Detection of Natural Resistance-Associated Substitutions by Ion Semiconductor Technology in HCV1b Positive, Direct-Acting Antiviral Agents-Naïve Patients
Abstract
:1. Introduction
2. Results
3. Discussion
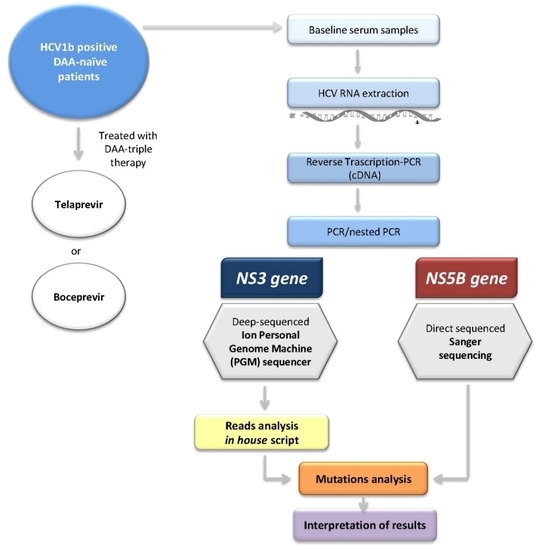
4. Materials and Methods
4.1. Ethic Statement
4.2. Study Population
4.3. HCV RNA Viral Load Determination
4.4. Liver Stiffness
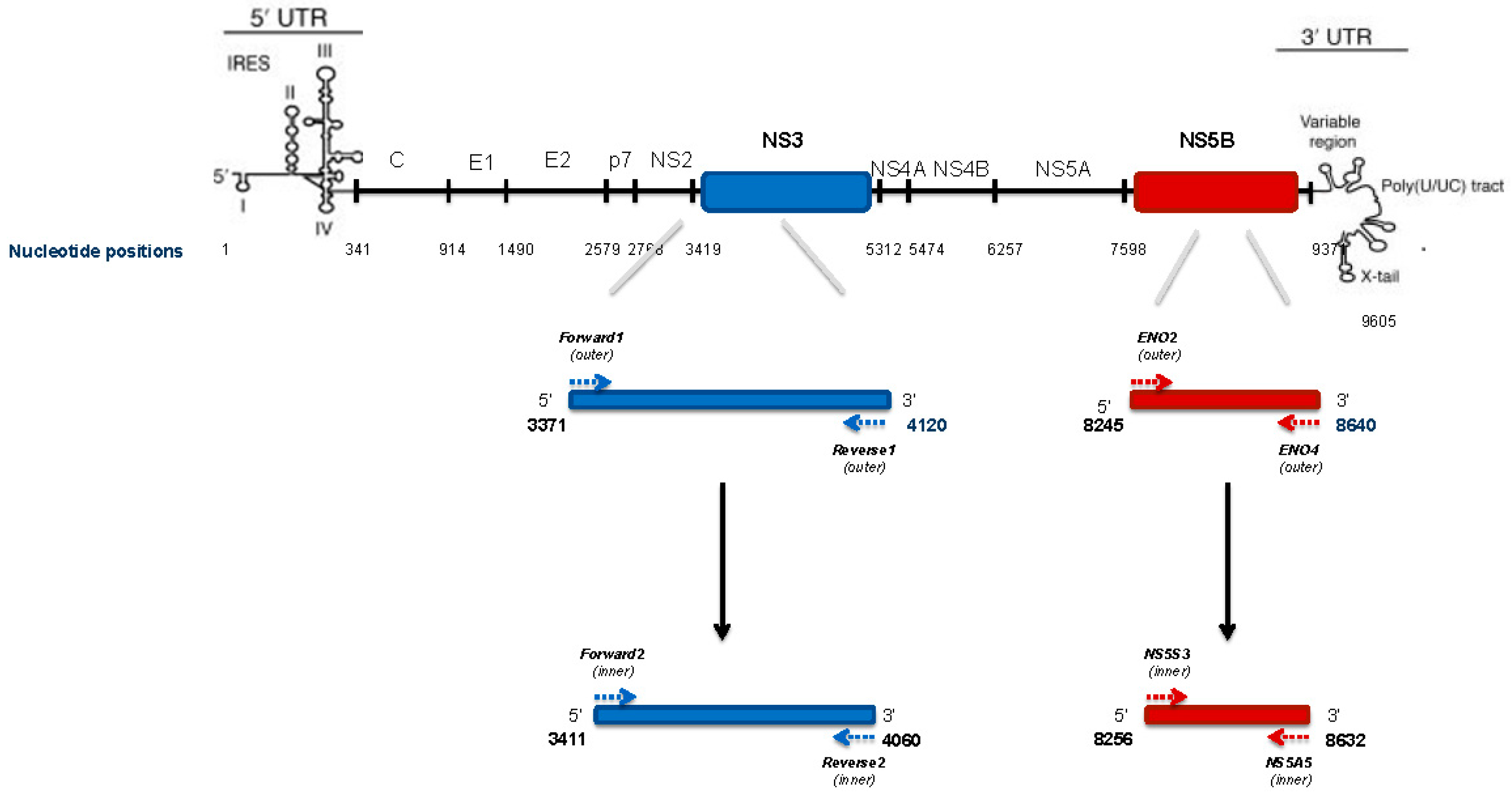
4.5. HCV NS3 Protease Deep Sequencing
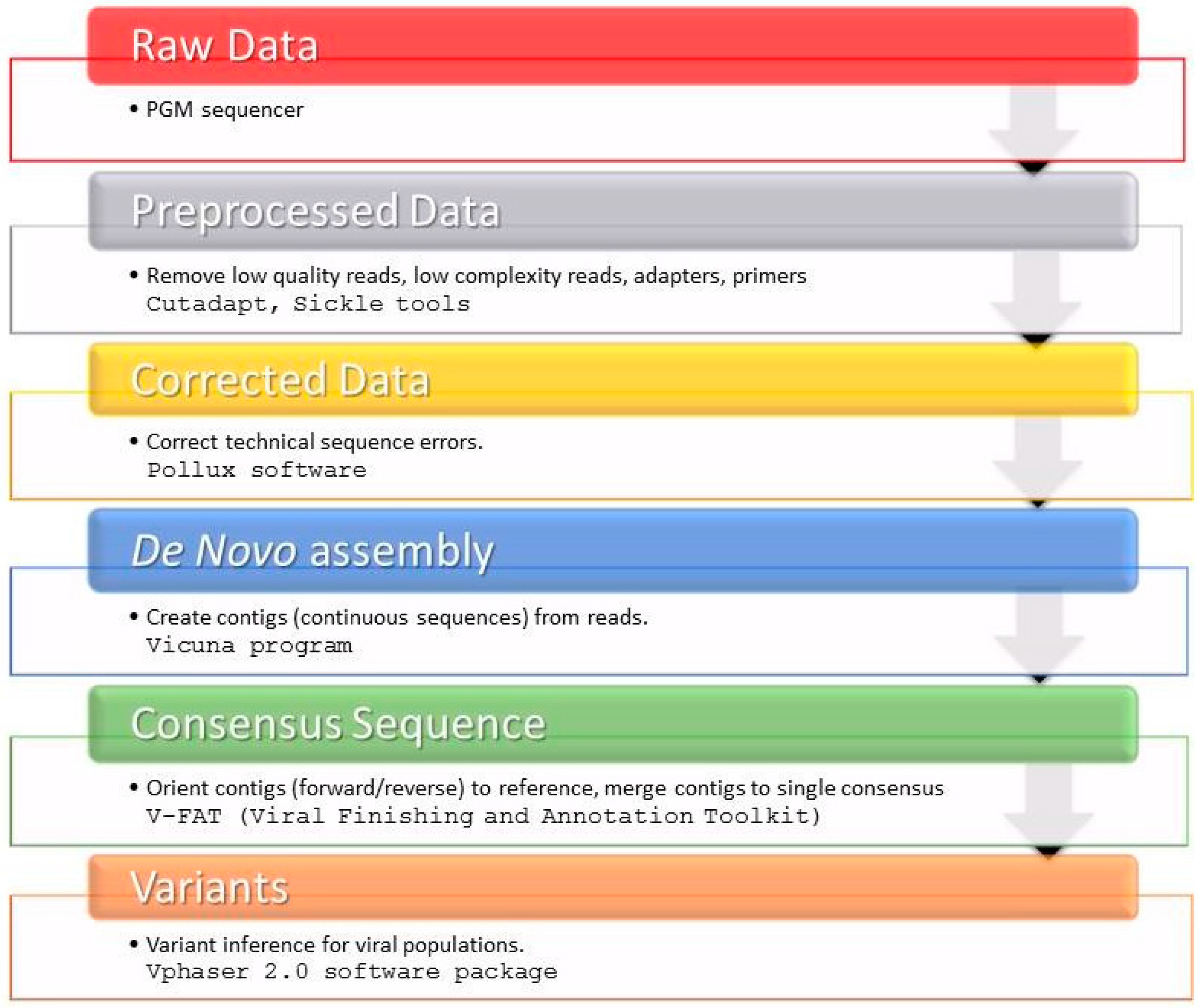
4.6. HCV NS3 NGS Read Analysis
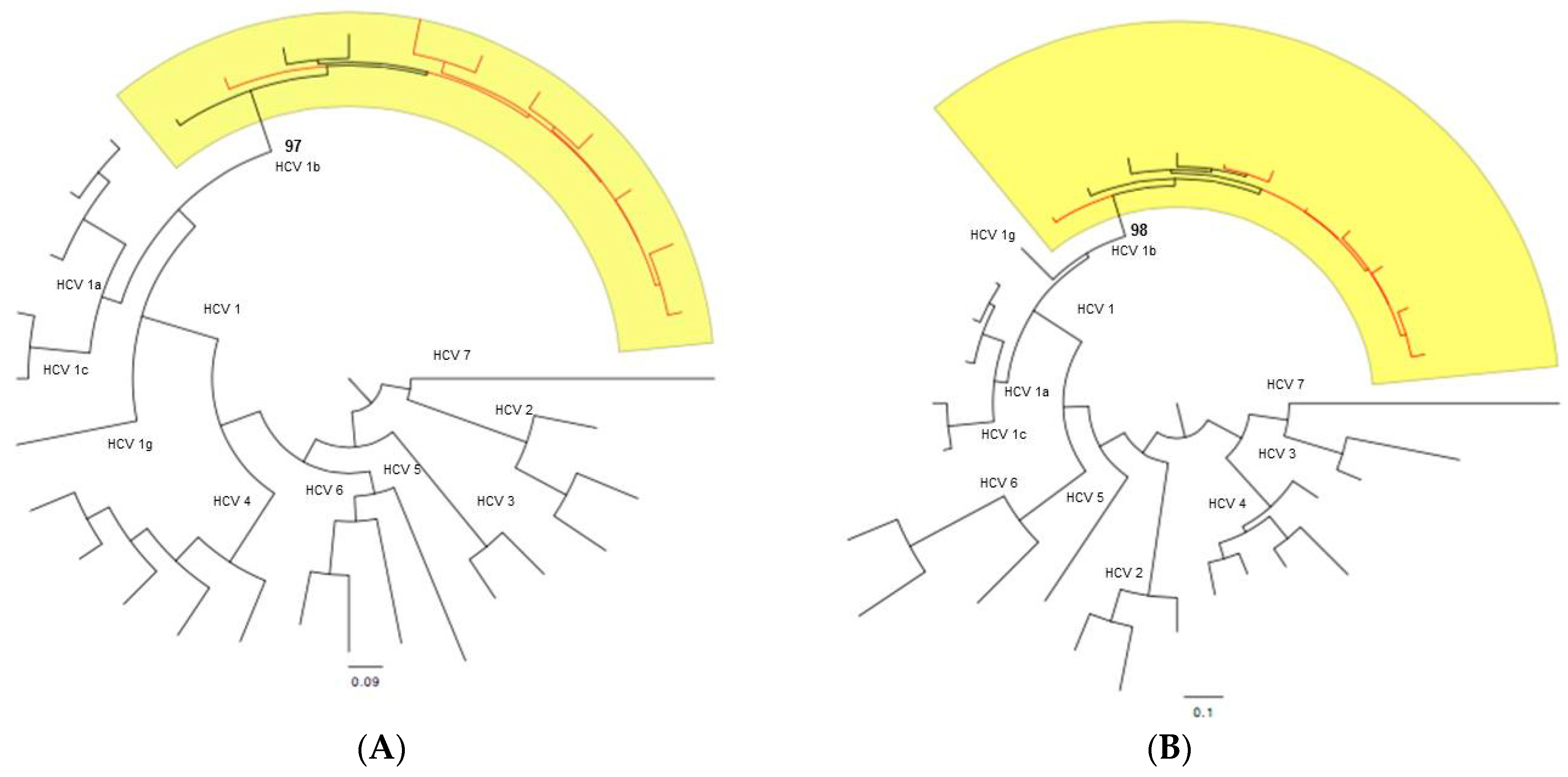
4.7. Subtyping Analysis
4.8. Genetic Variability Analysis
4.9. Public Availability of the Sequencing Data
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- European Association for the Study of the Liver. EASL recommendation on treatment of hepatitis C 2015. J. Hepatol. 2015, 63, 199–236. [Google Scholar]
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Goodman, Z.D.; Koury, K.; Ling, M.; Albrecht, J.K. Peginterferon α-2b plus ribavirin compared with interferon α-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar] [CrossRef]
- Feeney, E.R.; Chung, R.T. Antiviral treatment of hepatitis C. BMJ 2014. [Google Scholar] [CrossRef] [PubMed]
- Asselah, T.; Boyer, N.; Saadoun, D.; Martinot-Peignoux, M.; Marcellin, P. Direct-acting antivirals for the treatment of hepatitis C virus infection: Optimizing current IFN-free treatment and future perspectives. Liver Int. 2016, 36, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Lontok, E.; Harrington, P.; Howe, A.; Kieffer, T.; Lennerstrand, J.; Lenz, O.; McPhee, F.; Mo, H.; Parkin, N.; Pilot-Matias, T.; Miller, V. