu, S.; Shu, H.; Zhou, H.; Gu, J. MicroRNA-23a promotes neuroblastoma cell metastasis by targeting CDH1. Oncol. Lett. 2014, 7, 839–845. [Google Scholar] [PubMed]
- Ray, S.K. Emerging Roles of microRNAs in malignant neuroblastoma. J. Clin. Exp. Pharmacol. 2013, 3. [Google Scholar] [CrossRef]
- Das, S.; Bryan, K.; Buckley, P.G.; Piskareva, O.; Bray, I.M.; Foley, N.; Ryan, J.; Lynch, J.; Creevey, L.; Fay, J.; et al. Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs. Oncogene 2013, 32, 2927–2936. [Google Scholar] [CrossRef] [PubMed]
- Blumrich, A.; Zapatka, M.; Brueckner, L.M.; Zheglo, D.; Schwab, M.; Savelyeva, L. The FRA2C common fragile site maps to the borders of MYCN amplicons in neuroblastoma and is associated with gross chromosomal rearrangements in different cancers. Hum. Mol. Genet. 2011, 20, 1488–1501. [Google Scholar] [CrossRef] [PubMed]
- Alaminos, M.; Davalos, V.; Cheung, N.K.; Gerald, W.L.; Esteller, M. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J. Natl. Cancer Inst. 2004, 96, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.; Hofree, M.; Ideker, T. Genotype to phenotype via network analysis. Curr. Opin. Genet. Dev. 2013, 23, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Decock, A.; Ongenaert, M.; Cannoodt, R.; Verniers, K.; de Wilde, B.; Laureys, G.; van Roy, N.; Berbegall, A.P.; Bienertova-Vasku, J.; Bown, N.; et al. Methyl-CpG-binding domain sequencing reveals a prognostic methylation signature in neuroblastoma. Oncotarget. 2016, 7, 1960–1972. [Google Scholar] [PubMed]
- Decock, A.; Ongenaert, M.; de Wilde, B.; Brichard, B.; Noguera, R.; Speleman, F.; Vandesompele, J. Stage 4S neuroblastoma tumors show a characteristic DNA methylation portrait. Epigenetics 2016, 11, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Decock, A.; Ongenaert, M.; Hoebeeck, J.; de Preter, K.; van Peer, G.; van Criekinge, W.; Ladenstein, R.; Schulte, J.H.; Noguera, R.; Stallings, R.L.; et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol. 2012, 13, R95. [Google Scholar] [CrossRef] [PubMed]
- Decock, A.; Ongenaert, M.; van Criekinge, W.; Speleman, F.; Vandesompele, J. DNA methylation profiling of primary neuroblastoma tumors using methyl-CpG-binding domain sequencing. Sci. Data 2016, 3, 160004. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Castellano, G.; Mayol, G.; Queiros, A.; Martin-Subero, J.I.; Lavarino, C. DNA methylation fingerprint of neuroblastoma reveals new biological and clinical insights. Genom. Data 2015, 5, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.; Beck, S.; Kogner, P.; Martinsson, T.; Caren, H. Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics 2016, 11, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Hurlin, P.J. N-Myc functions in transcription and development. Birth Defects Res. C Embryo Today 2005, 75, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.T.; Tsanov, K.M.; Pearson, D.S.; Roels, F.; Spina, C.S.; Ebright, R.; Seligson, M.; de Soysa, Y.; Cahan, P.; Theissen, J.; et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016, 535, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Haug, B.H.; Henriksen, J.R.; Buechner, J.; Geerts, D.; Tomte, E.; Kogner, P.; Martinsson, T.; Flaegstad, T.; Sveinbjornsson, B.; Einvik, C. MYCN-regulated miRNA-92 inhibits secretion of the tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma. Carcinogenesis 2011, 32, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Samaraweera, L.; Grandinetti, K.B.; Huang, R.; Spengler, B.A.; Ross, R.A. MicroRNAs define distinct human neuroblastoma cell phenotypes and regulate their differentiation and tumorigenicity. BMC Cancer 2014, 14, 309. [Google Scholar] [CrossRef] [PubMed]
