Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy
Abstract
:1. Introduction
2. Arginine Metabolism and Bacterial Pathogenesis
2.1. Arginase Pathway and Bacterial Pathogenesis
2.1.1. Arginase Pathway
2.1.2. Regulation of Arginase Pathway
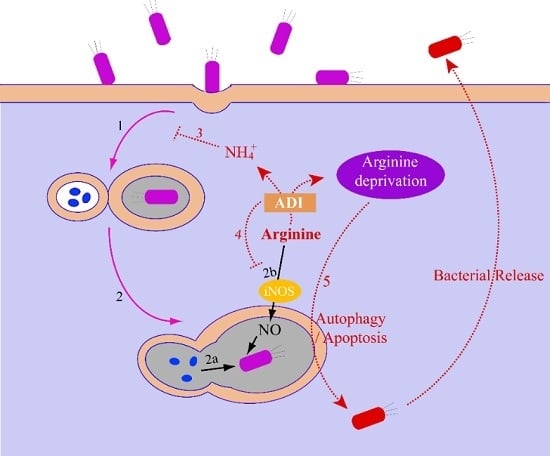
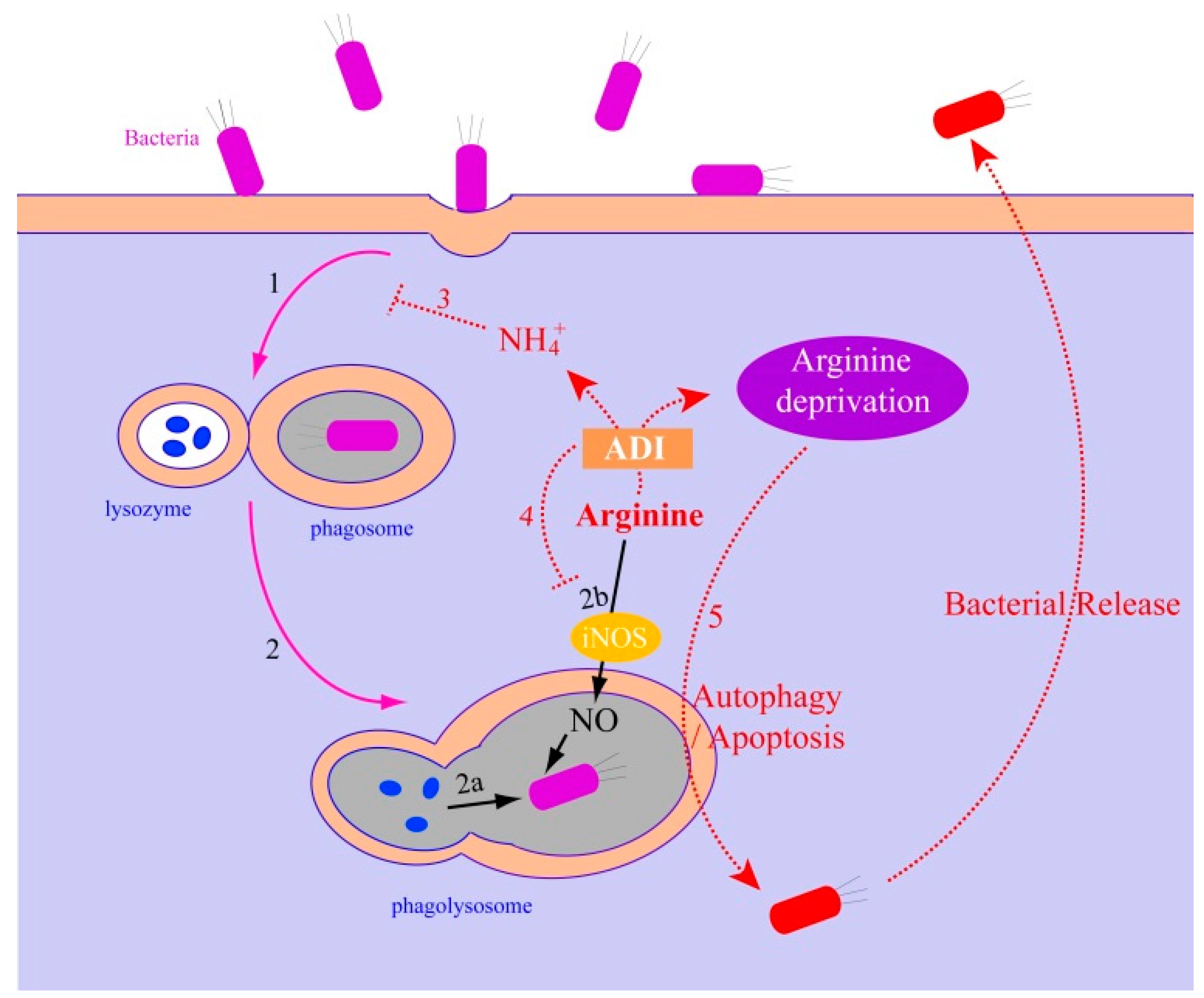
2.1.3. The Relationship between the Arginase Pathway and Bacterial Pathogenesis
2.2. Arginine Deiminase Pathway and Bacterial Pathogenesis
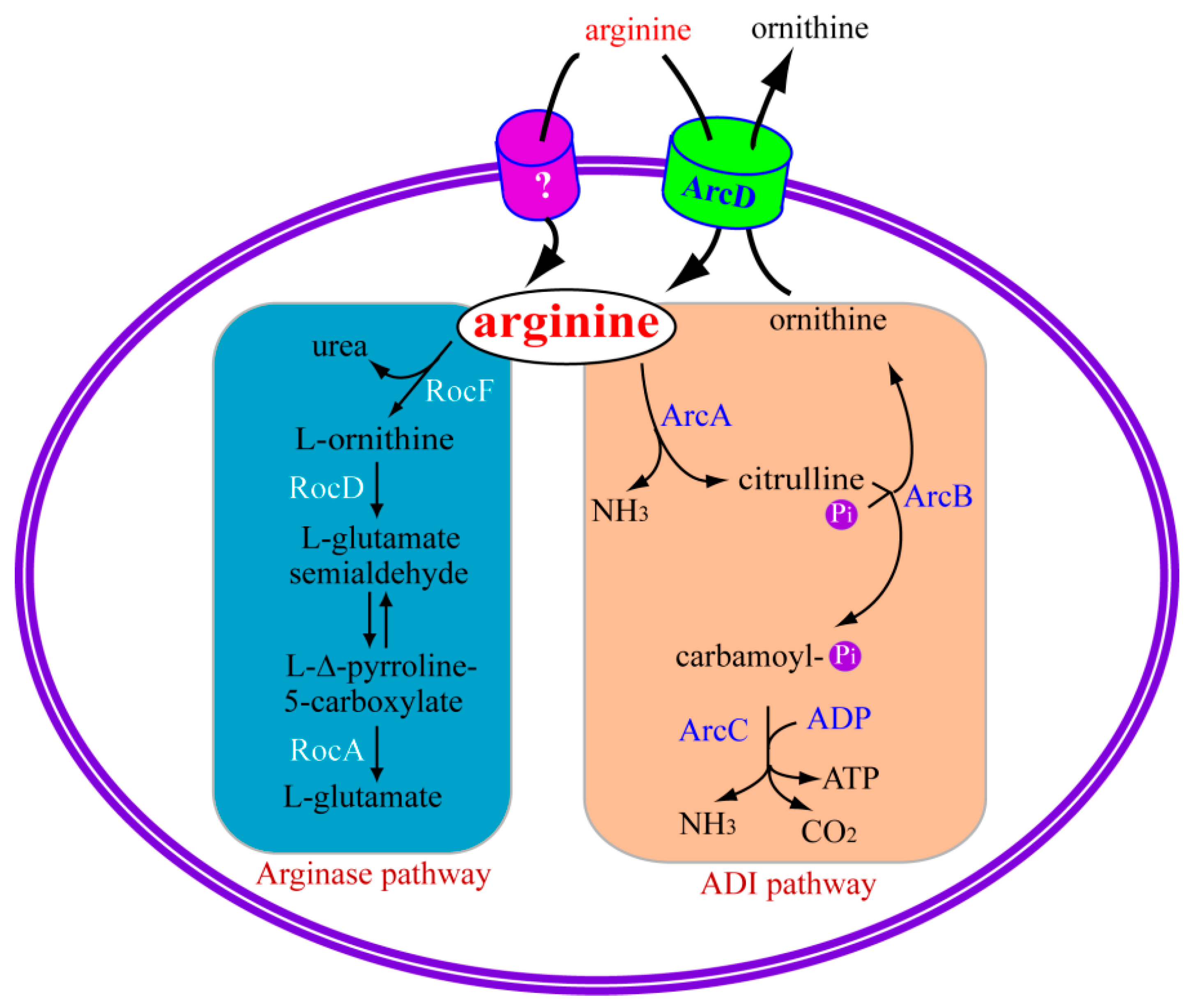
2.2.1. Arginine Deiminase Pathway
2.2.2. Regulation of Arginine Deiminase Pathway
2.2.3. The Relationship between the ADI Pathway and Bacterial Pathogenesis
3. Arginine Metabolism and Cancer Therapy
3.1. Arginine Deprivation and Cancer Therapy
3.2. Molecular Mechanisms of Arginine Depletion for Cancer Therapy
3.2.1. Arginine Deprivation Induces Autophagy in ASS-negative Cells
3.2.2. Arginine Deprivation Prompts Cell Death in ASS-negative Cells
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Egan, S.; Fernandes, N.D.; Kumar, V.; Gardiner, M.; Thomas, T. Bacterial pathogens, virulence mechanism and host defence in marine macroalgae. Environ. Microbiol. 2014, 16, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Hornef, M.W.; Wick, M.J.; Rhen, M.; Normark, S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 2002, 3, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.M.; Sheaff, M.T.; Szlosarek, P.W. Targeting arginine-dependent cancers with arginine-degrading enzymes: Opportunities and challenges. Cancer Res. Treat. 2013, 45, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.T.; Savaraj, N.; Feun, L.G. Targeted cellular metabolism for cancer chemotherapy with recombinant arginine-degrading enzymes. Oncotarget 2010, 1, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Feun, L.; You, M.; Wu, C.J.; Kuo, M.T.; Wangpaichitr, M.; Spector, S.; Savaraj, N. Arginine deprivation as a targeted therapy for cancer. Curr. Pharm. Des. 2008, 14, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Huang, J.; Sui, M. Targeting arginine metabolism pathway to treat arginine-dependent cancers. Cancer Lett. 2015, 364, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.D. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl. Microbiol. Biotechnol. 2006, 70, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M., Jr. Arginine metabolism: Boundaries of our knowledge. J. Nutr. 2007, 137, 1602s–1609s. [Google Scholar] [PubMed]
- Maghnouj, A.; de Sousa Cabral, T.F.; Stalon, V.; Vander Wauven, C. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 1998, 180, 6468–6475. [Google Scholar] [PubMed]
- Calogero, S.; Gardan, R.; Glaser, P.; Schweizer, J.; Rapoport, G.; Debarbouille, M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 1994, 176, 1234–1241. [Google Scholar] [PubMed]
- Gardan, R.; Rapoport, G.; Debarbouille, M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 1995, 249, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Belitsky, B.R.; Sonenshein, A.L. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1999, 96, 10290–10295. [Google Scholar] [CrossRef] [PubMed]
- Gardan, R.; Rapoport, G.; Debarbouille, M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 1997, 24, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Teng, J.L.; Watt, R.M.; Kan, B.; Lau, S.K.; Woo, P.C. Arginine deiminase pathway is far more important than urease for acid resistance and intracellular survival in Laribacter hongkongensis: A possible result of arc gene cassette duplication. BMC Microbiol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Begley, M.; Gahan, C.G.; Hill, C. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: Regulation and role in acid tolerance. Environ. Microbiol. 2009, 11, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Degnan, B.A.; Fontaine, M.C.; Doebereiner, A.H.; Lee, J.J.; Mastroeni, P.; Dougan, G.; Goodacre, J.A.; Kehoe, M.A. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 2000, 68, 2441–2448. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Choi, J.; Groisman, E.A.; Kang, D.H.; Shin, D.; Ryu, S. Expression of STM4467-encoded arginine deiminase controlled by the STM4463 regulator contributes to Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2012, 80, 4291–4297. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Willenborg, J.; Huber, C.; Hitzmann, A.; Willms, D.; Seitz, M.; Eisenreich, W.; Valentin-Weigand, P.; Goethe, R. The arginine-ornithine antiporter ArcD contributes to biological fitness of Streptococcus suis. Front. Cell. Infect. Microbiol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Herrgard, M.J.; Covert, M.W.; Palsson, B.O. Reconstruction of microbial transcriptional regulatory networks. Curr. Opin. Biotechnol. 2004, 15, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.L.; Brennan, R.G. Prokaryotic transcription regulators: More than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002, 12, 98–106. [Google Scholar] [CrossRef]
- Debarbouille, M.; Martin-Verstraete, I.; Kunst, F.; Rapoport, G. The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria. Proc. Natl. Acad. Sci. USA 1991, 88, 9092–9096. [Google Scholar] [CrossRef] [PubMed]
- Klingel, U.; Miller, C.M.; North, A.K.; Stockley, P.G.; Baumberg, S. A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism. Mol. Gen. Genet. 1995, 248, 329–340. [Google Scholar] [CrossRef] [PubMed]
- North, A.K.; Smith, M.C.; Baumberg, S. Nucleotide sequence of a Bacillus subtilis arginine regulatory gene and homology of its product to the Escherichia coli arginine repressor. Gene 1989, 80, 29–38. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C. Of microbes, macrophages and nitric oxide. Behring Inst. Mitt. 1997, 99, 58–72. [Google Scholar] [PubMed]
- Bogdan, C.; Rollinghoff, M.; Diefenbach, A. The role of nitric oxide in innate immunity. Immunol. Rev. 2000, 173, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Boucher, J.L.; Moali, C.; Tenu, J.P. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for l-arginine utilization. Cell. Mol. Life Sci. 1999, 55, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.P.; McGee, D.J.; Akhtar, M.; Mendz, G.L.; Newton, J.C.; Cheng, Y.; Mobley, H.L.; Wilson, K.T. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: A strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 2001, 98, 13844–13849. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Lahiri, A.; Lahiri, A.; Chakravortty, D. Modulation of the arginase pathway in the context of microbial pathogenesis: A metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 2010, 6, e1000899. [Google Scholar] [CrossRef] [PubMed]
- McGee, D.J.; Radcliff, F.J.; Mendz, G.L.; Ferrero, R.L.; Mobley, H.L. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 1999, 181, 7314–7322. [Google Scholar] [PubMed]
- Chaturvedi, R.; Asim, M.; Lewis, N.D.; Algood, H.M.; Cover, T.L.; Kim, P.Y.; Wilson, K.T. l-arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect. Immun. 2007, 75, 4305–4315. [Google Scholar] [CrossRef] [PubMed]
- Zabaleta, J.; McGee, D.J.; Zea, A.H.; Hernandez, C.P.; Rodriguez, P.C.; Sierra, R.A.; Correa, P.; Ochoa, A.C. Helicobacter pylori arginase inhibits T cell proliferation and reduces the expression of the TCR ζ-chain (CD3ζ). J. Immunol. 2004, 173, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Bussiere, F.I.; Chaturvedi, R.; Cheng, Y.; Gobert, A.P.; Asim, M.; Blumberg, D.R.; Xu, H.; Kim, P.Y.; Hacker, A.; Casero, R.A., Jr.; et al. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem. 2005, 280, 2409–2412. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, A.; Das, P.; Chakravortty, D. Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 2008, 10, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Talaue, M.T.; Venketaraman, V.; Hazbon, M.H.; Peteroy-Kelly, M.; Seth, A.; Colangeli, R.; Alland, D.; Connell, N.D. Arginine homeostasis in J774.1 macrophages in the context of Mycobacterium bovis BCG infection. J. Bacteriol. 2006, 188, 4830–4840. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, K.C.; Qualls, J.E.; Pesce, J.T.; Smith, A.M.; Thompson, R.W.; Henao-Tamayo, M.; Basaraba, R.J.; Konig, T.; Schleicher, U.; Koo, M.S.; et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008, 9, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.P.; Guinazu, N.L.; Pellegrini, A.V.; Gotoh, T.; Masih, D.T.; Gea, S. Cruzipain, a major Trypanosoma cruzi antigen, promotes arginase-2 expression and survival of neonatal mouse cardiomyocytes. Am. J. Physiol. Cell Physiol. 2004, 286, C206–C212. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Navarathna, D.H.; Roberts, D.D.; Cooper, J.T.; Atkin, A.L.; Petro, T.M.; Nickerson, K.W. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW264.7. Infect. Immun. 2009, 77, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Cunin, R.; Glansdorff, N.; Pierard, A.; Stalon, V. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 1986, 50, 314–352. [Google Scholar] [PubMed]