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology 2015, 61, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Felmlee, D.J. Mechanisms of hepatitis c viral resistance to direct acting antivirals. Viruses 2015, 7, 6716–6729. [Google Scholar] [CrossRef] [PubMed]
- Cento, V.; Chevaliez, S.; Perno, C.F. Resistance to direct-acting antiviral agents: Clinical utility and significance. Curr. Opin. HIV AIDS 2015, 10, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, L.; Ceccherini Silberstein, F.; Van Laethem, K.; Li, G.; Vandamme, A.M.; Rockstroh, J.K. Impact of HCV genotype on treatment regimens and drug resistance: A snapshot in time. Rev. Med. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, H.; Ren, H.; Hu, P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): Mining the GenBank HCV genome data. Sci. Rep. 2016, 6, 20310–20319. [Google Scholar] [CrossRef] [PubMed]
- Guelfo, J.R.; Macias, J.; Neukam, K.; di Lello, F.A.; Mira, J.A.; Merchante, N.; Mancebo, M.; Nunez-Torres, R.; Pineda, J.A.; Real, L.M. Reassessment of genotype 1 hepatitis C virus subtype misclassification by LiPA 2.0: Implications for direct-acting antiviral treatment. J. Clin. Microbiol. 2014, 52, 4027–4029. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.S. Laboratory assays for diagnosis and management of hepatitis C virus infection. J. Clin. Microbiol. 2002, 40, 4407–4412. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P. The origin of hepatitis C virus. Curr. Top. Microbiol. Immunol. 2013, 369, 1–15. [Google Scholar] [PubMed]