- Boloix, A.; Paris-Coderch, L.; Soriano, A.; Roma, J.; Gallego, S.; Sanchez de Toledo, J.; Segura, M.F. Novel micro RNA-based therapies for the treatment of neuroblastoma. Ann. Pediatr. 2016, 85, e1–e6. [Google Scholar] [CrossRef]
- Mei, H.; Lin, Z.Y.; Tong, Q.S. The roles of microRNAs in neuroblastoma. World J. Pediatr. 2014, 10, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Fatima, R.; Akhade, V.S.; Pal, D.; Rao, S.M. Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 2015, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Sayed, D.; Abdellatif, M. MicroRNAs in development and disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long noncoding RNA expression. Nucleic Acids Res. 2009, 37, D122–D126. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar]
- Amaral, P.P.; Clark, M.B.; Gascoigne, D.K.; Dinger, M.E.; Mattick, J.S. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res. 2011, 39, D146–D151. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An analysis of human microRNA and disease associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef] [PubMed]
- Koster, J. R2: Genomics Analysis and Visualization Center. Available online: http://hgserver1.amc.nl/cgi-bin/r2/main.cgi (accessed on 20 September 2016).
- Duijkers, F.A.; Gaal, J.; Meijerink, J.P.; Admiraal, P.; Pieters, R.; de Krijger, R.R.; van Noesel, M.M. Anaplastic lymphoma kinase (ALK) inhibitor response in neuroblastoma is highly correlated with ALK mutation status, ALK mRNA and protein levels. Cell. Oncol. 2011, 34, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, M.; Otani, K.; Hiyama, K.; Kamei, N.; Satoh, K.; Hiyama, E. A robust method for estimating gene expression states using Affymetrix microarray probe level data. BMC Bioinform. 2010, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Schramm, A.; Koster, J.; Assenov, Y.; Althoff, K.; Peifer, M.; Mahlow, E.; Odersky, A.; Beisser, D.; Ernst, C.; Henssen, A.G.; et al. Mutational dynamics between primary and relapse neuroblastomas. Nat. Genet. 2015, 47, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gong, B.; Bushel, P.R.; Thierry-Mieg, J.; Thierry-Mieg, D.; Xu, J.; Fang, H.; Hong, H.; Shen, J.; Su, Z.; et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol. 2014, 32, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Diskin, S.; Rappaport, E.; Attiyeh, E.; Mosse, Y.; Shue, D.; Seiser, E.; Jagannathan, J.; Shusterman, S.; Bansal, M.; et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006, 66, 6050–6062. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Liao, Y.F.; Juan, H.F.; Huang, H.C.; Wang, B.J.; Lu, Y.L.; Yu, I.S.; Shih, Y.Y.; Jeng, Y.M.; Hsu, W.M.; et al. Aryl hydrocarbon receptor downregulates MYCN expression and promotes cell differentiation of neuroblastoma. PLoS ONE 2014, 9, e88795. [Google Scholar] [CrossRef] [PubMed]
- Eleveld, T.F.; Oldridge, D.A.; Bernard, V.; Koster, J.; Daage, L.C.; Diskin, S.J.; Schild, L.; Bentahar, N.B.; Bellini, A.; Chicard, M.; et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 2015, 47, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Zhang, J.; Lu, C.; Parker, M.; Bahrami, A.; Tickoo, S.K.; Heguy, A.; Pappo, A.S.; Federico, S.; Dalton, J.; et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 2012, 307, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Stanke, J.; Lahti, J.M. The connections between neural crest development and neuroblastoma. Curr. Top. Dev. Biol. 2011, 94, 77–127. [Google Scholar] [PubMed]