- Dong, Y.; Chen, Y.Y.; Burne, R.A. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 2004, 186, 2511–2514. [Google Scholar] [CrossRef] [PubMed]
- Maas, W.K. The arginine repressor of Escherichia coli. Microbiol. Rev. 1994, 58, 631–640. [Google Scholar] [PubMed]
- Miller, C.M.; Baumberg, S.; Stockley, P.G. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: Novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 1997, 26, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Griswold, A.; Chen, Y.Y.; Snyder, J.A.; Burne, R.A. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 2004, 70, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Lu, C.D.; Abdelal, A.T. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 1997, 179, 5309–5317. [Google Scholar] [PubMed]
- Lu, C.D.; Winteler, H.; Abdelal, A.; Haas, D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 2459–2464. [Google Scholar] [PubMed]
- Fulde, M.; Willenborg, J.; de Greeff, A.; Benga, L.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 2011, 157, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Lu, C.D.; Abdelal, A.T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1997, 179, 5300–5308. [Google Scholar] [PubMed]
- Zuniga, M.; Miralles Md Mdel, C.; Perez-Martinez, G. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 2002, 68, 6051–6058. [Google Scholar] [CrossRef] [PubMed]
- Barcelona-Andres, B.; Marina, A.; Rubio, V. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 2002, 184, 6289–6300. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Que, T.L.; Yung, R.W.; Luk, W.K.; Lai, R.W.; Hui, W.T.; Wong, S.S.; Yau, H.H.; et al. Association of Laribacter hongkongensis in community-acquired gastroenteritis with travel and eating fish: A multicentre case-control study. Lancet 2004, 363, 1941–1947. [Google Scholar] [CrossRef]
- Yuen, K.Y.; Woo, P.C.; Teng, J.L.; Leung, K.W.; Wong, M.K.; Lau, S.K. Laribacter hongkongensis gen. Nov., sp. Nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 2001, 39, 4227–4232. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Teng, J.L.; Watt, R.M.; Liu, C.; Lau, S.K.; Woo, P.C. Molecular characterization of arginine deiminase pathway in Laribacter hongkongensis and unique regulation of arginine catabolism and anabolism by multiple environmental stresses. Environ. Microbiol. 2015, 17, 4469–4483. [Google Scholar] [CrossRef] [PubMed]
- Spiro, S. The FNR family of transcriptional regulators. Antonie van Leeuwenhoek 1994, 66, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Gamper, M.; Zimmermann, A.; Haas, D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 1991, 173, 4742–4750. [Google Scholar] [PubMed]
- Maghnouj, A.; Abu-Bakr, A.A.; Baumberg, S.; Stalon, V.; Vander Wauven, C. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 2000, 191, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Gruening, P.; Fulde, M.; Valentin-Weigand, P.; Goethe, R. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 2006, 188, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Eiglmeier, K.; Honore, N.; Iuchi, S.; Lin, E.C.; Cole, S.T. Molecular genetic analysis of FNR-dependent promoters. Mol. Microbiol. 1989, 3, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Spiro, S.; Guest, J.R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 1990, 6, 399–428. [Google Scholar] [PubMed]
- Makhlin, J.; Kofman, T.; Borovok, I.; Kohler, C.; Engelmann, S.; Cohen, G.; Aharonowitz, Y. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 2007, 189, 5976–5986. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, M.; Champomier-Verges, M.; Zagorec, M.; Perez-Martinez, G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 1998, 180, 4154–4159. [Google Scholar] [PubMed]