- Tong, Y.Q.; Liu, B.; Liu, H.; Zheng, H.Y.; Gu, J.; Liu, H.; Song, E.J.; Song, C.; Li, Y. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin. Microbiol. Infect. 2015, 21, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, V.C.; Cento, V.; di Paolo, D.; Aragri, M.; De Leonardis, F.; Tontodonati, M.; Micheli, V.; Bellocchi, M.C.; Antonucci, F.P.; Bertoli, A.; et al. HCV NS3 sequencing as a reliable and clinically useful tool for the assessment of genotype and resistance mutations for clinical samples with different HCV-RNA levels. J. Antimicrob. Chemother. 2016, 71, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Bergfors, A.; Leenheer, D.; Bergqvist, A.; Ameur, A.; Lennerstrand, J. Analysis of hepatitis C NS5A resistance associated polymorphisms using ultra deep single molecule real time (SMRT) sequencing. Antivir. Res. 2016, 126, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, B.; Lequime, S.; Theys, K.; Lemey, P. Covering all bases in HIV research: unveiling a hidden world of viral evolution. AIDS Rev. 2010, 12, 89–102. [Google Scholar] [PubMed]
- Zhao, Q.; Wen, Y.; Jiang, Y.; Zhang, C.; Li, Y.; Zhang, G.; Zhang, L.; Qiu, M. Next generation sequencing-based investigation of potential patient-to-patient hepatitis C virus transmission during hemodialytic treatment. PLoS ONE 2016, 11, e0147566. [Google Scholar] [CrossRef] [PubMed]
- Ogishi, M.; Yotsuyanagi, H.; Tsutsumi, T.; Gatanaga, H.; Ode, H.; Sugiura, W.; Moriya, K.; Oka, S.; Kimura, S.; Koike, K. Deconvoluting the composition of low-frequency hepatitis C viral quasispecies: Comparison of genotypes and NS3 resistance-associated variants between HCV/HIV coinfected hemophiliacs and HCV monoinfected patients in Japan. PLoS ONE 2015, 10, e0119145. [Google Scholar] [CrossRef] [PubMed]