- Beltran, H. The N-myc oncogene: Maximizing its targets, regulation, and therapeutic potential. Mol. Cancer Res. 2014, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Hansford, L.M.; Thomas, W.D.; Keating, J.M.; Burkhart, C.A.; Peaston, A.E.; Norris, M.D.; Haber, M.; Armati, P.J.; Weiss, W.A.; Marshall, G.M. Mechanisms of embryonal tumor initiation: Distinct roles for MycN expression and MYCN amplification. Proc. Natl. Acad. Sci. USA 2004, 101, 12664–12669. [Google Scholar] [CrossRef] [PubMed]
- Wartiovaara, K.; Barnabe-Heider, F.; Miller, F.D.; Kaplan, D.R. N-myc promotes survival and induces S-phase entry of postmitotic sympathetic neurons. J. Neurosci. 2002, 22, 815–824. [Google Scholar] [PubMed]
- Chanthery, Y.H.; Gustafson, W.C.; Itsara, M.; Persson, A.; Hackett, C.S.; Grimmer, M.; Charron, E.; Yakovenko, S.; Kim, G.; Matthay, K.K.; et al. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci. Transl. Med. 2012, 4, 115ra3. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, S.; Liu, Y.; Gu, J.; Gu, S.; Xu, Z.; Zhang, R.; Wang, Z.; Ma, H.; Chen, Y.; et al. Overexpression of MYCN promotes proliferation of non-small cell lung cancer. Tumour Biol. 2016, 37, 12855–12866. [Google Scholar] [CrossRef] [PubMed]
- Pession, A.; Tonelli, R. The MYCN oncogene as a specific and selective drug target for peripheral and central nervous system tumors. Curr. Cancer Drug Targets 2005, 5, 273–283. [Google Scholar] [PubMed]
- Williams, R.D.; Chagtai, T.; Alcaide-German, M.; Apps, J.; Wegert, J.; Popov, S.; Vujanic, G.; van Tinteren, H.; van den Heuvel-Eibrink, M.M.; Kool, M.; et al. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 2015, 6, 7232–7243. [Google Scholar] [CrossRef] [PubMed]
- Berry, T.; Luther, W.; Bhatnagar, N.; Jamin, Y.; Poon, E.; Sanda, T.; Pei, D.; Sharma, B.; Vetharoy, W.R.; Hallsworth, A.; et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell 2012, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Cazes, A.; Lopez-Delisle, L.; Tsarovina, K.; Pierre-Eugene, C.; de Preter, K.; Peuchmaur, M.; Nicolas, A.; Provost, C.; Louis-Brennetot, C.; Daveau, R.; et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget 2014, 5, 2688–2702. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.H.; Lindner, S.; Bohrer, A.; Maurer, J.; de Preter, K.; Lefever, S.; Heukamp, L.; Schulte, S.; Molenaar, J.; Versteeg, R.; et al. MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene 2013, 32, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Teitz, T.; Inoue, M.; Valentine, M.B.; Zhu, K.; Rehg, J.E.; Zhao, W.; Finkelstein, D.; Wang, Y.D.; Johnson, M.D.; Calabrese, C.; et al. Th-MYCN mice with caspase-8 deficiency develop advanced neuroblastoma with bone marrow metastasis. Cancer Res. 2013, 73, 4086–4097. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Sheng, J.; Gao, D.; Li, F.; Durrans, A.; Ryu, S.; Lee, S.B.; Narula, N.; Rafii, S.; Elemento, O.; et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015, 10, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Reinartz, S.; Finkernagel, F.; Adhikary, T.; Rohnalter, V.; Schumann, T.; Schober, Y.; Nockher, W.A.; Nist, A.; Stiewe, T.; Jansen, J.M.; et al. A transcriptome-based global map of signaling pathways in the ovarian cancer microenvironment associated with clinical outcome. Genome Biol. 2016, 17, 108. [Google Scholar] [CrossRef] [PubMed]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- DuBois, S.G.; Kalika, Y.; Lukens, J.N.; Brodeur, G.M.; Seeger, R.C.; Atkinson, J.B.; Haase, G.M.; Black, C.T.; Perez, C.; Shimada, H.; et al. Metastatic sites in stage IV and IVS neuroblastoma correlate with age, tumor biology, and survival. J. Pediatr. Hematol. Oncol. 