- Titgemeyer, F.; Hillen, W. Global control of sugar metabolism: A gram-positive solution. Antonie van Leeuwenhoek 2002, 82, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Winterhoff, N.; Goethe, R.; Gruening, P.; Rohde, M.; Kalisz, H.; Smith, H.E.; Valentin-Weigand, P. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 2002, 184, 6768–6776. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Zuniga, M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006, 32, 155–183. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, J.K.; Thomas, V.C.; Olson, M.E.; Chaudhari, S.S.; Nuxoll, A.S.; Schaeffer, C.R.; Lindgren, K.E.; Jones, J.; Zimmerman, M.C.; Dunman, P.M.; et al. Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J. Bacteriol. 2014, 196, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Casiano-Colon, A.; Marquis, R.E. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 1988, 54, 1318–1324. [Google Scholar] [PubMed]
- Conte, M.P.; Petrone, G.; Di Biase, A.M.; Ammendolia, M.G.; Superti, F.; Seganti, L. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb. Pathog. 2000, 29, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Aderem, A.; Underhill, D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef] [PubMed]
- Vieira, O.V.; Botelho, R.J.; Grinstein, S. Phagosome maturation: Aging gracefully. Biochem. J. 2002, 366, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Vergne, I.; Chua, J.; Singh, S.B.; Deretic, V. Cell biology of Mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 2004, 20, 367–394. [Google Scholar] [CrossRef] [PubMed]
- Pitt, A.; Mayorga, L.S.; Stahl, P.D.; Schwartz, A.L. Alterations in the protein composition of maturing phagosomes. J. Clin. Investig. 1992, 90, 1978–1983. [Google Scholar] [CrossRef] [PubMed]
- Bassoe, C.F.; Bjerknes, R. Phagocytosis by human leukocytes, phagosomal pH and degradation of seven species of bacteria measured by flow cytometry. J. Med. Microbiol. 1985, 19, 115–125. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, B.; Gahan, C.G.; Hill, C. Adaptive acid tolerance response in Listeria monocytogenes: Isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 1996, 62, 1693–1698. [Google Scholar] [PubMed]
- Myers, B.M.; Tietz, P.S.; Tarara, J.E.; LaRusso, N.F. Dynamic measurements of the acute and chronic effects of lysosomotropic agents on hepatocyte lysosomal pH using flow cytometry. Hepatology 1995, 22, 1519–1526. [Google Scholar] [PubMed]
- Ohkuma, S.; Poole, B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA 1978, 75, 3327–3331. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.S.; Cosio, G.; Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009, 7, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat. Rev. Microbiol. 2004, 2, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Gotoh, T. Arginine metabolic enzymes, nitric oxide and infection. J. Nutr. 2004, 134, 2820S–2825S. [Google Scholar] [PubMed]
- Yu, H.H.; Wu, F.L.; Lin, S.E.; Shen, L.J. Recombinant arginine deiminase reduces inducible nitric oxide synthase iNOS-mediated neurotoxicity in a coculture of neurons and microglia. J. Neurosci. Res. 2008, 86, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Pontes, M.H.; Groisman, E.A. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 2013, 154, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Shen, X.; Bryan, A.; Banga, S.; Swanson, M.S.; Luo, Z.Q. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010, 6, e1000822. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Bach, H.; Sun, J.; Hmama, Z.; Av-Gay, Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. USA 2011, 108, 19371–19376. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Gahan, C.G.; Hill, C. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 2000, 60, 137–146. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Savaraj, N.; You, M.; Wu, C.; Wangpaichitr, M.; Kuo, M.T.; Feun, L.G. Arginine deprivation, autophagy, apoptosis (AAA) for the treatment of melanoma. Curr. Mol. Med. 2010, 10, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Changou, C.A.; Chen, Y.R.; Xing, L.; Yen, Y.; Chuang, F.Y.; Cheng, R.H.; Bold, R.J.; Ann, D.K.; Kung, H.J. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 14147–14152. [Google Scholar] [CrossRef] [PubMed]