- Barzon, L.; Lavezzo, E.; Militello, V.; Toppo, S.; Palu, G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011, 12, 7861–7884. [Google Scholar] [CrossRef] [PubMed]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI Viral Genomes Resource. Nucleic Acids Res. 2014, 43, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Capobianchi, M.R.; Giombini, E.; Rozera, G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013, 19, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.F.; Swenson, L.C.; Dong, W.W.; Deng, W.; Kosakovsky Pond, S.L.; Brumme, Z.L.; Mullins, J.I.; Richman, D.D.; Harrigan, P.R.; Frost, S.D. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol. Biol. Evol. 2010, 27, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Zagordi, O.; Däumer, M.; Beisel, C.; Beerenwinkel, N. Read length versus depth of coverage for viral quasispecies reconstruction. PLoS ONE 2012, 7, e47046. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.W.; Oliveira, G.; Yuan, J.; Okulicz, J.F.; Levy, S.; Torbett, B.E. Rapid deep sequencing of patient-derived HIV with ion semiconductor technology. J. Virol. Methods 2013, 189, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Akuta, N.; Suzuki, F.; Seko, Y.; Kawamura, Y.; Sezaki, H.; Suzuki, Y.; Hosaka, T.; Kobayashi, M.; Hara, T.; Kobayashi, M.; et al. Emergence of telaprevir-resistant variants detected by ultra-deep sequencing after triple therapy in patients infected with HCV genotype 1. J. Med. Virol. 2013, 85, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.M.; Weber, J.; Winner, D.; Miller, M.D.; Quinones-Mateu, M.E. Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy. PLoS ONE 2014, 9, e104512. [Google Scholar] [CrossRef] [PubMed]