1999, 21, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Marrano, P.; Irwin, M.S.; Thorner, P.S. Heterogeneity of MYCN amplification in neuroblastoma at diagnosis, treatment, relapse, and metastasis. Genes Chromosomes Cancer 2016, 56, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Alabran, J.L.; Hooper, J.E.; Hill, M.; Smith, S.E.; Spady, K.K.; Davis, L.E.; Peterson, L.S.; Malempati, S.; Ryan, C.W.; Acosta, R.; et al. Overcoming autopsy barriers in pediatric cancer research. Pediatr. Blood Cancer 2013, 60, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Spunt, S.L.; Vargas, S.O.; Coffin, C.M.; Skapek, S.X.; Parham, D.M.; Darling, J.; Hawkins, D.S.; Keller, C. The clinical, research, and social value of autopsy after any cancer death: A perspective from the Children’s Oncology Group Soft Tissue Sarcoma Committee. Cancer 2012, 118, 3002–3009. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.S.; Shpak, M.; Townsend, J.P. Inferring the origin of metastases from cancer phylogenies. Cancer Res. 2015, 75, 4021–4025. [Google Scholar] [CrossRef] [PubMed]
- Aiello, N.M.; Stanger, B.Z. Echoes of the embryo: using the developmental biology toolkit to study cancer. Dis. Model. Mech. 2016, 9, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Cekanova, M.; Rathore, K. Animal models and therapeutic molecular targets of cancer: utility and limitations. Drug Des. Dev. Ther. 2014, 8, 1911–1921. [Google Scholar] [CrossRef] [PubMed]
- Jessy, T. Immunity over inability: The spontaneous regression of cancer. J. Natl. Sci. Biol. Med. 2011, 2, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, E.F.; Maris, J.M. Identifying rare events in rare diseases. Clin. Cancer Res. 2015, 21, 1782–1785. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L. Big biological data: Challenges and opportunities. Genom. Proteom. Bioinform. 2014, 12, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Group, U.S.C.S.W. States Cancer Statistics: 1999–2013 Incidence and Mortality Web-based Report. 2016. Available online: www.cdc.gov/uscs (accessed on 20 September 2016). [Google Scholar]
- Collins, C.L.; Malvar, J.; Hamilton, A.S.; Deapen, D.M.; Freyer, D.R. Case-linked analysis of clinical trial enrollment among adolescents and young adults at a National Cancer Institute-designated comprehensive cancer center. Cancer 2015, 121, 4398–4406. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. The ENCODE (encyclopedia of DNA elements) project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.V.; Kavcic, M.; Huang, Y.S.; Herbst, N.; Fisher, B.T.; Seif, A.E.; Li, Y.; Hennessy, S.; Aplenc, R.; Bagatell, R. Establishing a high-risk neuroblastoma cohort using the pediatric health information system database. Pediatr. Blood Cancer 2014, 61, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Guilhamon, P.; Butcher, L.M.; Presneau, N.; Wilson, G.A.; Feber, A.; Paul, D.S.; Schutte, M.; Haybaeck, J.; Keilholz, U.; Hoffman, J.; et al. Assessment of patient-derived tumour xenografts (PDXs) as a discovery tool for cancer epigenomics. Genome Med. 2014, 6, 116. [Google Scholar] [CrossRef] [PubMed]
- Pence, H.E.; Williams, A. ChemSpider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Amatruda, J.F.; Patton, E.E. Genetic models of cancer in zebrafish. Int. Rev. Cell Mol. Biol. 2008, 271, 1–34. [Google Scholar] [PubMed]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J.M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Thomas Look, A. Neuroblastoma and its zebrafish model. Adv. Exp. Med. Biol. 2016, 916, 451–478. [Google Scholar] [PubMed]
- He, S.; Mansour, M.R.; Zimmerman, M.W.; Ki, D.H.; Layden, H.M.; Akahane, K.; Gjini, E.; de Groh, E.D.; Perez-Atayde, A.R.; Zhu, S.; et al. Synergy between loss of NF1 and overexpression of MYCN in neuroblastoma is mediated by the GAP-related domain. eLIFE 2016, 5, e14713. [Google Scholar] [CrossRef] [PubMed]