- Tennant, D.A.; Duran, R.V.; Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 2010, 10, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, D.N.; Campbell, E. Arginine catabolism, liver extracts and cancer. Pathol. Oncol. Res. 2002, 8, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Chen, Y.R.; Liu, X.; Chu, C.Y.; Shen, L.J.; Xu, J.; Gaur, S.; Forman, H.J.; Zhang, H.; Zheng, S.; et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci. Signal. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Dillon, B.J.; Prieto, V.G.; Curley, S.A.; Ensor, C.M.; Holtsberg, F.W.; Bomalaski, J.S.; Clark, M.A. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: A method for identifying cancers sensitive to arginine deprivation. Cancer 2004, 100, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Bowles, T.L.; Kim, R.; Galante, J.; Parsons, C.M.; Virudachalam, S.; Kung, H.J.; Bold, R.J. Pancreatic cancer cell lines deficient in argininosuccinate synthetase are sensitive to arginine deprivation by arginine deiminase. Int. J. Cancer 2008, 123, 1950–1955. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Lamb, J.; Smith, S.; Wheatley, D.N. Single amino acid (arginine) deprivation: Rapid and selective death of cultured transformed and malignant cells. Br. J. Cancer 2000, 83, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, J.; Wu, Z.; Li, W.; Zhang, D.; Han, L.; Wang, F.; Reindl, K.M.; Wu, E.; Ma, Q. Arginine deiminase augments the chemosensitivity of argininosuccinate synthetase-deficient pancreatic cancer cells to gemcitabine via inhibition of NF-κB signaling. BMC Cancer 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Miraki-Moud, F.; Ghazaly, E.; Ariza-McNaughton, L.; Hodby, K.A.; Clear, A.; Anjos-Afonso, F.; Liapis, K.; Grantham, M.; Sohrabi, F.; Cavenagh, J.; et al. Arginine deprivation using pegylated arginine deiminase has activity against primary acute myeloid leukemia cells in vivo. Blood 2015, 125, 4060–4068. [Google Scholar] [CrossRef] [PubMed]
- Savaraj, N.; Wu, C.; Kuo, M.T.; You, M.; Wangpaichitr, M.; Robles, C.; Spector, S.; Feun, L. The relationship of arginine deprivation, argininosuccinate synthetase and cell death in melanoma. Drug Target Insights 2007, 2, 119–128. [Google Scholar] [PubMed]
- Lam, T.L.; Wong, G.K.; Chow, H.Y.; Chong, H.C.; Chow, T.L.; Kwok, S.Y.; Cheng, P.N.; Wheatley, D.N.; Lo, W.H.; Leung, Y.C. Recombinant human arginase inhibits the in vitro and in vivo proliferation of human melanoma by inducing cell cycle arrest and apoptosis. Pigment Cell Melanoma Res. 2011, 24, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, K.; Ohno, T.; Kusuyama, T.; Azuma, I. High sensitivity of human melanoma cell lines to the growth inhibitory activity of mycoplasmal arginine deiminase in vitro. Melanoma Res. 1992, 2, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Schwaneberg, U.; Sun, Z.H. Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 2008, 261, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.K.; Frankel, A.E.; Feun, L.G.; Ekmekcioglu, S.; Kim, K.B. Arginine deprivation therapy for malignant melanoma. Clin. Pharmacol. 2013, 5, 11–19. [Google Scholar] [PubMed]
- Ensor, C.M.; Holtsberg, F.W.; Bomalaski, J.S.; Clark, M.A. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res. 2002, 62, 5443–5450. [Google Scholar] [PubMed]
- Kim, R.H.; Bold, R.J.; Kung, H.J. ADI, autophagy and apoptosis: Metabolic stress as a therapeutic option for prostate cancer. Autophagy 2009, 5, 567–568. [Google Scholar] [PubMed]
- Kim, R.H.; Coates, J.M.; Bowles, T.L.; McNerney, G.P.; Sutcliffe, J.; Jung, J.U.; Gandour-Edwards, R.; Chuang, F.Y.; Bold, R.J.; Kung, H.J. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009, 69, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Li, Y.; Fan, J.; Zhao, H.; Xian, Z.; Sun, Y.; Wang, Z.; Wang, S.; Zhang, G.; Ju, D. Recombinant human arginase induced caspase-dependent apoptosis and autophagy in non-hodgkin’s lymphoma cells. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Delage, B.; Luong, P.; Maharaj, L.; O'Riain, C.; Syed, N.; Crook, T.; Hatzimichael, E.; Papoudou-Bai, A.; Mitchell, T.J.; Whittaker, S.J.; et al. Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Izzo, F.; Marra, P.; Beneduce, G.; Castello, G.; Vallone, P.; De Rosa, V.; Cremona, F.; Ensor, C.M.; Holtsberg, F.W.; Bomalaski, J.S.; et al. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: Results from phase I/II studies. J. Clin. Oncol. 2004, 22, 1815–1822. [Google Scholar] [CrossRef] [PubMed]