- Akuta, N.; Suzuki, F.; Fukushima, T.; Kawamura, Y.; Sezaki, H.; Suzuki, Y.; Hosaka, T.; Kobayashi, M.; Hara, T.; Kobayashi, M.; et al. Utility of detection of telaprevir-resistant variants for prediction of efficacy of treatment of hepatitis C virus genotype 1 infection. J. Clin. Microbiol. 2014, 52, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, L.; Faffe, D.S.; Lima, J.F.C.; Capitanio, T.A.; Cabral, B.C.A.; Urmenyi, T.P.; Coelho, H.S.; Rondinelli, E.; Villela-Nogueira, C.A.; Silva, R. No correspondence between resistance mutations in the HCV-NS3 protease at baseline and early telaprevir-based triple therapy. BBA Clin. 2015, 3, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Joshi, N.A.; Fass, J.N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). Available online: http://www.oalib.com/references/15483271 (accessed on 1 June 2016).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Marinier, E.; Brown, D.G.; McConkey, B.J. Pollux: Platform independent error correction of single and mixed genomes. BMC Bioinform. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Charlebois, P.; Gnerre, S.; Coole, M.G.; Lennon, N.J.; Levin, J.Z.; Qu, J.; Ryan, E.M.; Zody, M.C.; Henn, M.R. De novo assembly of highly diverse viral populations. BMC Genomics 2012, 13, 475. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, P.; Yang, X.; Newman, R.M.; Henn, M.R.; Zody, M.C. V-FAT: A post-assembly pipeline for the finishing and annotation of viral genomes. Available online: http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/v-fat (accessed on 1 June 2016).