- Houghton, P.J.; Morton, C.L.; Tucker, C.; Payne, D.; Favours, E.; Cole, C.; Gorlick, R.; Kolb, E.A.; Zhang, W.; Lock, R.; et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr. Blood Cancer 2007, 49, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Abdelfattah, N.S.; Blackburn, J.S.; Moore, J.C.; Martinez, S.A.; Moore, F.E.; Lobbardi, R.; Tenente, I.M.; Ignatius, M.S.; Berman, J.N.; et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods 2014, 11, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Moore, J.C.; Ignatius, M.S.; Tenente, I.M.; Hayes, M.N.; Garcia, E.G.; Torres Yordan, N.; Bourque, C.; He, S.; Blackburn, J.S.; et al. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 2016, 7, 10358. [Google Scholar] [CrossRef] [PubMed]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell. Stem Cell. 2008, 2, 183–189. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Rose, K.; Zon, L. Zebrafish cancer: The state of the art and the path forward. Nat. Rev. Cancer 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Caussinus, E.; Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005, 37, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Homem, C.C.; Knoblich, J.A. Drosophila neuroblasts: A model for stem cell biology. Development 2012, 139, 4297–4310. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. Cellosaurus—A Knowledge Resource on Cell Lines. 2016. Available online: http://web.expasy.org/cellosaurus/ (accessed on 20 September 2016).
- Thiele, C.J. Neuroblastoma cell lines. Hum. Cell 1998, 1, 21–53. [Google Scholar]
- Hall, J.M.; Friedman, L.; Guenther, C.; Lee, M.K.; Weber, J.L.; Black, D.M.; King, M.C. Closing in on a breast cancer gene on chromosome 17q. Am. J. Hum. Genet. 1992, 50, 1235–1242. [Google Scholar] [PubMed]
- Blessing, K.; McLaren, K.M. Histological regression in primary cutaneous melanoma: Recognition, prevalence and significance. Histopathology 1992, 20, 315–322. [Google Scholar]
- Board, P.P.T.E. PDQ Unusual Cancers of Childhood Treatment. 2016. Available online: https://www.cancer.gov/types/childhood-cancers/hp/unusual-cancers-childhood-pdq (accessed on 20 September 2016). [Google Scholar]
- Cajaiba, M.M.; Jennings, L.J.; Rohan, S.M.; Perez-Atayde, A.R.; Marino-Enriquez, A.; Fletcher, J.A.; Geller, J.I.; Leuer, K.M.; Bridge, J.A.; Perlman, E.J. ALK-rearranged renal cell carcinomas in children. Genes Chromosomes Cancer 2016, 55, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, R.; Schmid, A.; Eschle, D.; Baczko, K.; ter Meulen, V.; Billeter, M.A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 1988, 55, 255–265. [Google Scholar] [CrossRef]
- Dirks, W.G.; Fahnrich, S.; Lis, Y.; Becker, E.; MacLeod, R.A.; Drexler, H.G. Expression and functional analysis of the anaplastic lymphoma kinase (ALK) gene in tumor cell lines. Int. J. Cancer 2002, 100, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Eide, H.A.; Halvorsen, A.R.; Bjaanaes, M.M.; Piri, H.; Holm, R.; Solberg, S.; Jorgensen, L.; Brustugun, O.T.; Kiserud, C.E.; Helland, A. The MYCN-HMGA2-CDKN2A pathway in non-small cell lung carcinoma—differences in histological subtypes. BMC Cancer 2015, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, S.; Medeiros, L.J.; Kalhor, N.; Bueso-Ramos, C.E. Anaplastic large cell lymphoma: another entity in the differential diagnosis of small round blue cell tumors. Ann. Diagn. Pathol. 2009, 13, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Katowitz, J.A.; Rorke-Adams, L.B.; Fisher, M.J. Optic pathway gliomas: Neoplasms, not hamartomas. JAMA Ophthalmol. 2013, 131, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacol. Res. 2013, 68, 68–94. [Google Scholar] [CrossRef] [PubMed]