- Glazer, E.S.; Piccirillo, M.; Albino, V.; Di Giacomo, R.; Palaia, R.; Mastro, A.A.; Beneduce, G.; Castello, G.; De Rosa, V.; Petrillo, A.; et al. Phase II study of pegylated arginine deiminase for nonresectable and metastatic hepatocellular carcinoma. J. Clin. Oncol. 2010, 28, 2220–2226. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, J.A.; Lu, H.T.; Wu, K.C.; Knowles, S.K.; Thomson, J.A. Down-regulation of argininosuccinate synthetase is associated with cisplatin resistance in hepatocellular carcinoma cell lines: Implications for pegylated arginine deiminase combination therapy. BMC Cancer 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Daylami, R.; Muilenburg, D.J.; Virudachalam, S.; Bold, R.J. Pegylated arginine deiminase synergistically increases the cytotoxicity of gemcitabine in human pancreatic cancer. J. Exp. Clin. Cancer Res. 2014, 33. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, C.; Yoshino, K.; Yonezawa, K. mTOR integrates amino acid- and energy-sensing pathways. Biochem. Biophys. Res. Commun. 2004, 313, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Mussai, F.; Egan, S.; Higginbotham-Jones, J.; Perry, T.; Beggs, A.; Odintsova, E.; Loke, J.; Pratt, G.; U, K.P.; Lo, A.; et al. Arginine dependence of acute myeloid leukemia blast proliferation: A novel therapeutic target. Blood 2015, 125, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Zwaan, C.M.; Kolb, E.A.; Reinhardt, D.; Abrahamsson, J.; Adachi, S.; Aplenc, R.; De Bont, E.S.; De Moerloose, B.; Dworzak, M.; Gibson, B.E.; et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J. Clin. Oncol. 2015, 33, 2949–2962. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.; Langer, J.; Janczar, K.; Singh, P.; Lo Nigro, C.; Lattanzio, L.; Coley, H.M.; Hatzimichael, E.; Bomalaski, J.; Szlosarek, P.; et al. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblastoma. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Szlosarek, P.W.; Klabatsa, A.; Pallaska, A.; Sheaff, M.; Smith, P.; Crook, T.; Grimshaw, M.J.; Steele, J.P.; Rudd, R.M.; Balkwill, F.R.; et al. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin. Cancer Res. 2006, 12, 7126–7131. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.Y.; Shim, Y.J.; Kim, E.H.; Lee, J.H.; Won, N.H.; Kim, J.H.; Park, I.S.; Yoon, D.K.; Min, B.H. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. Int. J. Cancer 2007, 120, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P.; Jungbluth, A.A.; Wu, B.W.; Bomalaski, J.; Old, L.J.; Ritter, G. Arginine deiminase PEG20 inhibits growth of small cell lung cancers lacking expression of argininosuccinate synthetase. Br. J. Cancer 2012, 106, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, N.J.; Talbot, J.; Ward, S. pH-sensitive control of arginase by Mn(II) ions at submicromolar concentrations. Arch. Biochem. Biophys. 1991, 286, 217–221. [Google Scholar] [CrossRef]
- Cheng, P.N.; Lam, T.L.; Lam, W.M.; Tsui, S.M.; Cheng, A.W.; Lo, W.H.; Leung, Y.C. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007, 67, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.M.; Glazer, E.S.; Chantranupong, L.; Cherukuri, P.; Breece, R.M.; Tierney, D.L.; Curley, S.A.; Iverson, B.L.; Georgiou, G. Replacing Mn2+ with Co2+ in human arginase I enhances cytotoxicity toward l-arginine auxotrophic cancer cell lines. ACS Chem. Biol. 2010, 5, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Y.; Chen, J.; Cheng, B.; Hu, J.; Zhou, Y.; Gao, X.; Gao, L.; Mei, X.; Sun, M.; et al. An engineered arginase FC protein inhibits tumor growth in vitro and in vivo. Evid. Based Complement. Alternat. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, D.N. Arginine deprivation and metabolomics: Important aspects of intermediary metabolism in relation to the differential sensitivity of normal and tumour cells. Semin. Cancer Biol. 2005, 15, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Takaku, H.; Umeda, M.; Fujita, T.; Huang, W.D.; Kimura, T.; Yamashita, J.; Horio, T. Potent growth inhibition of human tumor cells in culture by arginine deiminase purified from a culture medium of a Mycoplasma-infected cell line. Cancer Res. 1990, 50, 4522–4527. [Google Scholar] [PubMed]