- Castera, L.; Vergniol, J.; Foucher, J.; Le Bail, B.; Chanteloup, E.; Haaser, M.; Darriet, M.; Couzigou, P.; de Ledinghen, V. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology 2005, 128, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.Y.; Black, S.; Curry, S.; Ludmerer, S.W.; Liu, R.; Barnard, R.J.; Newhard, W.; Hwang, P.M.; Nickle, D.; Gilbert, C. Virologic resistance analysis from a phase 2 study of MK-5172 combined with pegylated interferon/ribavirin in treatment-naive patients with hepatitis C virus genotype 1 infection. Clin. Infect. Dis. 2014, 59, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Zeuzem, S.; Ghalib, R.; Reddy, K.R.; Pockros, P.J.; Ben Ari, Z.; Zhao, Y.; Brown, D.D.; Wan, S.; di Nubile, M.J.; Nguyen, B.Y. Grazoprevir-Elbasvir Combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: A randomized trial. Ann. Intern. Med. 2015, 163, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, V.C.; Cento, V.; Mirabelli, C.; Artese, A.; Costa, G.; Alcaro, S.; Perno, C.F.; Ceccherini-Silberstein, F. Hepatitis C virus genetic variability and the presence of NS5B resistance-associated mutations as natural polymorphisms in selected genotypes could affect the response to NS5B inhibitors. Antimicrob. Agents Chemother. 2014, 58, 2781–2797. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, L.; Snoeck, J.; Vrancken, B.; Kerremans, L.; Vuagniaux, G.; Verbeeck, J.; Nevens, F.; Camacho, R.J.; Vandamme, A.M.; Van Dooren, S. A near-full length genotypic assay for HCV1b. J. Virol. Methods 2014, 209, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Zeuzem, S. What do We Need to Know about RAV’s Clinically, Oral presentation. In Presented at 14th European Meeting on HIV & Hepatitis-Treatment Strategies & Antiviral Drug Resistance, Rome, Italy, May 2016.
- Vrancken, B.; Trovao, N.S.; Baele, G.; van Wijngaerden, E.; Vandamme, A.M.; van Laethem, K.; Lemey, P. Quantifying next generation sequencing sample pre-processing bias in HIV-1 complete genome sequencing. Viruses 2016, 8, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Mantry, P.S.; Pathak, L. Dasabuvir (ABT333) for the treatment of chronic HCV genotype I: A new face of cure, an expert review. Expert Rev. Anti Infect. Ther. 2016, 14, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Ciccozzi, M.; Equestre, M.; Costantino, A.; Marascio, N.; Quirino, A.; Lo Presti, A.; Cella, E.; Bruni, R.; Liberto, M.C.; Foca, A.; et al. Hepatitis C virus genotype 4d in Southern Italy: Reconstruction of its origin and spread by a phylodynamic analysis. J. Med. Virol. 2012, 84, 1613–1619. [Google Scholar] [CrossRef] [PubMed]
- Marascio, N.; Ciccozzi, M.; Equestre, M.; Lo Presti, A.; Costantino, A.; Cella, E.; Bruni, R.; Liberto, M.C.; Pisani, G.; Zicca, E.; et al. Back to the origin of HCV2c subtype and spreading to the Calabria region (Southern Italy) over the last two centuries: A phylogenetic study. Infect. Genet. Evol. 2014, 26, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.C.; De Meyer, S.; Bartels, D.J.; Dierynck, I.; Zhang, E.Z.; Spanks, J.; Tigges, A.M.; Ghys, A.; Dorrian, J.; Adda, N.; et al. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin. Infect. Dis. 2013, 57, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, L.; Li, G.; Libin, P.; Piampongsant, S.; Vandamme, A.M.; Theys, K. Genetic diversity and selective pressure in hepatitis C virus genotypes 1–6: Significance for direct-acting antiviral treatment and drug resistance. Viruses 2015, 7, 5018–5039. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Hance, A.J. HIV drug resistance. N. Engl. J. Med. 2004, 350, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Torti, C.; Zazzi, M.; Abenavoli, L.; Trapasso, F.; Cesario, F.; Corigliano, D.; Cosco, L.; Costa, C.; Curia, R.L.; De Rosa, M.; et al. Future research and collaboration: The “SINERGIE” project on HCV (South Italian Network for Rational Guidelines and International Epidemiology). BMC Infect. Dis. 2012. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Charlebois, P.; Macalalad, A.; Henn, M.R.; Zody, M.C. V-Phaser 2: Variant inference for viral populations. BMC Genom. 2013. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, T.; Deforche, K.; Cassol, S.; Salminen, M.; Paraskevis, D.; Seebregts, C.; Snoeck, J.; van Rensburg, E.J.; Wensing, A.M.; van de Vijver, D.A.; et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 2005, 21, 3797–3800. [Google Scholar] [CrossRef] [PubMed]
- Struck, D.; Lawyer, G.; Ternes, A.M.; Schmit, J.C.; Bercoff, D.P. COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, C.; Hraber, P.; Thurmond, J.; Yusim, K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008, 36, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Kalaghatgi, P.; Sikorski, A.M.; Knops, E.; Rupp, D.; Sierra, S.; Heger, E.; Neumann-Fraune, M.; Beggel, B.; Walker, A.; Timm, J.; Walter, H.; et al. Geno2pheno[HCV]—A web-based interpretation system to support hepatitis C treatment decisions in the era of direct-acting antiviral agents. PLoS ONE 2016, 11, e0155869. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113–132. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2014, 42, 32–37. [Google Scholar] [CrossRef] [PubMed]
| Patient ID | Gender | Age | LiPA Genotyping | Liver Stiffness (KPa) * | Risk Factors # | Response Previous Therapy |
|---|---|---|---|---|---|---|
| HCV04 | female | 47 | 1b | 6.0 | Surgery and Cohabitation | Partial responder |
| HCV06 | male | 68 | 1b | 6.5 | Surgery and Cohabitation | Partial responder |
| HCV08 | male | 46 | 1b | 6.0 | Surgery | Relapser |
| HCV09 | male | 62 | 1b/4 | 6.1 | Surgery and Cohabitation | Relapser |
| HCV17 | female | 69 | 1b | 14.0 | Surgery | Naïve |
| HCV19 | male | 45 | 1b | 6.9 | Surgery and Tattoo | Relapser |
| HCV20 | male | 51 | 1b | 22.0 | Surgery | Relapser |
| HCV21 | male | 66 | 1b | 25.0 | Not Available | Partial responder |
| Patient ID | DAA Therapy * | HCV RNA (IU/mL) | Adverse Events | SVR 12 | |||||
|---|---|---|---|---|---|---|---|---|---|
| – | – | Baseline | 4 Weeks | 12 Weeks | 24 Weeks | 36 Weeks | 48 Weeks | – | – |
| HCV04 | TVR | 7,410,000 | TND | TND | TND | TND | TND | None | Yes |
| HCV06 | TVR | 1,580,000 | <15 | TND | TND | TND | TND | None | Yes |
| HCV08 | TVR | 2,090,000 | TND | TND | TND | TND | TND | None | Yes |
| HCV09 | TVR | 7,930,000 | <15 | TND | TND | TND | TND | Anemia | Yes |
| HCV17 | TVR | 2,170,000 | <15 | TND | TND | TND | TND | Anemia | Yes |
| HCV19 | TVR | 4,290,000 | <15 | TND | TND | TND | TND | None | Yes |