- Rushlow, D.E.; Mol, B.M.; Kennett, J.Y.; Yee, S.; Pajovic, S.; Theriault, B.L.; Prigoda-Lee, N.L.; Spencer, C.; Dimaras, H.; Corson, T.W.; et al. Characterisation of retinoblastomas without RB1 mutations: Genomic, gene expression, and clinical studies. Lancet Oncol. 2013, 14, 327–334. [Google Scholar] [CrossRef]
- Strum, D.P.; Eger, E.I., 2nd; Unadkat, J.D.; Johnson, B.H.; Carpenter, R.L. Age affects the pharmacokinetics of inhaled anesthetics in humans. Anesth. Analg. 1991, 73, 310–318. [Google Scholar]
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; Lee, W.; Obenauf, A.C.; Ran, L.; Murali, R.; Zhang, Q.F.; Wong, E.W.; Hu, W.; Scott, S.N.; Shah, R.H.; et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 2015, 526, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, B.; Wang, Y.; Gao, F.; Zhang, Z. Retinoblastoma spontaneous regression: Clinical and histopathologic analysis. Chin. J. Ophthalmol. 2014, 50, 729–732. [Google Scholar]
- Brookes, A.J.; Robinson, P.N. Human genotype-phenotype databases: Aims, challenges and opportunities. Nat. Rev. Genet. 2015, 16, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Freyhult, E.; Landfors, M.; Onskog, J.; Hvidsten, T.R.; Ryden, P. Challenges in microarray class discovery: A comprehensive examination of normalization, gene selection and clustering. BMC Bioinform. 2010, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Schaff, J.; Fink, C.C.; Slepchenko, B.; Carson, J.H.; Loew, L.M. A general computational framework for modeling cellular structure and function. Biophys. J. 1997, 73, 1135–1146. [Google Scholar] [CrossRef]
- Schaff, J.C.; Vasilescu, D.; Moraru, I.I.; Loew, L.M.; Blinov, M.L. Rule-based modeling with virtual cell. Bioinformatics 2016, 32, 2880–2882. [Google Scholar]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Fridley, B.L.; Lund, S.; Jenkins, G.D.; Wang, L. A Bayesian integrative genomic model for pathway analysis of complex traits. Genet. Epidemiol. 2012, 36, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Shin, H.; Song, Y.S.; Kim, J.H. Synergistic effect of different levels of genomic data for cancer clinical outcome prediction. J. Biomed. Inform. 2012, 45, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, E.R.; Dudek, S.M.; Frase, A.T.; Pendergrass, S.A.; Ritchie, M.D. ATHENA: The analysis tool for heritable and environmental network associations. Bioinformatics 2014, 30, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, S.P.; Mehra, A.; Brass, D.J.; Labianca, G. Network analysis in the social sciences. Science 2009, 323, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Creixell, P.; Reimand, J.; Haider, S.; Wu, G.; Shibata, T.; Vazquez, M.; Mustonen, V.; Gonzalez-Perez, A.; Pearson, J.; Sander, C.; et al. Pathway and network analysis of cancer genomes. Nat. Methods 2015, 12, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Najafi, A.; Bidkhori, G.; Bozorgmehr, J.H.; Koch, I.; Masoudi-Nejad, A. Genome scale modeling in systems biology: Algorithms and resources. Curr. Genom. 2014, 15, 130–159. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Tasan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Vandin, F.; Upfal, E.; Raphael, B.J. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 2011, 18, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Jensen, L.J. Protein-protein interaction databases. Methods Mol. Biol. 2015, 1278, 39–56. [Google Scholar] [PubMed]