- Takaku, H.; Takase, M.; Abe, S.; Hayashi, H.; Miyazaki, K. In vivo anti-tumor activity of arginine deiminase purified from Mycoplasma arginini. Int. J. Cancer 1992, 51, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Holtsberg, F.W.; Ensor, C.M.; Steiner, M.R.; Bomalaski, J.S.; Clark, M.A. Poly(ethylene glycol) (PEG) conjugated arginine deiminase: Effects of PEG formulations on its pharmacological properties. J. Control. Release 2002, 80, 259–271. [Google Scholar] [CrossRef]
- Dillon, B.J.; Holtsberg, F.W.; Ensor, C.M.; Bomalaski, J.S.; Clark, M.A. Biochemical characterization of the arginine degrading enzymes arginase and arginine deiminase and their effect on nitric oxide production. Med. Sci. Monit. 2002, 8, Br248–Br253. [Google Scholar] [PubMed]
- Feun, L.; Savaraj, N. Pegylated arginine deiminase: A novel anticancer enzyme agent. Expert Opin. Investig. Drugs 2006, 15, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.E.; Wu, F.L.; Wei, M.F.; Shen, L.J. Depletion of arginine by recombinant arginine deiminase induces nNOS-activated neurotoxicity in neuroblastoma cells. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Szlosarek, P.W.; Luong, P.; Phillips, M.M.; Baccarini, M.; Stephen, E.; Szyszko, T.; Sheaff, M.T.; Avril, N. Metabolic response to pegylated arginine deiminase in mesothelioma with promoter methylation of argininosuccinate synthetase. J. Clin. Oncol. 2013, 31, e111–e113. [Google Scholar] [CrossRef] [PubMed]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Brumell, J.H. Bacteria-autophagy interplay: A battle for survival. Nat. Rev. Microbiol. 2014, 12, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T.; Toth, M.L.; Kovacs, A.L. Janus-faced autophagy: A dual role of cellular self-eating in neurodegeneration? Autophagy 2007, 3, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; White, E. Role of autophagy in cancer: Management of metabolic stress. Autophagy 2007, 3, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Proud, C.G. mTOR-mediated regulation of translation factors by amino acids. Biochem. Biophys. Res. Commun. 2004, 313, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Whiteman, M.W.; Lian, H.; Wang, G.; Singh, A.; Huang, D.; Denmark, T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 2009, 284, 21412–21424. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zolzer, F.; von Recklinghausen, G.; Havers, W.; Schweigerer, L. Arginine deiminase inhibits proliferation of human leukemia cells more potently than asparaginase by inducing cell cycle arrest and apoptosis. Leukemia 2000, 14, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Szlosarek, P.W. Arginine deprivation and autophagic cell death in cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 14015–14016. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef] [PubMed]
 and
and  ) means the reactions are dependent on the availability of enzymes (panel A) or the reactions have not yet confirmed by experiments (panel B). MEK: mitogen-activated protein kinase, also known as extracellular signal-regulated kinase kinase; ERK: extracellular signal-regulated kinase.
) means the reactions are dependent on the availability of enzymes (panel A) or the reactions have not yet confirmed by experiments (panel B). MEK: mitogen-activated protein kinase, also known as extracellular signal-regulated kinase kinase; ERK: extracellular signal-regulated kinase.
 and
and  ) means the reactions are dependent on the availability of enzymes (panel A) or the reactions have not yet confirmed by experiments (panel B). MEK: mitogen-activated protein kinase, also known as extracellular signal-regulated kinase kinase; ERK: extracellular signal-regulated kinase.