| HCV20 | TVR | 1,520,000 | TND | TND | TND | TND | TND | Anemia and Neutropenia | Yes |
| – | – | Baseline | 4 Weeks | 8 Weeks | 12 Weeks | 24 Weeks | 36 Weeks | – | – |
| HCV21 | BOC | 1,030,000 | 11,900 | <15 | TND | TND | TND | Anemia | Yes |
| Patient ID | NS3 Protease | NS5B Polymerase | ||
|---|---|---|---|---|
| – | RASs | Polymorphisms | RASs | Polymorphisms |
| HCV04 | I132V | S7A | none | V338A |
| D30E | ||||
| P86Q | ||||
| M94L | ||||
| V114I | ||||
| S122N | ||||
| I170V | ||||
| HCV06 | I132V | L14F | C316N | R254K |
| D30E | – | S300T | ||
| V48I | V338A | |||
| P86Q | – | |||
| P89S | ||||
| M94L | ||||
| V114I | ||||
| S122N | ||||
| V150A | ||||
| I170V | ||||
| HCV08 | I132V | D30E | none | V338A |
| T72I | – | |||
| P86Q | ||||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV09 | none | D30E | none | L266M |
| V48I | V338A | |||
| T72I | – | |||
| P89S | ||||
| M94L | ||||
| H110Y | ||||
| S122N | ||||
| V150A | ||||
| I170V | ||||
| HCV17 | V36L | L14I | none | A252V |
| D30E | Q309R | |||
| V48I | V338A | |||
| S61A | – | |||
| P86Q | ||||
| A87S | ||||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV19 | I132V | L14F | C316N | Q309R |
| D30E | – | S335N | ||
| S61T | V338A | |||
| P86Q | – | |||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV20 | V170I | D30E | none | Q309R |
| S61A | S335N | |||
| P86Q | V338A | |||
| M94L | – | |||
| V114I | ||||
| V150A | ||||
| HCV21 | none | L14F | none | V338A |
| D30E | – | |||
| V48I | ||||
| I71V | ||||
| T95A | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marascio, N.; Pavia, G.; Strazzulla, A.; Dierckx, T.; Cuypers, L.; Vrancken, B.; Barreca, G.S.; Mirante, T.; Malanga, D.; Oliveira, D.M.; et al. Detection of Natural Resistance-Associated Substitutions by Ion Semiconductor Technology in HCV1b Positive, Direct-Acting Antiviral Agents-Naïve Patients. Int. J. Mol. Sci. 2016, 17, 1416. https://doi.org/10.3390/ijms17091416
Marascio N, Pavia G, Strazzulla A, Dierckx T, Cuypers L, Vrancken B, Barreca GS, Mirante T, Malanga D, Oliveira DM, et al. Detection of Natural Resistance-Associated Substitutions by Ion Semiconductor Technology in HCV1b Positive, Direct-Acting Antiviral Agents-Naïve Patients. International Journal of Molecular Sciences. 2016; 17(9):1416. https://doi.org/10.3390/ijms17091416
Chicago/Turabian StyleMarascio, Nadia, Grazia Pavia, Alessio Strazzulla, Tim Dierckx, Lize Cuypers, Bram Vrancken, Giorgio Settimo Barreca, Teresa Mirante, Donatella Malanga, Duarte Mendes Oliveira, and et al. 2016. "Detection of Natural Resistance-Associated Substitutions by Ion Semiconductor Technology in HCV1b Positive, Direct-Acting Antiviral Agents-Naïve Patients" International Journal of Molecular Sciences 17, no. 9: 1416. https://doi.org/10.3390/ijms17091416
APA StyleMarascio, N., Pavia, G., Strazzulla, A., Dierckx, T., Cuypers, L., Vrancken, B., Barreca, G. S., Mirante, T., Malanga, D., Oliveira, D. M., Vandamme, A.-M., Torti, C., Liberto, M. C., Focà, A., & The SINERGIE-UMG Study Group. (2016). Detection of Natural Resistance-Associated Substitutions by Ion Semiconductor Technology in HCV1b Positive, Direct-Acting Antiviral Agents-Naïve Patients. International Journal of Molecular Sciences, 17(9), 1416. https://doi.org/10.3390/ijms17091416