- Brohee, S.; Faust, K.; Lima-Mendez, G.; Vanderstocken, G.; van Helden, J. Network Analysis Tools: From biological networks to clusters and pathways. Nat. Protoc. 2008, 3, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, E.L.; Ung, C.Y.; McGehee, C.D.; Correia, C.; Li, H. NetDecoder: A network biology platform that decodes context-specific biological networks and gene activities. Nucleic Acids Res. 2016, 44, e100. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alcalde, F.; Garcia-Lopez, F.; Dopazo, J.; Conesa, A. Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics 2011, 27, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Wang, P.; Li, M.J.; Qin, J.; Wang, X.; Zhang, M.Q.; Wang, J. EpiRegNet: Constructing epigenetic regulatory network from high throughput gene expression data for humans. Epigenetics 2011, 6, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Marbach, D.; Costello, J.C.; Kuffner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Consortium, D.; Kellis, M.; Collins, J.J.; et al. Wisdom of crowds for robust gene network inference. Nat. Methods 2012, 9, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef] [PubMed]
- Cantone, I.; Marucci, L.; Iorio, F.; Ricci, M.A.; Belcastro, V.; Bansal, M.; Santini, S.; di Bernardo, M.; di Bernardo, D.; Cosma, M.P. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell 2009, 137, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Dalla Favera, R.; Califano, A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef] [PubMed]
- McKinney-Freeman, S.; Cahan, P.; Li, H.; Lacadie, S.A.; Huang, H.T.; Curran, M.; Loewer, S.; Naveiras, O.; Kathrein, K.L.; Konantz, M.; et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell 2012, 11, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Altrock, P.M.; Liu, L.L.; Michor, F. The mathematics of cancer: integrating quantitative models. Nat. Rev. Cancer 2015, 15, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Kolch, W.; Halasz, M.; Granovskaya, M.; Kholodenko, B.N. The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer 2015, 15, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ung, C.Y.; Ma, X.H.; Liu, X.H.; Li, B.W.; Low, B.C.; Chen, Y.Z. Pathway sensitivity analysis for detecting pro-proliferation activities of oncogenes and tumor suppressors of epidermal growth factor receptor-extracellular signal-regulated protein kinase pathway at altered protein levels. Cancer 2009, 115, 4246–4263. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Carey, V.J.; Chen, X.W.; Romero, R.; Draghici, S. Machine learning and its applications to biology. PLoS Comput. Biol. 2007, 3, e116. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.Y.; Alipanahi, B.; Lee, L.J.; Bretschneider, H.; Merico, D.; Yuen, R.K.; Hua, Y.; Gueroussov, S.; Najafabadi, H.S.; Hughes, T.R.; et al. RNA splicing—The human splicing code reveals new insights into the genetic determinants of disease. Science 2015, 347, 1254806. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sheridan, R.P.; Liaw, A.; Dahl, G.E.; Svetnik, V. Deep neural nets as a method for quantitative structure-activity relationships. J. Chem. Inf. Model. 2015, 55, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Rusu, A.A.; Veness, J.; Bellemare, M.G.; Graves, A.; Riedmiller, M.; Fidjeland, A.K.; Ostrovski, G.; et al. Human-level control through deep reinforcement learning. Nature 2015, 518, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Saeys, Y.; Inza, I.; Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.; Krause, S.; Li, H.; Kowalski, M.; Goldberg, M.S.; Collins, J.J.; Ingber, D.E. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Sci. Transl. Med. 2014, 6, 217ra2. [Google Scholar] [CrossRef] [PubMed]