) means the reactions are dependent on the availability of enzymes (panel A) or the reactions have not yet confirmed by experiments (panel B). MEK: mitogen-activated protein kinase, also known as extracellular signal-regulated kinase kinase; ERK: extracellular signal-regulated kinase.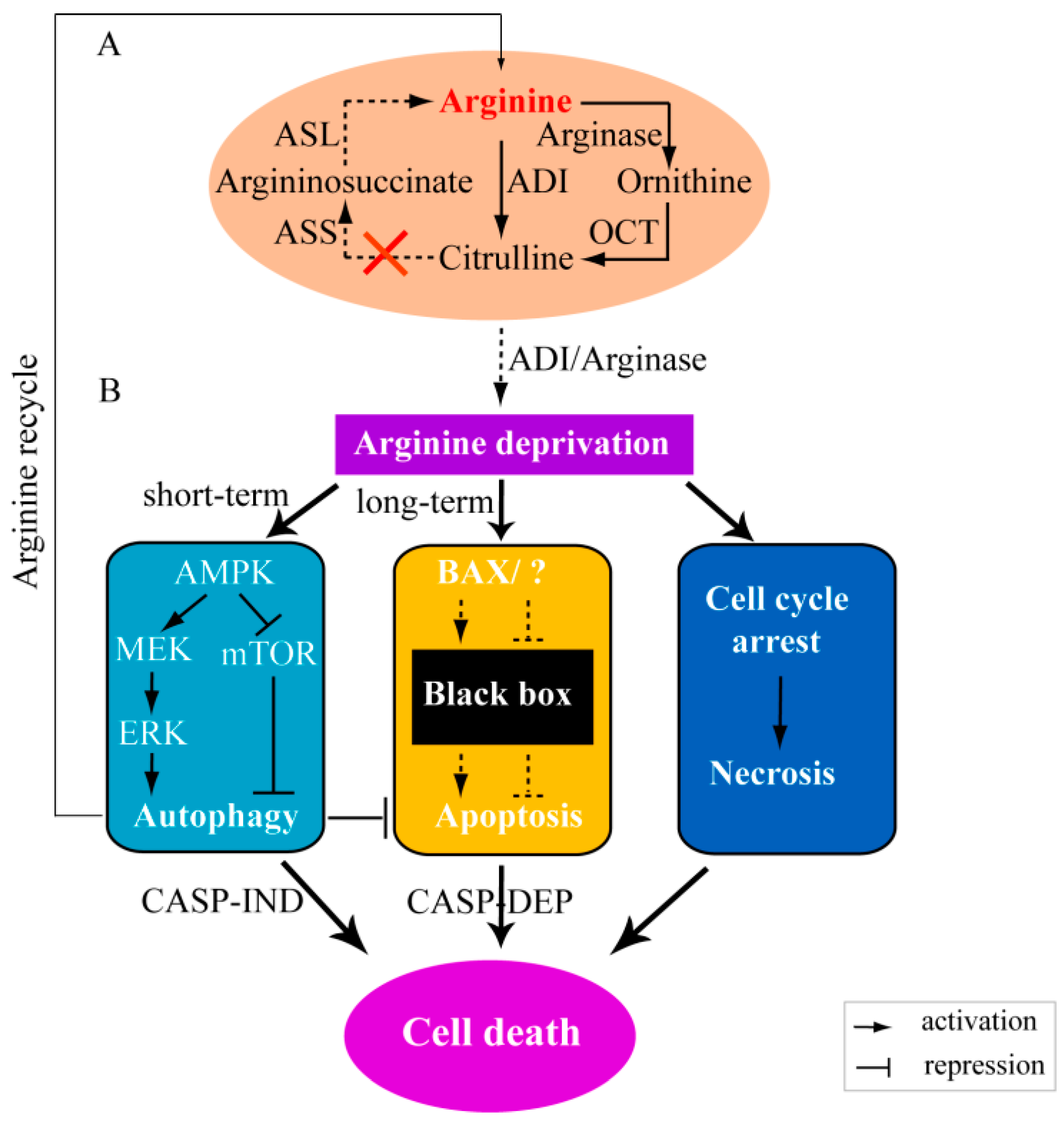
| Pathway (Genes) | Products | Counterparts in Mammalian Cells (with Similar Function or Homologue) | Source/Reference |
|---|---|---|---|
| The arginase pathway | |||
| rocA | Pyrroline-5-carboxylate dehydrogenase | Pyrroline-5-carboxylate dehydrogenase | [10] |
| rocB | Probable citrullinase | - | [13] |
| rocC | Arginine permease | Arginine permease-like | [11,13] |
| rocD | Ornithine aminotransferase (OAT) | Ornithine aminotransferase | [11] |
| rocE | Arginine permease | Arginine permease-like | [11,13] |
| rocF | Arginase | Arginase I and II | [11] |
| The ADI pathway | |||
| arcA | Arginine deiminase | Nitric oxide synthase (NOS) | [14,15,16,17,18] |
| arcB | Ornithine carbamoyltransferase | Ornithine carbamoyltransferase | [14,18] |
| arcC | Carbamate kinase | Carbamate kinase-like | [15,16] |
| arcD | Arginine-ornithine antiporter | - | [16,17,18] |
| Cancer Cell Types | Source or Reference |
|---|---|
| Melanoma | [5,85,90,93,95,96,97,98,99,100] |
| Breast cancer cells | [89] |
| Prostate cancer cells | [4,86,90,101,102] |
| Lymphoma | [89,103,104] |
| Hepatocellular carcinoma (HCC) | [5,90,93,98,100,105,106,107] |
| Pancreatic cancer cells | [91,93,108] |
| Leukemia | [94,109,110,111] |
| Glioma | [89,112] |
| Mesothelioma cell lines | [5,113] |
| Renal cell carcinoma | [4,93,114] |
| Lung cancer | [93,115] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, L.; Teng, J.L.L.; Botelho, M.G.; Lo, R.C.; Lau, S.K.P.; Woo, P.C.Y. Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. Int. J. Mol. Sci. 2016, 17, 363. https://doi.org/10.3390/ijms17030363
Xiong L, Teng JLL, Botelho MG, Lo RC, Lau SKP, Woo PCY. Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. International Journal of Molecular Sciences. 2016; 17(3):363. https://doi.org/10.3390/ijms17030363
Chicago/Turabian StyleXiong, Lifeng, Jade L. L. Teng, Michael G. Botelho, Regina C. Lo, Susanna K. P. Lau, and Patrick C. Y. Woo. 2016. "Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy" International Journal of Molecular Sciences 17, no. 3: 363. https://doi.org/10.3390/ijms17030363
APA StyleXiong, L., Teng, J. L. L., Botelho, M. G., Lo, R. C., Lau, S. K. P., & Woo, P. C. Y. (2016). Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. International Journal of Molecular Sciences, 17(3), 363. https://doi.org/10.3390/ijms17030363