- Cahan, P.; Li, H.; Morris, S.A.; Lummertz da Rocha, E.; Daley, G.Q.; Collins, J.J. CellNet: Network biology applied to stem cell engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.A.; Cahan, P.; Li, H.; Zhao, A.M.; San Roman, A.K.; Shivdasani, R.A.; Collins, J.J.; Daley, G.Q. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 2014, 158, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Chen, M.; Wang, H.; Zheng, X. A network flow-based method to predict anticancer drug sensitivity. PLoS ONE 2015, 10, e0127380. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Cunningham, J.J.; Brown, J.S. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat. Commun. 2014, 5, 5499. [Google Scholar] [CrossRef] [PubMed]
- Louis, C.U.; Shohet, J.M. Neuroblastoma: Molecular pathogenesis and therapy. Annu. Rev. Med. 2015, 66, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Speleman, F.; Park, J.R.; Henderson, T.O. Neuroblastoma: A tough nut to crack. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e548–e557. [Google Scholar] [CrossRef] [PubMed]
- Blatt, J.; Corey, S.J. Drug repurposing in pediatrics and pediatric hematology oncology. Drug Discov. Today 2013, 18, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, S.; Chen, B.; Butte, A.J.; Swamidass, S.J.; Lu, Z. A survey of current trends in computational drug repositioning. Brief. Bioinform. 2016, 17, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Cairns, J.; Ung, C.Y.; da Rocha, E.L.; Zhang, C.; Correia, C.; Weinshilboum, R.; Wang, L.; Li, H. A network-based phenotype mapping approach to identify genes that modulate drug response phenotypes. Sci. Rep. 2016, 6, 37003. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Sanderson, P.E.; Zheng, W. Drug combination therapy increases successful drug repositioning. Drug Discov. Today 2016, 21, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Lehar, J.; Krueger, A.S.; Avery, W.; Heilbut, A.M.; Johansen, L.M.; Price, E.R.; Rickles, R.J.; Short, G.F., 3rd; Jin, X.; et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 2009, 27, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Lehar, J.; Krueger, A.S.; Zimmermann, G.R.; Borisy, A.A. Therapeutic selectivity and the multi-node drug target. Discov. Med. 2009, 8, 185–190. [Google Scholar] [PubMed]
- Ryall, K.A.; Tan, A.C. Systems biology approaches for advancing the discovery of effective drug combinations. J. Cheminform. 2015, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins Hospital; Kahl, L.; Hughes, H.K. The Harriet Lane Handbook: Mobile Medicine Series, 21st ed.; Elsevier Health Sciences: Baltimore, Maryland.
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Ratcliffe, P.; Tomlinson, I. The mini-driver model of polygenic cancer evolution. Nat. Rev. Cancer 2015, 15, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Koldobskiy, M.A.; Gondor, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.I.; van de Geijn, B.; Raj, A.; Knowles, D.A.; Petti, A.A.; Golan, D.; Gilad, Y.; Pritchard, J.K. RNA splicing is a primary link between genetic variation and disease. Science 2016, 352, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Fu, Y.; Chakravarty, D.; Demichelis, F.; Rubin, M.A.; Gerstein, M. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016, 17, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.R.; Sherr, D.H.; Hu, Z.; DeLisi, C. A network based approach to drug repositioning identifies plausible candidates for breast cancer and prostate cancer. BMC Med. Genom. 2016, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Bellini, A.; Bernard, V.; Leroy, Q.; Rio Frio, T.; Pierron, G.; Combaret, V.; Lapouble, E.; Clement, N.; Rubie, H.; Thebaud, E.; et al. Deep sequencing reveals occurrence of subclonal ALK Mutations in neuroblastoma at diagnosis. Clin. Cancer Res. 2015, 21, 4913–4921. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).